Abstract
Visible light is an important source of energy and information for much of life on this planet. Though fungi are neither photosynthetic nor capable of observing adjacent objects, it is estimated that the majority of fungal species display some form of light response, ranging from developmental decision-making to metabolic reprogramming to pathogenesis. As such, advances in our understanding of fungal photobiology will likely reach the broad fields impacted by these organisms, including agriculture, industry and medicine. In this review, we will first describe the mechanisms by which fungi sense light and then discuss the selective advantages likely imparted by their ability to do so.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Molecular basis of light sensing in the Kingdom Fungi
Fungal species have been shown to respond to light qualities ranging from ~450 nm (blue) to ~700 nm (red), approximating the span of the human visible spectrum. This capability is genetic in basis, mediated by photoresponsive proteins that are highly conserved across the entire fungal kingdom. While some proteins mediate a single biochemical reaction in response to light, e.g., photolyases that repair DNA, others activate a more complicated signaling cascade that leads to broad changes in gene expression. Both types of proteins share in common the requirement to interact with a distinct chromophore that, upon photon absorption, undergoes a physico-chemico and structural change that is sensed by the interacting protein (Losi 2007). We will briefly discuss the biochemistry and signaling mechanisms of the major fungal photoreceptors, an understanding of which stems largely from the model fungus Neurospora crassa and to a lesser extent Aspergillus nidulans and Phycomyces blakesleeanus. Notes concerning the conservation of these genes across other fungi will also be made. Though we have attempted to simplify the nomenclature in text, a list of photoreceptor orthologs across several important species are provided in Table 1. In addition, the reader is referred to several excellent and recent reviews that cover these topics further (Idnurm et al. 2010, Idnurm and Heitman 2005b, Dunlap and Loros 2006, Bayram et al. 2010, Rodriguez-Romero et al. 2010).
Blue light and UV
All fungi that are photoresponsive appear to be sensitive to light at the blue to near-UV-A (~400–495 nm) range. Several classes of blue light receptors have been identified in fungi and will be briefly discussed; starting with the most well understood fungal photoreceptor class, the white collars.
White collar orthologs
The first cloned fungal photoreceptor gene was wc-1 (encoding white collar-1) of the bread mold Neurospora (Ballario et al. 1996). Test-tube slants of wild-type Neurospora appear completely orange following incubation in light due to the induction of carotenoid pigment biosynthesis in the hyphae. In contrast, wc-1 mutants display a non-pigmented mycelial border underneath the constitutively pigmented conidia at the top of the slant, resulting in a “white collar” appearance from which the gene derives its name. Though all known light responses in Neurospora are lost in a wc-1 mutant, its identity as a bona fide photoreceptor was not truly established until demonstration that the WC-1 protein interacts with the chromophore flavin-adenine dinucleotide (FAD) and that the action spectrum of the physiological response (~465 nm) matched the in vitro WC-1 protein response (Froehlich et al. 2002; He et al. 2002). Since then, WC-1-mediated light signaling in Neurospora has emerged as the paradigm for fungal photobiology both at the biochemical and molecular level.
The Neurospora WC-1 protein is a GATA-like transcription factor that contains a Zn-finger DNA binding domain as well as three PAS (Per-Arnt-Sim) domains. The N-terminal most PAS domain is of a special subclass called the LOV domain (for light, oxygen and voltage), which noncovalently binds a single molecule of FAD (Froehlich et al. 2002). WC-1 interacts with a second protein, WC-2, to form a heterodimer known as the white collar complex (WCC). WC-2 also contains a Zn-finger domain, but lacks a LOV domain for direct light sensing. Nevertheless, WC-2 is essential for the Neurospora light response as its loss-of-function mutants yield the same “blind” phenotype as those within wc-1 (Linden et al. 1997; Linden and Macino 1997). Indeed, it is the Zn-finger domain of WC-2 that is required for binding of the WCC to DNA (Froehlich et al. 2002; Collett et al. 2002; Liu and Cheng 2003).
A simple model for WCC-mediated signaling is as follows: (1) the WCC sits on specific sequences (called light response elements, LREs) in the promoters of light-regulated genes (Froehlich et al. 2002); (2) upon absorption of a blue light photon by FAD, a cysteinyl adduct is formed with the LOV domain of WC-1 (Zoltowski et al. 2007); and (3) a conformational change in WC-1 promotes the transcriptional activity of the WCC and gene expression is induced (Froehlich et al. 2002; He et al. 2002). This model is likely applicable to WCC-mediated signaling in other fungi based on genetic inferences, i.e., loss of light-induced gene expression upon deletion of the wc-1 and wc-2 orthologs. However, in contrast to Neurospora in which the WCC is thought to serve almost exclusively as a transcriptional activator, it is thought to have repressive roles in other species including A. nidulans (Bayram et al. 2010), Alternaria alternata (Pruß et al. 2014), Trichoderma reesei (Tisch and Schmoll 2013), Trichoderma atroviride (Sanchez-Arreguin et al. 2012), Fusarium graminearum (Kim et al. 2014a) and Cercospora zae-maydis (Kim et al. 2011a). Interestingly, deletion of the wc-1 ortholog in T. atroviride leads to a broad down-regulation of proteins in the dark, whereas deletion of wc-2 leads predominantly to protein up-regulation, indicating independent functions for the two proteins (Sanchez-Arreguin et al. 2012). Moreover, other aspects of the Neurospora model have yet to be described in other organisms, including chromatin modifications that are, in part, mediated by histone acetyltransferases that interact directly with WC-1 (Brenna et al. 2012).
Genome-wide transcriptional analyses have estimated that approximately 6 % of the Neurospora genome is regulated within 90 min of light exposure, all of which are lost upon deletion of wc-1 or wc-2 (Chen et al. 2009). However, not all such genes are influenced directly by the WCC. Instead, the WCC regulates a subset of genes that display induction between 5 and 30 min after light exposure (early light-induced genes). Among the early-induced genes are transcription factors, e.g., SUB-1, that regulate their own unique targets displaying induction between 45 and 90 min post-light exposure (late-light induced genes) (Chen et al. 2009). There is evidence that this hierarchical transcriptional cascade is conserved among fungal species. For example, a sub-1 ortholog involved in the light response of the plant pathogen Botrytis cinerea has been recently characterized (Schumacher et al. 2014). Additionally, a whole-genome transcriptional analysis of the A. fumigatus light response has revealed an early- and late-induction of genes across a 120-min time course (Fuller et al. 2013).
With orthologs found in essentially all fungal lineages, including species in the anciently diverged chytrids, WC-1 is among the most highly conserved fungal photoreceptors in terms of evolutionary depth (Idnurm et al. 2010; Dunlap and Loros 2006). There are a few organisms that notably lack wc-1 as well as most other photoreceptor genes, likely as a result of gene loss in that species or lineage. These include most Saccharomycotina yeasts, including Saccharomyces and Candida, as well as species of the dermatophytic fungi, e.g., Malassezia, Trichophyton and Microsporum. As many of these organisms are obligate pathogens with a close association with their host, it is possible that genes involved in environmental light signaling no longer provided a selective advantage and were lost. In contrast, other human pathogens, such as Aspergillus fumigatus, Cryptococcus, Paracoccidioides and Histoplasma all have an environmental life cycle as well as a wc-1 ortholog (Idnurm et al. 2010).
In theory, WC-1 should serve alone as a photoreceptor since it has domains for both signal input (LOV domain) and output (Zn-finger domain). Interestingly, the essential interaction between WC-1 and WC-2 appears to have emerged early in the fungi, as a wc-2 ortholog is present in all species that contain wc-1. Indeed, many WC-1 proteins in the basidiomycete fungi, including Cryptococcus, Ustilago maydis, Phanerochaete chrysosporium and Coprinus cinereus, have lost the Zn-finger domain altogether (Idnurm and Heitman 2005a). Interestingly, the wc-1 orthologs of the Schizosaccharomyces japonicus and Yarrowia lipolytica, the only ascomycete yeasts known to harbor white collar genes, also lack the Zn-finger domain (Okamoto et al. 2013).
Small LOV domain proteins: VIVID and ENVOY
Several ascomycete species encode a small protein that is essentially a single LOV domain with an N-terminal cap (Zoltowski et al. 2007). The first member of this family to be identified was the 186-amino acid protein in Neurospora, VIVID (encoded by the gene vvd) (Heintzen et al. 2001). The name stems from its loss-of-function mutants which display bright orange conidia and mycelia when grown in constant light, attributed to the persistent synthesis of carotenoid pigments (Heintzen et al. 2001). Like WC-1, VIVID binds FAD as a chromophore (Schwerdtfeger and Linden 2003). The expression of vvd is strongly induced by the WCC upon exposure to light and, upon seeing light for itself, VIVID interacts with the LOV domain of WC-1 to block the transcriptional activity of the WCC (Chen et al. 2010a; Hunt et al. 2010; Malzahn et al. 2010). This negative feedback on the WCC is termed photoadaptation and allows for the fungus to be responsive to increasing light intensities over the course of the day. Ultimately, the amount of WCC activity is dependent upon the amount of VIVID in the system, which itself is a function of the WCC-mediated light response. Current models suggest that VIVID mediates its effects only through its feedback on the WCC (Gin et al. 2013).
An apparent ortholog of VIVID is the 207-amino acid protein ENVOY in T. reeseii (Hypocrea jecorina). ENVOY is similarly induced by the WCC but, in contrast to VIVID, appears to provide positive output from the WCC rather than acting simply as an inhibitor of WCC activity (Tisch and Schmoll 2013). For example, ENVOY is involved in the light regulation of various genes, including those involved in cellulose metabolism, sexual development and G-protein signaling (Schuster et al. 2007; Schmoll et al. 2009; Seibel et al. 2012). Moreover, ENVOY appears to positively regulate genes that are not affected upon loss of the WCC, including several polyketide synthases, hydrophobins, and sulphur metabolism genes (Tisch and Schmoll 2013). In spite of the fact that both VIVID and ENVOY contain only a single LOV domain, the two proteins cannot cross-complement one another, suggesting a complicated mechanism of regulation for these small proteins that is not yet understood (Schmoll et al. 2005).
Cryptochromes and photolyases
Cryptochromes and photolyases are members of blue/UV-A light receptors, both of which contain an N-terminal domain [photolyase-related (PHR) region] that binds noncovalently to two chromophores, FAD and 5,10 methyltetrahydrofolate (MTHF) (Losi 2007). The photolyases are characterized by their ability to use light energy to catalyze the repair of DNA (e.g., pyrimidine dimers) that arise from UV exposure. The cryptochromes likely emerged from a photolyase ancestor, but have typically lost DNA repair capability and instead serve as bona fide light-signaling proteins (Kim et al. 2014b). Comparative genomics and functional analyses can place the photolyase/cryptochrome proteins into several families that are sporadically distributed among species in the Ascomycota, Basidiomycota and Mucormycotina (Idnurm et al. 2010). Generally, the photolyases (CPD photolyases) are found in essentially all species where they serve as light-activated DNA repair proteins; however, we will briefly describe those proteins with a photo-signaling function (i.e., the cryptochromes). Specific roles of selected proteins will be discussed in latter sections of this review.
The CRY-DASH (Cryptochrome-Drosophila, Arabidopsis, Synechocystis, Human) family is thought to represent the earliest divergence of cryptochromes from the photolyases as it is the only family with an ortholog in bacteria. The CRY-DASH proteins display minimal to no photorepair activity against double-stranded DNA, but may display activity towards cyclobutane adducts on single-stranded DNA. In Neurospora, the CRY-DASH ortholog (CRY) displays a major absorption at 375 nm, corresponding to the peak absorption of the MTHF chromophore and minor peaks at 445 and 470 nm, corresponding to FAD. Interestingly, CRY displays binding to both single and double-stranded DNA and RNA by electrophoretic mobility shift assays (EMSA) (Froehlich et al. 2010). However, whole-genome microarray analysis suggests that the Neurospora transcriptional response to light is unaltered upon deletion of cry, as are all overt light-responsive phenotypes (Chen et al. 2009; Froehlich et al. 2010). However, CRY-DASH mediated signaling has been observed in several fungal species, including Sclerotinia sclerotiorum where it partially controls sclerotium biomass (sexual reproduction) in response to UV-A (Veluchamy and Rollins 2008), and Fusarium fujikuroi where it serves as a repressor of secondary metabolism (Castrillo et al. 2013).
As mentioned, essentially all species encode a member of the CPD photolyases that have DNA repair capability. Interestingly, the ortholog in A. nidulans displays not only repair activity, but also a role in suppressing sexual development (Bayram et al. 2008a). It is unclear whether the dual function of this protein, CryA, reflects a photolyase that has gained signaling function, or rather an ancestral form of the photolyase/cryptochrome proteins that displayed both functions. Similarly, the CPD photolyase of T. atroviride, PHR1, regulates its own expression, possibly through feedback on the WC orthologs (Berrocal-Tito et al. 2007).
The corn gray leaf spot pathogen, C. zeae-maydis, encodes both a member of the CPD photolyases (called CPD1) and a member of the so-called 6-4 photolyases (called PHL1). Despite the annotation and activity as a photolyase, deletion of phl1 also leads to defects in the light induction of cpd1 and other DNA repair genes, suggesting that PHL1 has also a signaling (cryptochrome) function (Bluhm and Dunkle 2008). Along the same lines, overexpression of the 6-4 photolyase in T. reesei was shown to lead to increased DNA repair capability in the dark, suggesting that the photolyase was also regulating the expression of other DNA repair systems (Guzmán-Moreno et al. 2014).
In summary, much less is known about cryptochrome-mediated light signaling in fungi compared to the white-collar orthologs. However, recent reports have begun to uncover important roles for this diverse group of proteins in various species. Moreover, cryptochromes may play important, but yet to be identified, roles in fungi that have persistent blue light responses in a white collar deletion background, such as UV resistance in A. fumigatus (Fuller et al. 2013), melanization in B. cinerea (Canessa et al. 2013), and carotenogenesis in F. fujikuroi (Estrada and Avalos 2008).
Red light
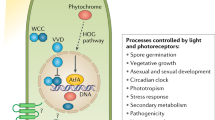
Although blue light responses are most conserved among fungi, many species respond to wavelengths between 600 and 850 nm (red/far-red light). The detection of red light is mediated by a class of proteins called the phytochromes, which are widely distributed among bacteria and plants. Phytochromes display a modular architecture consisting of an N-terminal photosensory module (GIF and PHY domains), which binds a porphyrin bilin (a tetrapyrrole) as a chromophore (Idnurm and Heitman 2005b). The C-terminal output domains vary considerably among species, but will always contain a histidine kinase domain (Schaller et al. 2011; Blumenstein et al. 2005). Phytochromes convert between two conformational states, the red-light absorbing Pr confirmation and the far-red light absorbing Pfr confirmation, with the ratio of the two determining the signaling state within the cell. Both PHY-2 of Neurospora and FphA of A. nidulans are capable of binding biliverdin as a chromophore in vitro and display a Pr absorption peak at ~700 nm (Froehlich et al. 2005; Brandt et al. 2008). Conversion of the Pfr form of PHY-2 to the Pr form occurs spontaneously after 100 min in the dark (Froehlich et al. 2005).
Like the cryptochromes, the phytochromes are sporadically distributed among fungal lineages. Though predominately found among the Ascomycota and Basidiomycota, putative orthologs have been identified in the more distantly related chytrids (Idnurm et al. 2010, Idnurm and Heitman 2005b). Interestingly, the presence of a phytochrome is not a strict indicator of a red-light response in fungi. For example, neither Cryptococcus neoformans nor Neurospora displays an overt response to red light, in spite of both organisms expressing one and two phytochrome genes, respectively (Idnurm and Heitman 2005a, Froehlich et al. 2005). Yet, several other fungi do indeed display red light responses, including Alternaria solani (Lukens 1965), B. cinerea (Tan 1974), T. atroviride (Casas-Flores et al. 2004), Magnaporthe oryzae (Lee et al. 2006), Puccinia graminis (Schneider and Murray 1979), A. nidulans (Bayram et al. 2010; Mooney and Yager 1990; Röhrig et al. 2013), and A. fumigatus (Fuller et al. 2013). Among these fungi, however, only the phytochrome ortholog of A. nidulans and A. fumigatus have an established role in the light response based on the analysis of deletion mutants.
Green light
Rhodopsins are a family of seven-transmembrane receptors that bind retinal as a chromophore. The so-called type II rhodopsins serve as the primary photoreceptor in animal vision, where they ultimately promote membrane hyperpolarization through the regulation of cyclic-GMP gated-sodium ion channels (Avelar et al. 2014). A highly divergent group of rhodopsins (Type I) are found among the Archaea, Bacteria and lower eukaryotes, including the fungi. Functionally, rhodopsin has been linked to the green light phototaxis of two blastocladiomycete fungi, Allomyces reticulatus and Blastocladiella emersonii, with a peak action spectrum of ~536 nm (Saranak and Foster 1997; Avelar et al. 2014). Interestingly, the rhodopsin ortholog in these early branching fungi have a C-terminal gaunylyl cyclase (GC) domain, representing a novel gene fusion that imparts cGMP regulation similarly to, although mechanistically distinct from, the type II proteins of animals. Conversely, the rhodopsin orthologs found in more recently diverged fungi (e.g., the Dikarya) lack a GC domain and, consequently, are likely more similar to the bacteriorhodposins that function as light-regulated proton pumps (Avelar et al. 2014).
Despite the presence of a rhodopsin gene in numerous ascomycete and basidiomycete species, few green light-specific responses have been observed for these organisms. In several species, including Neurospora (Chen et al. 2009; Bieszke et al. 1999), F. fujikuroi (Estrada and Avalos 2009) and C. neoformans (Idnurm and Heitman 2005a), the expression of the rhodopsin gene is regulated by white light, yet their deletion does not lead to an overt photoresponsive defect. Deletion of nop-1 in Neurospora does, however, lead to light-dependent morphological difference in the presence of the mitochondrial ATPase inhibitor oligomycin, suggesting it may play a role as a light-dependent proton pump rather than a signaling protein (Bieszke et al. 1999).
An understanding of fungal opsins is complicated by the fact that most species contain a family of opsin-related proteins (ORPs), distinguished based on the absence of a conserved lysine residue required for retinal binding (Brown 2004). These proteins are widely distributed and are even present in the multiple copies in the genomes of the Saccharomycotina yeasts, which typically lack orthologs to all other photoreceptor genes (Idnurm et al. 2010). A functional role for these ORPs has yet to be described.
Conclusions
Though the various photoreceptor proteins were discussed separately for the sake of exposition, it is important to consider that the light response for most fungi undoubtedly involves the integration of multiple light inputs by discrete photosensory proteins. This is perhaps best illustrated in A. nidulans, which responds to both red and blue light signals by distinct proteins (WCC orthologs and phytochrome) that interact physically within a large complex along with a key regulator of development and secondary metabolism (Velvet) (Purschwitz et al. 2008, 2009; Bayram et al. 2008b). Even Neurospora, which only responds overtly to blue light via the WCC, likely integrates input from additional photoreceptors. For example, a recent report has demonstrated that the light induction of the con-10 gene, involved in conidiation, occurs earlier and to a larger extent in cry, nop-1, or phy-2 deletion backgrounds (Olmedo et al. 2010). Therefore, while WC-1 may be the only essential photoreceptor in driving the light response, the other proteins may play an ancillary role that modulate WCC activity.
Furthermore, the relative spectral qualities and intensities change over the course of a day, with blue/near-UV-A peaks near mid-day and red/far-red peaks at morning and near dusk. Nevertheless, most laboratory experiments are performed under constant light or dark conditions. As such, photoreceptors that have been reported to have no function to an organism’s light response (e.g., opsin in Cryptococcus) may indeed have subtle, yet important functions in a more natural light environment.
With a brief understanding of the fungal photoreceptors, we will now shift our focus to the physiological consequences of the light detection and why such systems likely provide a selective advantage for fungi that employ them.
Visible light as a signal for stress
Genotoxic and oxidative stress
Ultraviolet radiation represents a major source of stress for the cell. Direct genomic damage is largely in the form cyclobutane formation, e.g., pyrimidine dimerization, caused by UV-B (~315–280 nm) (Schuch et al. 2013). As such, the first photosensitive proteins to emerge in the prokaryotes were likely the photolyases, which utilize light energy in the blue/near-UV-A range to catalyze the repair of such adducts (Hitomi et al. 2000). Additionally, UV-B and UV-A (~315–400 nm) wavelengths are capable of inducing the formation of reactive oxygen species (ROS) in the form of superoxide, hydrogen peroxide or hydroxyl radicals, which can then damage various macromolecules including lipids, proteins and DNA (Schuch et al. 2013; Shiu and Lee 2005). Even visible light itself can be a source of oxidative stress for the fungal cell as was shown most recently in B. cinerea, in which a white light (400–720 nm)-dependent reduction in growth rate could be rescued simply by the addition of the antioxidant ascorbate to the medium (Canessa et al. 2013). A similarly slight reduction in mycelial growth by light has also been noted in numerous fungi including, A. fumigatus (Fuller et al. 2013), Cordyceps militaris (Yang and Dong 2014), F. graminearum (Kim et al. 2014a) and Colletotrichum acutatum (Yu et al. 2013).
The ability to utilize less harmful wavelengths of light to signal the presence of, and promote resistance to, harmful UV is likely the initial evolutionary basis for photoperception. This premise has been shown through multiple lines of evidence among diverse fungal species. In A. fumigatus, for example, exposure of dark-grown fungus to blue light leads to resistance to subsequent exposure to either UV or hydrogen peroxide, relative to samples kept in the dark prior to exposure (Fuller et al. 2013). The ability of visible light to promote UV resistance was also shown in C. neoformans (Verma and Idnurm 2013). Similarly, the insect pathogen Metarhizium robertsii grown under visible light produces conidia that are more tolerant to UV-B radiation compared to conidia produced in the dark (Rangel et al. 2011). The importance of light in promoting stress resistance has also been shown less directly through the analysis of photoreceptor mutants. For example, white collar-1 deletion mutants of C. neoformans and B. cinerea are hyper-sensitive to UV and oxidative stress, respectively (Idnurm and Heitman 2005a; Canessa et al. 2013). Interestingly in A. fumigatus, only upon deletion of lreA (white collar-1) along with a phytochrome ortholog (fphA) does the light-mediated resistance to oxidative stress drop out (Fuller et al. 2013), suggesting a complicated interplay between the two photoreceptors. In summary, it appears that light serves as a conserved signal among fungi to promote resistance to UV-mediated damage, namely direct DNA damage and oxidative stress. The mechanisms for such resistance will be described next.
One of the most common fungal light responses is the induced expression of pigments, which may be protective either directly absorbing UV photons or by scavenging UV-generated ROS. One class of such pigments is the orange carotenoids, which are induced in a WC-1 dependent manner in Neurospora and hyper-accumulate in the vvd mutants as mentioned previously. Photo-carotenogenesis is described in numerous fungi including Fusarium spp. (Kim et al. 2014a; Estrada and Avalos 2008; Ruiz-Roldan et al. 2008; Avalos and Estrada 2010), Aspergillus giganteus (El-Jack et al. 1988), P. blakesleeanus (Corrochano and Cerdá-Olmedo 1992; Corrochano and Garre 2010), Mucor circinelloides (Silva et al. 2006), Blakeslea trispora (Quiles-Rosillo et al. 2005), and even numerous freshwater yeasts, e.g., Rhodotorula (Libkind et al. 2004). For those species in which it has been tested, this response is typically mediated by blue light specifically and is largely dependent upon the WC-1 ortholog. Interestingly, deletion of the opsin ortholog, opsA, in F. fujikuroi leads to an attenuated photoinduction of carotenoid genes although no pigment phenotype is observed in the mutant (Estrada and Avalos 2009).
A second class of fungal pigments is the melanins which, depending on the species, range from bluish-green to black in color. Photo-induced melanization has been observed in numerous ascomycetes including A. fumigatus (Fuller et al. 2013), M. oryzae (Kim et al. 2011b), Cercospora kikuchii (Bluhm et al. 2010), B. cinerea (Canessa et al. 2013) and Colletotrichum acutatum (Yu et al. 2013), as well as in the basidiomycete mushroom Lentinula edodes (Sano et al. 2009). The WCC is presumed to play a major role in melanization for these fungi based on the action spectrum for the response and, in the case of A. fumigatus, has been demonstrated directly through deletion analysis. Surprisingly, however, a blue light-to-dark- driven melanin banding pattern in B. cinerea persists in the wc-1 deletion mutant, indicating that additional blue-light receptors are operative (Canessa et al. 2013). Moreover, red light, in addition to blue light, was shown to have an inducing effect in C. acutatum, but the role of a phytochrome in this response is unknown (Yu et al. 2013).
A photoinduction of DNA repair enzymes has also been noted in various fungal species. The induction of photolyases and/or UV endonucleases by blue light has been shown in the ascomycetes Neurospora, A. fumigatus (Fuller et al. 2013), A. nidulans (Ruger-Herreros et al. 2011), Fusarium oxysporum (Ruiz-Roldan et al. 2008) and C. zeae-maydis (Yu et al. 2013). In C. neoformans, endonuclease (Uve1) induction is dependent upon the WCC as is light-induced UV resistance (Verma and Idnurm 2013). However, in C. zae-maydis, the induction of repair genes and UV resistance is also dependent upon PHL1, a cryptochrome (Bluhm and Dunkle 2008). Similarly, an lreA (white collar-1) deletion mutant of A. fumigatus is defective for photolyase induction, but still displays light induction of additional repair genes as well as light-induced UV resistance. This suggests that other blue light receptors that play a role in UV resistance, likely the cryptochrome ortholog, are important in that fungus.
In addition to the pigmentation responses described above, light-mediated resistance to ROS comes predictably in the form of antioxidant enzyme regulation. For example, the photoinduction of catalase, superoxide dismutase (SOD) and glutathione peroxidase has been demonstrated in both Neurospora (Chen et al. 2009) and A. nidulans (Ruger-Herreros et al. 2011). Perhaps less obvious is the regulation of certain metabolic pathways that may promote a ROS resistant state. For example, the heme biosynthetic pathway is characterized by several porphyrin intermediates, such as those that may serve as the phytochrome chromophore, that are highly photoreactive and may produce toxic ROS upon light exposure. Interestingly, genes involved in heme biosynthesis, particularly the terminal enzyme ferrochelatase, are light-induced in numerous fungal species including the ascomycetes Neurospora (Chen et al. 2009; Idnurm and Heitman 2010) and B. cinerea (Canessa et al. 2013), the basidiomycete C. neoformans (Idnurm and Heitman 2010), and the Mucormycotina species P. blakesleeanus and Rhizopus oryzae (Idnurm and Heitman 2010). Deletion of the ferrochelatase gene in C. neoforman leads to an apparent hyper-accumulation of porphyrins and an inability to grow in light, suggesting that the up-regulation of heme biosynthesis is important for depleting the porphyrin pool in the cell and mitigating porphyrin-mediated phototoxicity (Idnurm and Heitman 2010).
Heat stress
For many organisms, visible light may also serve as a signal for increasing daytime temperatures and/or a direct source of thermal stress (thermal radiation), which may lead to protein and membrane destabilization. As such, it is not surprising that genes involved in the temperature stress (heat-shock) response are also light-regulated in several fungal species. This has been most thoroughly described in P. blakesleeanus, in which hspA transcript, encoding heat-shock protein100, is induced tenfold upon blue light exposure (Rodriguez-Romero and Corrochano 2004). Hsp100, like other heat-shock proteins, are ATP-binding protein chaperones with the ability to repair misfolded and aggregated proteins. Consistent with a true light response, the photoinduction of hspA is dependent upon the wc-1 ortholog, madA; however, the hspA photoinduction is less sensitive to light than other mycelial light responses (e.g., phototropism), suggesting that the regulation of hspA involves alternative regulatory mechanisms or proteins (Rodriguez-Romero and Corrochano 2004, 2006). Indeed, in addition to madA, the photoinduction of hspA is also dependent upon two genes involved in beta-carotene biosynthesis, carB and carR, suggesting that beta-carotene itself may act as secondary chromophore required for full hspA light-activation (Rodriguez-Romero and Corrochano 2006). Beyond Phycomyces, the light induction of heat-shock proteins has been noted in both Neurospora (Chen et al. 2009) and A. nidulans (Ruger-Herreros et al. 2011) based on transcriptional arrays.
In addition to protein chaperones, the disaccharide trehalose suppresses aggregation of heat-denatured proteins and may protect the cell from oxidative damage by free radicals. The trehalose synthase gene, ccg-9, of Neurospora is both heat-shock and light-induced, suggesting an additional mechanism for responding to light and heat-mediated stress in the fungal cell (Shinohara et al. 2002). This strategy may be conserved across fungi as the ccg-9 ortholog was also light induced on the A. nidulans arrays (Ruger-Herreros et al. 2011).
Regulation of growth by light
As mentioned above, light can be a direct source of stress that leads to reductions in radial growth rates. Similarly, both blue and/or red light have been shown to inhibit the germination kinetics of spores in A. nidulans (Röhrig et al. 2013), A. fumigatus (Fuller et al. 2013), P. graminis (Lucas et al. 1975) and C. acutatum (Yu et al. 2013). In the case of both A. nidulans and A. fumigatus, the phytochrome ortholog is involved in the inhibition in response to both red and blue light, whereas the wc-1 ortholog is dispensable. More specifically, the germination rates of the phytochrome mutants in the dark mirror the germination rates of wild-type strains in the light, supporting a model in which light inhibits phytochrome-mediated germination (Fuller et al. 2013; Röhrig et al. 2013). Beyond blue and red light, it was recently shown that green light can inhibit the conidial germination kinetics of B. cinerea, although the contribution of an opsin ortholog in the response is unknown (Zhu et al. 2013). Importantly, these data demonstrate that the influence of light on growth can be a photoreceptor-mediated process, rather than a simple consequence of ROS production. From an evolutionary standpoint, delaying germination by some hours may be advantageous for the fungus by increasing the likelihood that growth is initiated under more favorable conditions (i.e., lower light intensity).
Light as a signal for space
Fungi reproduce and spread through the environment through the release of clonal (conidia) and/or sexually derived spores. As the morphogenic switch from vegetative growth to development may be energetically costly, there is likely an important selective advantage for fungi that produce spores only at the surface-to-air interface, where dispersal is optimal (Rodriguez-Romero et al. 2010). To this end, it is likely that light serves as an important positional marker that signals the open air. This is strikingly illustrated by the bending of sporangiophores (asexual developmental structures) towards light in several Mucormycotina species, such as P. blakesleeanus (Corrochano and Garre 2010), M. circenilloides (Silva et al. 2006) and Pilobolus crystallinus (Page 1962). Similarly, Neurospora orients its protoperithecial beaks containing the sexual ascospores towards blue light in a WC-1/2 dependent manner (Linden et al. 1997; Chen et al. 2010b).
More broadly, light serves as a signal to regulate the induction of developmental pathways in numerous fungi. Perhaps the most well-conserved response, and consistent with the idea of light signaling the open air, is the blue light-mediated induction of conidiation observed in numerous species, including Neurospora (Chen et al. 2010b), T. atroviride (Sanchez-Arreguin et al. 2012; Casas-Flores et al. 2004), C. miltaris (Yang and Dong 2014), F. graminearum (Kim et al. 2014a), Coprinopsis cinerea (Kamada et al. 2010), and others. The influence of red light on conidiation has been noted in some organisms and is best described in A. nidulans, in which both blue and red light promote conidiation via the WC-1 ortholog, LreA, and the phytochrome, FphA. The red and blue light responses appear to be additive in A. nidulans, as neither monochromatic treatment can achieve the level of induction seen by white light (Bayram et al. 2010).
Conversely, blue and/or red light represses asexual development in Aspergillus oryzae (blue and red) (Hatakeyama et al. 2007), A. alternata (blue, and to a lesser extent, red) (Pruß et al. 2014; Lukens 1965), M. oryzae (blue) (Lee et al. 2006) and C. zeae-maydis (blue) (Kim et al. 2011a). In the case of M. oryzae, A. alternata and C. zeae-maydis, loss of the wc-1 ortholog leads to a de-repression of conidiation in light. In these organisms it is possible that the stress signal by light dominates, and conidiation is repressed in favor of the induction of stress resistance pathways, e.g., pigmentation. Interestingly, in P. blakesleeanus the blue light signal is more complex, inducing macrophorogenesis while inhibiting microphorogenesis, both of which are asexual developmental programs (Corrochano and Cerdá-Olmedo 1992). Still, other species must likely integrate oppositional light signals in order to mediate a proper response. In Fusarium verticilloides, for example, white light promotes conidiation even though red and short-wave blue light are inducing while longer-wave length blue light is repressive (Fanelli et al. 2012).
In addition to asexual sporulation, light also serves to regulate sexual reproduction. For example, light clearly promotes fruiting body formation in a variety of fungi including Neurospora (Chen et al. 2010b), F. graminearum (Kim et al. 2014a) and various mushroom producing basidiomycetes, including C. cinerea (Kamada et al. 2010), Schizophyllum commune (Perkins and Gordon 1969), Pleurotus ostreatus (Richartz and MacLellan 1987) and Favolus arcularius (Kitamoto et al. 1972). In contrast, however, both red and blue light repress cleistothecium (fruiting body) formation in A. nidulans (Bayram et al. 2010). This response appears to involve a complicated interplay between LreA (WC-1) and FphA (phytochrome). Deletion of lreA leads to reduced cleistothecial formation in both dark and light, suggesting it is serving as a general activator of sexual development. In contrast, deletion of fphA leads to enhanced cleistothecial development, suggesting FphA acts as a repressor (Blumenstein et al. 2005). Therefore, the FphA-mediated repression appears to dominate in response to both red and blue light, the mechanism of which is poorly understood. In addition, the cryptochrome, CryA, is also involved in the A. nidulans response as cryA deletion strains display enhanced sexual development (Bayram et al. 2008a). Sexual differentiation is also repressed in the basidiomycetous yeast C. neoformans (Idnurm and Heitman 2005a) and in Phycomyces (Corrochano and Cerdá-Olmedo 1992), but this occurs only in response to blue light and is dependent upon the white collar-1/2 orthologs. The light repression of sexual development in these species is thought to be advantageous since the meiotic progeny will be formed away from mutagenic UV.
Light as a signal for time
Due to the day/night cycles of the rotating Earth, most organisms experience predictable 24 h oscillations in their light and temperature environment. This has led to the evolution of circadian rhythms, which are endogenous cellular oscillators that allow an organism to anticipate, rather than simply respond to, these daily changes. For example, many plants will position their leaves and upregulate photosynthetic machinery before dawn and down-regulate photosynthetic genes and upregulate cold-resistance proteins prior to nightfall (McClung and Gutiérrez 2010). Such environmental synchronization likely optimizes energy utilization and stress resistance, thus providing a selective advantage in organisms ranging from bacteria to humans (Bell-Pedersen et al. 2005). Circadian behavior has been described in Neurospora since 1959 in the form of rhythmic conidiation patterns (Pittendrigh et al. 1959). Since then, Neurospora has emerged as a premiere model for circadian biology due to its simple circadian output, tractability and easy exploitable genetics (Collopy et al. 2010; Colot et al. 2006).
The Neurospora clock
A biological rhythm must meet several criteria to be considered circadian: (1) it must cycle approximately every 24 h; (2) it must persist in the absence of external timing cues, such as light or temperature (free running); (3) it must maintain the capacity to be reset by such cues (entrainable); and (4) the period length should be resistant to temperature changes (compensated) such that the rhythm serves as a clock rather than a thermometer. When grown across an agar surface in constant darkness, Neurospora will produce conidia every ~22 h, resulting in discrete conidial bands separated by stretches of undifferentiated hyphae (Baker et al. 2012) (Fig. 1). The periodicity of this rhythm is maintained across a range of temperatures (temperature compensated) and is reset/abolished upon blue light exposure. Therefore, in addition to being a stress and developmental signal for Neurospora, light also serves to synchronize the clock to the external environment.
a Light induction of pigmentation. Incubation of Aspergillus fumigatus in constant white light leads to the production of blue-green melanin in the hyphae. Plate bottoms were photographed following 48 h incubation at 37 °C. Deletion of the Neurospora wc-1 ortholog, lreA, abolishes this response (not shown) (Fuller et al. 2013). b Light regulation of asexual development. Top Light induces conidiation in a clinical A. fumigatus strain. Conidia were point-inoculated at the center of a petri plate and the top was photographed following incubation in an alternating light (12 h)/dark (12 h) environment for several days. The dark conidial bands correspond to the light portion of the photocycle (white bars). Bottom:Light represses conidiation in a clinical Fusarium strain. Conidia were point-inoculated at the center of the petri plate and the top was photographed following several days in an alternating light (12 h)/dark (12 h) environment that was interrupted by 24 h of constant light for one day (the wider white band). The lighter, fluffy bands correspond to the dark portions of the photocycle (black bars). The schematic proposes two models of developmental regulation by light in this Fusarium species, in which the WCC is presumed to be a major regulator. c WCC in the Neurospora clock. In constant darkness, the WCC interacts with the c-box of the frq promoter and drives expression of frq gene. The FRQ protein interacts with FRH to negatively feedback on the WCC, thereby leading to reduced WCC activity and frq expression. Together with FRQ turnover, this feedback loop leads to observable oscillations in FRQ protein that are discernible when the protein is fused to the luciferase reporter. Beyond FRQ, the WCC also drives output from the clock as it regulates other genes whose products do not participate in the feedback loop (ccgs). Included among the ccgs are those involved in asexual development, resulting in the rhythmic conidiation pattern formed when the fungus grows across race tubes in constant darkness
A transcription/translation feedback loop makes up the circadian oscillator in all eukaryotes. In Neurospora, the positive arm is the WCC, consisting of WC-1 and WC-2, which drives the expression of genes that provide the physiological output of the clock (the so-called clock-controlled genes, ccgs). In addition, the WCC directly induces the expression of a gene called frequency (frq), the protein of which, FRQ, in complex with a second protein, FRH, then inhibits the activity of WCC (Hurley et al. 2013) (Fig. 1). This negative feedback loop comprises the core clock and operates with a time constant determined by the phosphorylation of FRQ that eventually leads to its turnover. Thus, WC-1 leads a dual life: it is a photoreceptor that mediates the blue light response, and it is the positive arm of the circadian clock that functions in the dark by driving ccgs and frq. Not surprisingly then, there is considerable overlap between light- and clock-controlled genes in Neurospora and the photo-induced expression of frq via the WCC accounts for the resetting of the clock by light. It is estimated that between 7 and 15 % of the Neurospora genome is circadianly regulated, depending on the method of experimentation (Dunlap and Loros 2004; Dong et al. 2008, Hurley et al. 2014, personal communication) and includes genes involved in the metabolism, development and stress resistance. For example, the above-mentioned ccg-9 (encoding trehalose synthase) is circadianly regulated, likely allowing the fungus to prepare for oxidative and heat stresses associated with the light portion of the diurnal cycle (Shinohara et al. 2002).
Although WC-1 is the only photoreceptor to play an essential role in the Neurospora circadian rhythm, additional blue light photoreceptors are involved in fine-tuning the light input into the clock. For example, the small LOV domain protein VIVID (described above) is not only induced by the WCC in light, but it is also circadianly regulated. As a consequence, the levels the VIVID in the dark are sufficient to inactivate any WCC activity induced by moonlight, thereby keeping the clock on pace on bright nights and directing it to take its principle light cues at dawn (Heintzen et al. 2001; Malzahn et al. 2010). Transcript levels of the CRY-Dash gene, cry, are also circadianly regulated and the deletion mutants displays a delay in clock phase, defined by the time at which the first conidial band emerges after transition into constant darkness (Froehlich et al. 2010). Similarly, the phytochrome gene, phy-1, is circadianly regulated, yet deletion of the gene leads to no observable change in clock period or phase (Froehlich et al. 2005).
Circadian rhythms in other fungi
The identification of an overt phenotypic rhythm that meets each of the criteria to be called “circadian” (i.e., free running, entrainable, compensated) has been notably lacking for fungal species beyond Neurospora. Some of the few exceptions include rhythmic conidiospore formation in Pilobolus (Bruce et al. 1960), sclerotia formation in Aspergillus flavus (Greene et al. 2003), and melanization in C. kikuchii (Bluhm et al. 2010). At the molecular level, the rhythmic expression of gpdA transcript, encoding the glyceraldehyde-3-phosphate dehydrogenase, in A. nidulans has been reported with a free-running period of 28–32 h, yet no developmental rhythm is obvious in this organism (Greene et al. 2003).
It is important to note that the robust conidiation rhythms of Neurospora are not observed in wild-type cultures unless gentle airflow is applied to the tubes or an oxidant such as menadione is added to the medium. Instead, most circadian studies in Neurospora are performed in strains harboring a mutation in the ras-1 gene (called the band, bd, strain) which have slower growth rates and enhanced conidiation that is refractory to the accumulation of CO2 in the culture tubes (Sargent et al. 1966; Belden et al. 2007). Therefore, the lack of observed developmental rhythms in other fungi may largely stem from suboptimal culturing conditions or genetic backgrounds.
At the genomic level, however, the conservation of core-clock components across the fungi has been well investigated (Dunlap and Loros 2006; Lombardi and Brody 2005; Salichos and Rokas 2010). As mentioned, wc-1 and wc–2 orthologs are highly conserved among the fungi. In contrast, frq orthologs appear to be present only within the Sordariomycetes, Dothidiomycetes and Leotiomycetes, suggesting that clocks in other fungal lineages (e.g., Aspergillus) operate with a different core molecular feedback loop. In this regard, gpdA expression in A. nidulans is reported as rhythmic even in an lreA (wc-1) deletion background, suggesting positive arm components of the clock may also differ (Greene et al. 2003).
Summary and perspectives
This is a particularly exciting time for fungal photobiology as studies move beyond the model organisms and become applied to species of industrial, agricultural and medical relevance. As genetic and molecular tools improve in these organisms, an understanding of the molecular basis for these responses will undoubtedly come to light and important and interesting questions may be addressed.
One exciting area of research concerns the involvement of light/photoreceptors in fungal pathogenesis. In the causative agent of pepper anthracnose, C. acutatum, lesion size of infected plants is considerably larger during incubation in white, blue, red and green light, relative to incubation in the dark (Yu et al. 2013). It will be interesting to see how the photoreceptor pathways in this fungus contribute to this response. Conversely, light represses infectivity in the rice blast pathogen M. oryzae and this repression is dependent upon the wc-1 ortholog (Kim et al. 2011b). Moreover, the deletion of the wc-1 ortholog leads to attenuated virulence in the gray mold pathogen B. cinerea (Canessa et al. 2013) and the maize leaf pathogen C. zae-maydis (Kim et al. 2011a). Similarly, and perhaps more surprisingly, deletion of the wc-1 ortholog also attenuates virulence in two human fungal pathogens, the basidiomycete yeast C. neoformans (Idnurm and Heitman 2005a) and the ascomycete mold F. oxysporum (Ruiz-Roldan et al. 2008). As it seems that the perception of light within the host is negligible, it is possible that the WCC is regulating virulence associated genes (e.g., toxins, stress resistance genes) in the dark. Taken together, these data suggest that photoreceptors are conservatively linked to fungal pathogenesis and may represent novel targets for antifungal therapy in both plants and humans.
A second interesting area for future research involves the circadian clocks in other fungal species. As mentioned, developmental rhythms are notably lacking in other fungi, potentially due to unknown variables in culturing conditions, entrainment regimens, and/or genetic background. However, these difficulties can be bypassed or mitigated by following the molecular rhythms of candidate ccgs, i.e., homologs of genes that are known to be rhythmic in Neurospora. While real-time PCR and northern blot analyses remain an option for genetically intractable species, the optimal approach seems to be the employment of firefly luciferase that is either driven by candidate ccg promoters or translationally fused to the candidate ccg proteins (Gooch et al. 2008; Larrondo et al. 2012). In this way, gene expression can be followed in real time under a variety of conditions without time-consuming RNA/protein time courses that must be followed by variable blotting/PCR analyses. Moreover, the emergence of new genome sequences and optimized transformation protocols will allow for the influence of putative core-clock components, e.g., the wc-1/wc-2, to be analyzed through gene deletion.
In conclusion, light is a pervasive environmental signal that regulates a myriad of fungal processes that may impact human health and agriculture and industry. Only time will tell to what extent understanding fungal visual systems will influence the fields that are impacted by these important organisms.
References
Avalos J, Estrada AF (2010) Regulation by light in Fusarium. Fungal Genet Biol 47:930–938
Avelar GM, Schumacher RI, Zaina PA, Leonard G, Richards TA, Gomes SL (2014) A rhodopsin-guanylyl cyclase gene fusion functions in visual perception in a fungus. Curr Biol 24:1234–1240
Baker C, Loros J, Dunlap J (2012) The circadian clock of Neurospora crassa. FEMS Microbiol Rev 36:95–110
Ballario P, Vittorioso P, Magrelli A, Talora C, Cabibbo A, Macino G (1996) White collar-1, a central regulator of blue light responses in Neurospora, is a zinc finger protein. EMBO J 15:1650–1657
Bayram Ö, Biesemann C, Krappmann S, Galland P, Braus GH (2008a) More than a repair enzyme: Aspergillus nidulans photolyase-like CryA Is a regulator of sexual development. Mol Biol Cell 19:3254–3262
Bayram Ö, Krappmann S, Ni M, Bok JW, Helmstaedt K, Valerius O, Braus-Stromeyer S, Kwon N, Keller NP, Yu J, Braus GH (2008b) VelB/VeA/LaeA complex coordinates light signal with fungal development and secondary metabolism. Science 320:1504–1506
Bayram Ö, Braus GH, Fischer R, Rodriguez-Romero J (2010) Spotlight on Aspergillus nidulans photosensory systems. Fungal Genet Biol 47:900–908
Belden WJ, Larrondo LF, Froehlich AC, Shi M, Chen C, Loros JJ, Dunlap JC (2007) The band mutation in Neurospora crassa is a dominant allele of ras-1 implicating RAS signaling in circadian output. Genes Dev 21:1494–1505
Bell-Pedersen D, Cassone V, Earnest D, Golden S, Hardin P, Thomas T, Zoran M (2005) Circadian rhythms from multiple oscillators: lessons from diverse organisms. Nat Rev Genet 6:544–556
Berrocal-Tito GM, Esquivel-Naranjo EU, Horwitz BA, Herrera-Estrella A (2007) Trichoderma atroviride PHR1, a fungal photolyase responsible for DNA repair, autoregulates its own photoinduction. Eukaryot Cell 6:1682–1692
Bieszke JA, Braun EL, Bean LE, Kang S, Natvig DO, Borkovich KA (1999) The nop-1 gene of Neurospora crassa encodes a seven transmembrane helix retinal-binding protein homologous to archaeal rhodopsins. Proc Natl Acad Sci USA 96:8034–8039
Bluhm BH, Dunkle LD (2008) PHL1 of Cercospora zeae-maydis encodes a member of the photolyase/cryptochrome family involved in UV protection and fungal development. Fungal Genet Biol 45:1364–1372
Bluhm BH, Burnham AM, Dunkle LD (2010) A circadian rhythm regulating hyphal melanization in Cercospora kikuchii. Mycologia 102:1221–1228
Blumenstein A, Vienken K, Tasler R, Purschwitz J, Veith D, Frankenberg-Dinkel N, Fischer R (2005) The Aspergillus nidulans phytochrome FphA represses sexual development in red light. Curr Biol 15:1833–1838
Brandt S, von Stetten D, Günther M, Hildebrandt P, Frankenberg-Dinkel N (2008) The fungal phytochrome FphA from Aspergillus nidulans. J Biol Chem 283:34605–34614
Brenna A, Grimaldi B, Filetici P, Ballario P (2012) Physical association of the WC-1 photoreceptor and the histone acetyltransferase NGF-1 is required for blue light signal transduction in Neurospora crassa. Mol Biol Cell 23:3863–3872
Brown L (2004) Fungal rhodopsins and opsin-related proteins: eukaryotic homologues of bacteriorhodopsin with unknown functions. Photochem Photobiol Sci 3:555–565
Bruce V, Weight F, Pittendrigh C (1960) Resetting the sporulation rhythm in Pilobolus with short light flashes of high intensity. Science 131:728–730
Canessa P, Schumacher J, Hevia M, Tudzynski P, Larrondo L (2013) Assessing the effects of light on differentiation and virulence of the plant pathogen Botrytis cinerea: characterization of the white collar complex. PLoS One 8:e84223
Casas-Flores S, Rios-Momberg M, Bibbins M, Ponce-Noyola P, Herrera-Estrella A (2004) BLR-1 and BLR-2, key regulatory elements of photoconidiation and mycelial growth in Trichoderma atroviride. Microbiology 150:3561–3569
Castrillo M, García-Martínez J, Avalos J (2013) Light-dependent functions of the Fusarium fujikuroi CryD DASH cryptochrome in development and secondary metabolism. Appl Environ Microbiol 79:2777–2788
Chen CH, Ringelberg CS, Gross RH, Dunlap JC, Loros JJ (2009) Genome-wide analysis of light-inducible responses reveals hierarchical light signalling in Neurospora. EMBO J 28:1029–1042
Chen CH, DeMay BS, Gladfelter AS, Dunlap JC, Loros JJ (2010a) Physical interaction between VIVID and white collar complex regulates photoadaptation in Neurospora. Proc Natl Acad Sci USA 107:16715–16720
Chen CH, Dunlap JC, Loros JJ (2010b) Neurospora illuminates fungal photoreception. Fungal Genet Biol 47:922–929
Collett MA, Garceau N, Dunlap JC, Loros JJ (2002) Light and clock expression of the Neurospora clock gene frequency Is differentially driven by but dependent on WHITE COLLAR-2. Genetics 160:149–158
Collopy P, Colot H, Park G, Ringelberg C, Crew C, Borkovich K, Dunlap J (2010) High-throughput construction of gene deletion cassettes for generation of Neurospora crassa knockout strains. Methods Mol Biol 638:33–40
Colot HV, Park G, Turner GE, Ringelberg C, Crew CM, Litvinkova L, Weiss RL, Borkovich KA, Dunlap JC (2006) A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc Natl Acad Sci USA 103:10352–10357
Corrochano LM, Cerdá-Olmedo E (1992) Sex, light and carotenes: the development of Phycomyces. Trends Genet 8:268–274
Corrochano LM, Garre V (2010) Photobiology in the Zygomycota: Multiple photoreceptor genes for complex responses to light. Fungal Genet Biol 47:893–899
Dong W, Tang X, Yu Y, Nilsen R, Kim R, Griffith J, Arnold J, Schuttler H (2008) Systems biology of the clock in Neurospora crassa. PLoS One 3:e3105
Dunlap J, Loros J (2004) The Neurospora circadian system. J Biol Rhythms 19:414–424
Dunlap JC, Loros JJ (2006) How fungi keep time: circadian system in Neurospora and other fungi. Curr Opin Microbiol 9:579–587
El-Jack M, Mackenzie M, Bramley P (1988) The photoregulation of carotenoid biosynthesis in Aspergillus giganteus mut. alba. Planta 174:59–66
Estrada AF, Avalos J (2008) The white collar protein WcoA of Fusarium fujikuroi is not essential for photocarotenogenesis, but is involved in the regulation of secondary metabolism and conidiation. Fungal Genet Biol 45:705–718
Estrada AF, Avalos J (2009) Regulation and targeted mutation of opsA, coding for the NOP-1 opsin orthologue in Fusarium fujikuroi. J Mol Biol 387:59–73
Fanelli F, Schmidt-Heydt M, Haidukowski M, Susca A, Geisen R, Logrieco A, Mulè G (2012) Influence of light on growth, conidiation and fumonisin production by Fusarium verticillioides. Fungal Biol 116:241–248
Froehlich AC, Liu Y, Loros JJ, Dunlap JC (2002) White Collar-1, a circadian blue light photoreceptor, binding to the frequency promoter. Science 297:815–819
Froehlich AC, Noh B, Vierstra RD, Loros J, Dunlap JC (2005) Genetic and molecular analysis of phytochromes from the filamentous fungus Neurospora crassa. Eukaryot Cell 4:2140–2152
Froehlich AC, Chen C, Belden WJ, Madeti C, Roenneberg T, Merrow M, Loros JJ, Dunlap JC (2010) Genetic and molecular characterization of a cryptochrome from the filamentous fungus Neurospora crassa. Eukaryot Cell 9:738–750
Fuller KK, Ringelberg CS, Loros JJ, Dunlap JC (2013) The fungal pathogen Aspergillus fumigatus regulates growth, metabolism, and stress resistance in response to light. MBio 4:pii.e00142–13
Gin E, Diernfellner A, Brunner M, Höfer T (2013) The Neurospora photoreceptor VIVID exerts negative and positive control on light sensing to achieve adaptation. Mol Syst Biol 9:667
Gooch VD, Mehra A, Larrondo LF, Fox J, Touroutoutoudis M, Loros JJ, Dunlap JC (2008) Fully codon-optimized luciferase uncovers novel temperature characteristics of the Neurospora clock. Eukaryot Cell 7:28–37
Greene AV, Keller N, Haas H, Bell-Pedersen D (2003) A circadian oscillator in Aspergillus spp. regulates daily development and gene expression. Eukaryot Cell 2:231–237
Guzmán-Moreno J, Flores-Martínez A, Brieba LG, Herrera-Estrella A (2014) The Trichoderma reesei Cry1 protein is a member of the cryptochrome/photolyase family with 6-4 photoproduct repair activity. PLoS One 9:e100625
Hatakeyama R, Nakahama T, Higuchi Y, Kitamoto K (2007) Light represses conidiation in koji mold Aspergillus oryzae. Biosci Biotechnol Biochem 71:1844–1849
He Q, Cheng P, Yang Y, Wang L, Gardner KH, Liu Y (2002) White collar-1, a DNA binding transcription factor and a light sensor. Science 297:840–843
Heintzen C, Loros JJ, Dunlap JC (2001) The PAS protein VIVID defines a clock-associated feedback loop that represses light input, modulates gating, and regulates clock resetting. Cell 104:453–464
Hitomi K, Okamoto K, Daiyasu H, Miyashita H, Iwai S, Toh H, Ishiura M, Todo T (2000) Bacterial cryptochrome and photolyase: characterization of two photolyase-like genes of Synechocystis sp. PCC6803. Nucleic Acids Res 28:2353–2362
Hunt SM, Thompson S, Elvin M, Heintzen C (2010) VIVID interacts with the WHITE COLLAR complex and FREQUENCY-interacting RNA helicase to alter light and clock responses in Neurospora. Proc Natl Acad Sci USA 107:16709–16714
Hurley JM, Larrondo LF, Loros JJ, Dunlap JC (2013) Conserved RNA helicase FRH acts nonenzymatically to support the intrinsically disordered Neurospora clock protein FRQ. Mol Cell 52:832–843
Hurley H, Dasgupta A, Emerson J, Zhou X, Ringelberg C, Knabe N, Lipzen A, Lindquist E, Daum C, Barry K, Grigoriev I, Smith K, Galagan J, Bell-Pedersen D, Freitag M, Cheng C, Loros J, Dunlap J (2014) Analysis of clock-regulated genes in Neurospora reveals widespread post-transcriptional control of metabolic potential. Proc Natl Acad Sci USA (in Press)
Idnurm A, Heitman J (2005a) Light controls growth and development via a conserved pathway in the fungal kingdom. PLoS Biol 3:e95
Idnurm A, Heitman J (2005b) Photosensing fungi: phytochrome in the spotlight. Curr Biol 15:R829–R832
Idnurm A, Heitman J (2010) Ferrochelatase is a conserved downstream target of the blue light-sensing white collar complex in fungi. Microbiology 156:2393–2407
Idnurm A, Verma S, Corrochano LM (2010) A glimpse into the basis of vision in the kingdom Mycota. Fungal Genet Biol 47:881–892
Kamada T, Sano H, Nakazawa T, Nakahori K (2010) Regulation of fruiting body photomorphogenesis in Coprinopsis cinerea. Fungal Genet Biol 47:917–921
Kim H, Ridenour J, Dunkle L, Bluhm B (2011a) Regulation of stomatal tropism and infection by light in Cercospora zeae-maydis: evidence for coordinated host/pathogen responses to photoperiod? PLoS Pathog 7:e1002113
Kim S, Singh P, Park J, Park S, Friedman A, Zheng T, Lee Y, Lee K (2011b) Genetic and molecular characterization of a blue light photoreptor MGWC-1 in Magnaporthe oryzae. Fungal Genet Biol 48:400–407
Kim H, Son H, Lee Y- (2014a) Effects of light on secondary metabolism and fungal development of Fusarium graminearum. J Appl Microbiol 116:380–389
Kim Y, Choi J, Lee H, Lee G, Lee Y, Choi D (2014b) dbCRY: a Web-based comparative and evolutionary genomics platform for blue-light receptors. Database (Oxford) 2014:bau037
Kitamoto A, Suzuki S, Furukawa S (1972) An action spectrum for light-induced primordium formation in a basidiomycete, Favolus arcularius (FR) Ames. Plant Physiol 49:338–340
Larrondo L, Loros J, Dunlap J (2012) High resolution spatiotemporal analysis of gene expression in real time: in vivo analysis of circadian rhythms in Neurospora crassa using a FREQUENCY-luciferase translational reporter. Fungal Genet Biol 49:681–683
Lee K, Singh P, Chung W, Ash J, Kim TS, Hang L, Park S (2006) Light regulation of asexual development in the rice blast fungus, Magnaporthe oryzae. Fungal Genet Biol 43:694–706
Libkind D, Pérez P, Sommaruga R, Diéguez delC M, Brizzio S, Zagarese H, van Broock MF (2004) Constitutive and UV-inducible synthesis of photoprotective compounds (carotenoids and mycosporines) by freshwater yeasts. Photochem Photobiol Sci 3:281–286
Linden H, Macino G (1997) White collar 2, a partner in blue-light signal transduction, controlling expression of light-regulated genes in Neurospora crassa. EMBO J 16:98–109
Linden H, Ballario P, Macino G (1997) Blue light regulation in Neurospora crassa. Fungal Genet Biol 22:141–150
Liu Y, Cheng P (2003) Photoreception in Neurospora: a tale of two white collar proteins. Cell Mol Life Sci 60:2131–2138
Lombardi LM, Brody S (2005) Circadian rhythms in Neurospora crassa: clock gene homologues in fungi. Fungal Genet Biol 42:887–892
Losi A (2007) Flavin-based blue-light photosensors: a photobiophysics update. Photochem Photobiol 83:1283–1300
Lucas J, Kendrick R, Givan C (1975) Photocontrol of fungal spore germination. Plant Physiol 56:847–849
Lukens RJ (1965) Reversal by red light of blue light inhibition of sporulation in Alternaria solani. Phytopathology 55:1032
Malzahn E, Ciprianidis S, Kaldi K, Schafmeier T, Brunner M (2010) Photoadaptation in Neurospora by competitive interaction of activating and inhibitory LOV domains. Cell 142:762–772
McClung CR, Gutiérrez RA (2010) Network news: prime time for systems biology of the plant circadian clock. Curr Opin Genet Dev 20:588–598
Mooney JL, Yager LN (1990) Light is required for conidiation in Aspergillus nidulans. Genes Dev 4:1473–1482
Okamoto S, Furuya K, Nozaki S, Aoki K, Niki H (2013) Synchronous activation of cell division by light or temperature stimuli in the dimorphic yeast Schizosaccharomyces japonicus. Eukaryot Cell 12:1235–1243
Olmedo M, Ruger-Herreros C, Luque EM, Corrochano LM (2010) A complex photoreceptor system mediates the regulation by light of the conidiation genes con-10 and con-6 in Neurospora crassa. Fungal Genet Biol 47:352–363
Page R (1962) Light and the asexual reproduction of Pilobolus. Science 138:1238–1245
Perkins J, Gordon S (1969) Morphogenesis in Schizophyllum commune II. Effects of monochromatic light. Plant Physiol 44:1712–1716
Pittendrigh C, Bruce V, Rosensweig N, Rubin M (1959) Growth patterns in Neurospora: a biological clock in Neurospora. Nature 184:169–170
Pruß S, Fetzner R, Seither K, Herr A, Pfeiffer E, Metzler M, Lawrence CB, Fischer R (2014) Role of the Alternaria alternata blue-light receptor LreA (white-collar 1) in spore formation and secondary metabolism. Appl Environ Microbiol 80:2582–2591
Purschwitz J, Müller S, Kastner C, Schöser M, Haas H, Espeso EA, Atoui A, Calvo AM, Fischer R (2008) Functional and physical interaction of blue- and red-light sensors in Aspergillus nidulans. Curr Biol 18:255–259
Purschwitz J, Müller S, Fischer R (2009) Mapping the interaction sites of Aspergillus nidulans phytochrome FphA with the global regulator VeA and the white collar protein LreB. Mol Genet Genomics 281:35–42
Quiles-Rosillo M, Ruiz-Vázquez R, Torres-Martínez S, Garre V (2005) Light induction of the carotenoid biosynthesis pathway in Blakeslea trispora. Fungal Genet Biol 42:141–153
Rangel DEN, Fernandes EKK, Bragu GUL, Roberts DW (2011) Visible light during mycelial growth and conidiation of Metarhizum robertsii produces conidia with increased stress tolerance. FEMS Microbiol Lett 315:81–86
Richartz G, MacLellan A (1987) Action spectra for hyphal aggregation, the first stage of fruiting, in the basidiomycete Pleurotus ostreatus. Photochem Photobiol 45:815–820
Rodriguez-Romero J, Corrochano L (2004) The gene for the heat-shock protein HSP100 is induced by blue light and heat-shock in the fungus Phycomyces blakesleeanus. Curr Genet 46:295–303
Rodriguez-Romero J, Corrochano L (2006) Regulation by blue light and heat shock of gene transcription in the fungus Phycomyces: proteins required for photoinduction and mechanism for adaptation to light. Mol Microbiol 61:1049–1059
Rodriguez-Romero J, Hedtke M, Kastner C, Müller S, Fischer R (2010) Fungi, hidden in soil or up in the air: light makes a difference. Annu Rev Microbiol 64:585–610
Röhrig J, Kastner C, Fischer R (2013) Light inhibits spore germination through phytochrome in Aspergillus nidulans. Curr Genet 59:55–62
Ruger-Herreros C, Rodríguez-Romero J, Fernández-Barranco R, Olmedo M, Fischer R, Corrochano LM, Canovas D (2011) Regulation of conidiation by light in Aspergillus nidulans. Genetics 188:809–822
Ruiz-Roldan MC, Garre V, Guarro J, Marine M, Roncero MI (2008) Role of the White Collar 1 photoreceptor in carotenogenesis, UV resistance, hydrophobicity, and virulence of Fusarium oxysporum. Eukaryot Cell 7:1227–1230
Salichos L, Rokas A (2010) The diversity and evolution of circadian clock proteins in fungi. Mycologia 102:269–278
Sanchez-Arreguin A, Perez-Martinez A, Herrera-Estrella A (2012) Proteomic analysis of Trichoderma atroviride reveals independent roles for transcription factors BLR-1 and BLR-2 in light and darkness. Eukaryot Cell 11:30–41
Sano H, Kaneko S, Sakamoto Y, Sato T, Shishido K (2009) The basidiomycetous mushroom Lentinula edodes white collar-2 homolog PHRB, a partner of putative blue-light photoreceptor PHRA, binds to a specific site in the promoter region of the L. edodes tyrosinase gene. Fungal Genet Biol 46:333–341
Saranak J, Foster KW (1997) Rhodopsin guides fungal phototaxis. Nature 387:465–466
Sargent M, Briggs W, Woodward D (1966) Circadian nature of a rhythm expressed by an invertaseless strain of Neurospora crassa. Plant Physiol 41:1343–1349
Schaller GE, Shiu SH, Armitage JP (2011) Two-component systems and their co-option for eukaryotic signal transduction. Curr Biol 21:R320–R330
Schmoll M, Franchi L, Kubicek CP (2005) Envoy, a PAS/LOV Domain Protein of Hypocrea jecorina (anamorph Trichoderma reesei), modulates cellulase gene transcription in response to light. Eukaryot Cell 4:1998–2007
Schmoll M, Schuster A, Silva Rdo N, Kubicek CP (2009) The G-alpha protein GNA3 of Hypocrea jecorina (anamorph Trichoderma reesei) regulates cellulase gene expression in the presence of light. Eukaryot Cell 8:410–420
Schneider MJ, Murray BJ (1979) Phytochrome mediation of uredospore germination in the fungus Puccinia graminis. Photochem Photobiol 29:1051–1052
Schuch AP, Garcia CC, Makita K, Menck CF (2013) DNA damage as a biological sensor for environmental sunlight. Photochem Photobiol Sci 12:1259–1272
Schumacher J, Simon A, Cohrs K, Viaud M, Tudzynski P (2014) The transcription factor BcLTF1 regulates virulence and light responses in the nectrotrophic plant pathogen Botrytis cinerea. PLoS Genet 10:e1004040
Schuster A, Kubicek CP, Friedl MA, Druzhinina IS, Schmoll M (2007) Impact of light on Hypocrea jecorina and the multiple cellular roles of ENVOY in this process. BMC Genom 8:449
Schwerdtfeger C, Linden H (2003) VIVID is a flavoprotein and serves as a fungal blue light photoreceptor for photoadaptation. EMBO J 22:4846–4855
Seibel C, Tisch D, Kubicek CP, Schmoll M (2012) ENVOY Is a major determinant in regulation of sexual development in Hypocrea jecorina (Trichoderma reesei). Eukaryot Cell 11:885–895
Shinohara ML, Correa A, Bell-Pedersen D, Dunlap JC, Loros JJ (2002) Neurospora clock-controlled gene 9 (ccg-9) encodes trehalose synthase: circadian regulation of stress responses and development. Eukaryot Cell 1:33–43
Shiu C, Lee T (2005) Ultraviolet-B-induced oxidative stress and responses of the ascorbate–glutathione cycle in a marine macroalga Ulva fasciata. J Exp Bot 56:2851–2865
Silva F, Torres-Martínez S, Garre V (2006) Distinct white collar-1 genes control specific light responses in Mucor circinelloides. Mol Microbiol 61:1023–1037
Tan KK (1974) Red-far-red reversible photoreaction in the recovery from blue-light inhibition of sporulation in Botrytis cinerea. J Gen Microbiol 8a:201–202
Tisch D, Schmoll M (2013) Targets of light signalling in Trichoderma reesei. BMC Genom 14:657
Veluchamy S, Rollins JA (2008) A CRY-DASH-type photolyase/cryptochrome from Sclerotinia sclerotiorum mediates minor UV-A-specific effects on development. Fungal Genet Biol 45:1265–1276
Verma S, Idnurm A (2013) The Uve1 endonuclease is regulated by the white collar complex to protect Cryptococcus neoformans from UV damage. PLoS Genet 9:e1003769
Yang T, Dong C (2014) Photo morphogenesis and photo response of the blue-light receptor gene Cmwc-1 in different strains of Cordyceps militaris. FEMS Microbiol Lett 352:190–197
Yu S-, Ramkumar G, Lee YH (2013) Light quality influences the virulence and physiological responses of Colletotrichum acutatum causing anthracnose in pepper plants. J Appl Microbiol 115:509–516
Zhu P, Zhang C, Xiao H, Wang Y, Toyoda H, Xu L (2013) Exploitable regulatory effects of light on growth and development of Botrytis cinerea. J Plant Path 95:499–507
Zoltowski BD, Schwerdtfeger C, Widom J, Loros JJ, Bilwes AM, Dunlap JC, Crane BR (2007) Conformational switching in the fungal light sensor Vivid. Science 316:1054–1057
Acknowledgments
The authors would like to than Jillian Emerson for her contributions to the figure. Our own work was supported by the National Institute of General Medical Sciences of the National Institute of Health to J.J.L (Grant RO1 GM083336) and J.C.D (Grants RO1 GM34985 and PO1 GM68087). This review article is supported, in part, by a grant from the São Paulo Research Foundation (FAPESP) of Brazil #2014/01229-4.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Communicated by D.E.N. Rangel.
This article is part of the Special Issue “Fungal Stress Responses”.
Rights and permissions
About this article
Cite this article
Fuller, K.K., Loros, J.J. & Dunlap, J.C. Fungal photobiology: visible light as a signal for stress, space and time. Curr Genet 61, 275–288 (2015). https://doi.org/10.1007/s00294-014-0451-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00294-014-0451-0