Abstract
The evolutionarily conserved Dim1 proteins belong to the TRX fold superfamily. An EST showing high identity values with genes coding for Dim1 proteins was selected from an EST library collection of Trichoderma virens T59. Here, we report the cloning, characterization, and functional analysis of a T. virens T59 TvDim1 gene. The TvDim1 gene, with a sequence size of 614 bp, was PCR-amplified and found to contain three introns. The TvDim1 gene was present as a single copy in the T. virens genome and was also present in another five Trichoderma strains investigated. Increased levels of expression and redox-activity were detected when the fungus was grown in the presence of H2O2. The overexpression and silencing of TvDim1 in T. harzianum T34 gave rise to transformants, with higher and lower growth, redox-activity, and quantities of biomass, respectively, than the wild-type strain after culture under oxidative stress.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Most species of Trichoderma have been linked to biocontrol and biotechnological applications. The antagonistic properties of Trichoderma species are well known and have been related to mechanisms of action, such as mycoparasitism (Benítez et al. 2004; Lorito 1998), antibiosis (Vinale et al. 2006), and competition for nutrients (Sivan and Chet 1989), and their capacity to promote plant growth (Harman et al. 2004) and induce plant defense (Alfano et al. 2007; Djonovic et al. 2006, 2007; Shoresh and Harman 2008; Viterbo and Chet 2006). Nevertheless, little is known about aspects related to the development and cell cycle of this fungus (Brunner et al. 2008; Esquivel-Naranjo and Herrera-Estrella 2007; Rocha-Ramírez et al. 2002).
The thioredoxin (TRX)-like fold superfamily is a group of proteins that has been related to a large number of functions in the cell, such as the regulation of redox homeostasis (Grant 2001), impaired cell cycles and sexual development (Berry and Gould 1997; Malagnac et al. 2007), pre-mRNA splicing (Simeoni and Divita 2007), and protein folding (Ferrari et al. 1998). These proteins show similar three-dimensional structures, the so-called TRX fold (Martin 1995), which is present in several classes of proteins, such as TRX, glutaredoxin, the Dsb protein from Escherichia coli, glutathione peroxidase, glutathione S-transferase, peroxiredoxin, disulfide isomerase, cytochrome c oxidase, iodothyronine selenodeiodinases, and the evolutionarily conserved Dim1 protein (Stefankova et al. 2005). In these proteins the TRX fold can be modified by additional amino acid residues at the N and/or C terminus of the sequence and some insertion in the specific regions of this fold (Stefankova et al. 2005). In this sense, although Dim1 proteins structurally belong to the TRX superfamily, the most important feature of TRXs—the canonical CxxC motif—is replaced by a DxxC motif (Reuter et al. 1999). The importance of the two cysteines in the reduction of protein disulfide bonds, and hence in protecting the cell from oxidative stress, is not clear (Fomenko and Gladyshev 2003).
A TRX study including several Dim1 proteins revealed high homology and similarity of protein secondary structure, although such proteins also showed a highly conserved C-terminal extension, important for its biological function (Zhang et al. 1999). These proteins have been identified as part of the spliceosome, a complex machine comprising several elements involved in the removal of introns from nuclear mRNA precursors. Dim-encoding genes have been identified in Homo sapiens, hDim1; in Saccharomyces cerevisiae, Dib1p/Snu16p (Reuter et al. 1999); and in Schizosaccharomyces pombe, Dim1p (Berry and Gould 1997). Their role in pre-mRNA splicing is not clear, but this protein appears to bind other U5-specific proteins, such as Prp1p/Prp6p (Carnahan et al. 2005).
In work with mutants, other functions have also been assigned to Dim proteins. Thus, Dim1p has been shown to be essential for G2/M progression and for chromosome segregation during mitosis in S. pombe (Berry and Gould 1997). Gene deletions—of Dim1p in S. pombe and of Dib1p in S. cerevisiae—produce a lethal G2 arrest (Carnahan et al. 2005), and Dim1 deletion also induces lethality during embryogenesis in metazoans, such as Caenorhabditis elegans (Zhang et al. 2000).
Highly conserved Dim sequences are present in all genomes but to date no Dim1 proteins have been described in filamentous fungi. In the present work, we explored the presence of Dim1-encoding genes in our collection of Trichoderma expressed sequence tags (ESTs), generated in a functional genomics project carried out on eight Trichoderma species (Rey et al. 2004). An EST showing high identity with Dim1 proteins was identified in a Trichoderma virens T59 cDNA library. Here, we report the cloning and characterization of the TvDim1 gene in the biocontrol fungus T. virens T59. Using a heterologous expression approach, the TvDim1 gene from T. virens T59 was overexpressed and silenced in T. harzianum T34, a strain commonly used in basic research studies, to investigate the relationship of this gene with the regulation of redox homeostasis.
Materials and methods
Bacterial and fungal strains
Escherichia coli DH5α (Invitrogen Life Technologies, Carlsbad, CA, USA) was used as host for plasmid construction and propagation. This bacterial strain was grown in Luria–Bertani (LB) broth or on LB plates, which were supplemented with ampicillin (0.1 mg/ml), X-gal (40 μg/ml), and IPTG (10 μg/ml) when required.
Six Trichoderma strains were used in this study: T. atroviride T11, T. asperellum T25, T. longibrachiatum T52, and T. virens T59 from the NBT collection (Newbiotechnic S.A., Sevilla, Spain), T. harzianum T22 (ATCC 20847, American Type Culture Collection, Manassas, USA), and T. harzianum T34 (CECT 2413, Spanish Type Culture Colletion, Burjasot, Spain). Four pathogenic fungal strains were used in the antifungal assays: Botrytis cinerea 98, isolated from diseased strawberry plants; Fusarium oxysporum f. sp. lycopersici (CECT 2866); Rhizoctonia solani (CECT 2815), and Colletotrichum acutatum 73 (IMI 364856, CABI Bioscience, Egham, UK), isolated from diseased oranges. All fungal strains were grown on Potato Dextrose Agar (PDA, Difco Becton Dickinson, Sparks, MD, USA) and were maintained at −80°C in a 20% glycerol solution, with the exception of the R. solani strain, which was maintained at 25°C in sterilized wheat grains and B. cinerea strain, which was grown on Malt Extract Agar (MEA).
EST database
As much as 26 cDNA libraries were previously constructed for the TrichoEST project using different mRNA populations from 10 strains belonging to eight Trichoderma species expressed under mycoparasitic and nutrient stress conditions (Rey et al. 2004). Expressed sequence tags (ESTs) were generated by sequencing cDNA clones from the 5′ end, and an EST database was compiled with 13,814 unique ESTs (Vizcaíno et al. 2006, 2007).
cDNA library
L20, a cDNA library constructed with RNA from T. virens T59 for the TrichoEST project, was used (Vizcaíno et al. 2007). Biomass was obtained following a two-step liquid culture procedure. First, the fungus was grown in minimal medium (MM) (Penttilä et al. 1987) containing 2% glucose at 28°C for 36 h. Mycelia were harvested and transferred to MM without glucose under the following substrates/conditions in separate cultures: 1.5% crab chitin for 8 h; nitrogen starvation for 8 h; 1.5% crab chitin and pH 3.5 for 4 h, and 2% plant cell wall polymers for 96 h. Nitrogen starvation conditions included a 100-fold decrease in the concentration of ammonium sulfate in the medium (50 mg/l). RNA was extracted using the TRIZOL® reagent (Invitrogen Life Technologies, Carlsbad, CA, USA) according to the manufacturer’s instructions. After total RNA extraction, equal amounts of RNA from the four different growth conditions were mixed and used for mRNA purification, employing Dynabeads (Dynal, Oslo, Norway). The cDNA library was constructed using the Uni-ZAP XR Vector System (Stratagene, La Jolla, CA, USA), following the manufacturer’s instructions.
DNA and RNA manipulations
Standard molecular techniques were performed throughout these studies (Sambrook and Russel 2001). Mycelia for DNA extractions were obtained from Potato Dextrose Broth (PDB, Difco) cultures incubated at 28°C and 200 rpm for 2 days. Then, they were collected by filtration, washed with distilled water, frozen, and lyophilized. Fungal genomic DNA was isolated according to previously described protocols (Raeder and Broda 1985).
For Southern analysis, 10 μg of genomic DNA was digested with restriction enzymes, electrophoresed on 0.7% agarose gels, and transferred to a Hybond-N+ membrane (Amersham Biosciences AB, Uppsala, Sweden). The probe was labeled with 32P using Ready-To-Go DNA labeling beads (32P-dCTP) kit (Amersham). Hybridizations were carried out at 65°C for 16 h. Membranes were washed under high-stringency conditions.
Mycelia for RNA extraction and qRT-PCR expression analyses were obtained following a two-step liquid culture procedure, as described above, except that the following substrates/conditions were used in separate cultures: glucose starvation (0% glucose), 1 mM H2O2, 0.5 mM Paraquat® (Afrasa S.A., Valencia, Spain), and 10% NaCl. Cultures were maintained at 25°C on a rotary shaker at 150 rpm for 4, 8, or 24 h. Mycelia were collected by filtration, thoroughly washed with sterile water, lyophilized, and kept at −80°C until RNA extraction. Fungal RNA was obtained as described above.
PCR procedures
The TvDim1 gene was isolated by PCR from strain T59 genomic DNA using the degenerated primer 614-4 (5′-ATGGGT/CTCC/TGTCGTTCTCCC-3′), designed on the 5′ end from an alignment of Dim1 genes from databases, and the primer 614-3 (5′-GTAATCCTTGGGGCTGACC-3′), designed on the 3′ end from the EST 614. The TvDim1 cDNA was PCR-amplified using phagemid DNA from the cDNA library L20 as template and the TRX-1 (5′-ATGGGCTCTGTCGTTCTC-3′) and TRX-3 (5′-CTA GTATCTGTGTCTCGT-3′) primers.
The primer pair TRX-1 and 614-3 was used to amplify the TvDim1 gene, using the genomic DNA from T. virens T59 as template. This PCR product was used as probe in Southern blot experiment.
Screening of the T. harzianum T34 overexpressed transformants was carried out by PCR with the primers pKi-614 (5′-GGAGCTTGACGAAGACCTG-3′) and 614-3, described above. The screening of T. harzianum T34 silenced transformants was also carried out by PCR with the primer pair pta-p (5′-GATGCACTGCAGTCCACATTG-3′) and 614-3.
PCR amplifications were carried out for 35 cycles of 1 min at 94°C, 1 min at 55°C, and 1 min 30 s at 72°C, with the Pfu or Taq (for the screening of transformants) polymerase system (Biotools, Edmonton, Canada), following the manufacturer’s instructions.
qRT-PCR analysis
For the expression analysis of TvDim1 gene in T. virens T59, total RNA was extracted using the TRIZOL® reagent (Invitrogen) and 1 μg of total RNA was used for a RT reaction using reverse transcription system (Promega, Madison, WI, USA) following the manufacturer’s instructions. The real-time PCR was performed using the Stratagene’s Brilliant SYBR® Green QPCR master mix (Stratagene, La Jolla, CA, USA). The reaction mix was prepared according the manufacturer’s instructions. The primer pair TRX-5 (5′-GAAGAGGATCGTCTCGTCGTC-3′) and TRX-3 (5′-TCAGGAACCTCGTCAATGTCG-3′) was used to amplify a 143-bp fragment of TvDim1 gene and the β-tubulin gene was used as an internal reference. Amplifications were carried out in the ABI PRISM® 7000 (Applied Biosystems, Foster City, CA, USA) and the thermal cycler was programmed as follows: one cycle of 10 min at 95°C, 40 two-step cycles of 15 s at 95°C, and 1 min at 60°C.
In a similar way, the relative expression of TvDim1 gene in the overexpressed transformants T. harzianum TvDim1-1, TvDim1-2, and TvDim1-3 respecting to ThDim1 (the endogenous gene) was analyzed by qRT-PCR using the primer pair TRX-5 and TRX-3.
Plasmid constructions and Trichoderma transformation procedure
For gene overexpression, plasmid pLMRS3, which contained the pki1 (pyruvate kinase I) gene promoter and the cbh2 (cellobiohydrolase II) terminator region, both from T. reesei (Mach et al. 1994), was digested with XbaI, treated with a Klenow fragment, and dephosphorylated with calf intestine alkaline phosphatase (CIAP). Then, it was ligated to the TvDim1 ORF. An NsiI–Klenow–EcoRI fragment, containing the pki promoter and the TvDim1 gene, was ligated into plasmid pJL43b (Gutiérrez et al. 1997), to construct the plasmid designated pSPDim1-1 (4.43 kb), which contained the expression cassette (1465 bp). This cassette, containing the pki promoter, the TvDim1 gene, and the CYC1 terminator from Saccharomyces cerevisiae, was isolated by digestion with XhoI and ligated to plasmid pJL43b1 linearized with XhoI. This vector, designated pJLTvDim1 (5.95 kb), contained the phleomycin resistance gene (ble) from Streptoalloteichus hindustanus under the control of the Aspergillus nidulans gpdA gene promoter. The pJLTvDim1 plasmid was used to transform T. harzianum T34 using the previously described protoplast method (Cardoza et al. 2006). Transformants were selected for phleomycin resistance.
For gene silencing, the plasmid pSIL (Sousa 2004) was linearized with BamHI, treated with Klenow fragment, and dephosphorylated with CIAP. Then, it was ligated to a dephosphorylated 432 bp fragment, corresponding to the TvDim1 ORF. This fragment was “in sense” relative to the tss1 promoter. This construction (pSILDim-0) was later linearized with EcoRV and treated with CIAP, after which the ORF was ligated in “anti-sense” with respect to the tss1 promoter. An intron of 150 bp was introduced between both the “sense” and “anti-sense” ORF of the TvDim1 gene. This plasmid also contained the terminator region of the cbh2 gene from T. reesei and was designated pSILTvDim1 (6,114 bp). Plasmids pSILTvDim1 and pJL43b1, which contained phleomycin resistance gene (Gutiérrez and Martín unpublished), were used at a proportion of 9:1 to co-transform T. harzianum T34 using a previously described protoplast method (Cardoza et al. 2006). Transformants were selected as described above.
TRX activity assays
TRX activity was tested in a 1,000-μl reaction volume using the insulin-disulfide reduction assay according to the method of Holmgren (1979). Briefly, the assay was monitored by the addition of a master mix of the following reaction components at the indicated end concentration—63 mM sodium phosphate buffer, pH 7.0, 2 mM EDTA, 0.1% (w/v) insulin solution, and 1 mM DTT—to intracellular protein extracts from Trichoderma cultures. The reactions were incubated at 25°C for 20 min. Then, measurements were performed at OD650 nm every 5 min over a total of 20 min on a spectrophotometer. TRX from Escherichia coli (Sigma-Aldrich, San Luis, MO, USA) at an end concentration of 1 μg/ml was used as a positive control, and the master mix without TRX was used as a negative control. Total activity corresponds to μmol formed in 1 min, and specific activity corresponds to μmol formed in 1 min per mg of protein. Quantitative protein determination was carried out with the Bradford assay (Bradford 1976). Tests were performed in triplicate and data represent mean values with standard deviations.
Oxidative stress resistance assays
The wild-type, three TvDim1-overexpressing transformant strains and three TvDim1-silenced transformant strains were tested for resistance to oxidative stress by measuring colony diameters after incubation under different growth conditions. As much as 200 conidia from each strain were inoculated onto plates containing MM supplemented or not (control) with 1 mM or 3 mM H2O2 and incubated at 28°C for 1 week.
The growth of strain T34, one overexpressing transformant and two silenced transformants was also tested in liquid media. A 250-Erlenmeyer flask containing 100 ml of MM 1% glucose supplemented or not (control) with 1 mM H2O2 was inoculated with 106 conidia and maintained at 28°C for 48 h. Fungal growth was determined by measuring the accumulation of biomass dry weight according to Montero-Barrientos et al. (2007). Both growth assays—in liquid medium and on plates—were carried out in triplicate.
Direct confrontation assays
In vitro confrontation assays between Trichoderma strains and the pathogens F. oxysporum f. sp. lycopersici, C. acutatum, and R. solani (on PDA plates), and B. cinerea (on MEA plates) were carried out at 25°C as previously described (Rubio et al. 2009). These assays were performed in triplicate, and single cultures of Trichoderma strains and pathogens were used as controls.
Sequence analyses
Sequences were analyzed using the DNAstar package (Lasergene). For phylogenetic analysis, gene sequences were aligned using the CLUSTAL X algorithm (Thompson et al. 1997). The NJ tree was constructed with the MEGA software (Kumar et al. 2001) and a bootstrap analysis of 1,000 replicates was performed. The nucleotide sequence of TvDim1 was deposited in the GenBank database under accession number FJ788527.
Results
The TvDim1 gene of T. virens T59
The 421-bp EST 614 was selected for further characterization, based on its high degree of similarity with Dim1 proteins, using the BLASTX algorithm. EST 614 was obtained from the T. virens T59 cDNA library L20, which was constructed after growing the T59 strain under simulated mycoparasitic and plant interaction conditions. The TvDim1 gene was obtained by PCR with the primers 614-4 and 614-3 and T59 genomic DNA as template, and was found to be 614-bp long. The TvDim1 gene contained three introns of 52, 53, and 77 bp. To obtain the ORF of the TvDim1 gene by PCR, the TRX-1 and 614-3 primer pair was designed using the TvDim1 genomic sequence and a cDNA of T59 strain as template. The ORF of the TvDim1 gene was 432-bp long and encoded a protein of 143 amino acids, with a theoretical molecular mass of 16.78 kDa and a deduced isoelectric point of 5.56 units.
The highest degree of similarity (97% amino acid sequence identity) was found with a conserved hypothetical protein from Chaetomium globosum (accession number XP_001229514). The analysis of the 143 amino acids of the predicted sequence revealed the presence of a conserved DXXC domain located at positions 36 to 39, and the carboxy-terminal extension located beyond the TRX homology region, from amino acid 130 to end of the protein. As a result of the TvDim1 structural analysis using the SMART database (http://www.smart.embl.de), a Dim1 domain (pfam 02966) extending from amino acid 5 to 137 was found.
A NJ tree (Fig. 1) was constructed after an alignment of TvDim1 gene with other 13 GenBank-retrieved sequences, representing genes encoding Dim1 proteins from ascomycetes, yeasts, animals and plants, as well as other encoding thioredoxin and peroxiredoxin proteins. Three independent clades containing Dim1, thioredoxin, and peroxiredoxin genes could be identified, supported by bootstrap values of 100%. Dim1 genes were separated according to their evolutionary relationships. The TvDim1 gene from T. virens T59 was located with the other three fungi sequences and particularly, it formed a subclade with a hypothetical Dim1 gene from T. atroviride, separated from animal, plant, and yeast sequences, supported by a bootstrap value of 99%.
Neighbor-joining (NJ) tree of TvDim1 gene from T. virens T59 and other 13 GenBank-retrieved sequences, representing genes encoding Dim1, thioredoxin, and peroxiredoxin proteins. Nucleotide sequences were obtained from T. atroviride (protein id: 83143, http://www.genome.jgi-psf.org/), Neosartorya fischeri, Aspergillus nidulans, Schizosaccharomyces pombe, Kluyveromyces lactis, Drosophyla melanogaster, Homo sapiens, Populus trichocarpa, Arabidopsis thaliana, Aspergillus terreus, and Aspergillus fumigatus. Sequences were aligned using the CLUSTAL X algorithm (Thompson et al. 1997) and the NJ tree was constructed with the MEGA software (Kumar et al. 2001). Sequences are indicated using their GenBank accession numbers. Bar represents one substitution per 10 nucleotides
A Southern blot analysis was performed to determine the copy number of the TvDim1 gene in the T. virens T59 genome and the presence of homologs of this gene in other Trichoderma species (Fig. 2). A single band was detected in the T59 genomic DNA digested with the endonuclease XhoI, which does not cut inside the probe, indicating that the TvDim1 gene exists as a single copy in the genome of T. virens T59. In addition, when the TvDim1 probe was used to hybridize XhoI-digested genomic DNA from another five strains belonging to four different Trichoderma species, hybridization signals were also observed: a single signal in the DNA from T. harzianum T34, T. atroviride T11, and T. asperellum T25 strains, and two signals in the DNA from T. harzianum T22 and T. longibrachiatum T52, indicating that a homologous gene was also present in these strains.
Southern blot analysis of the TvDim1 gene in different Trichoderma species. Genomic DNA from T. atroviride T11, T. harzianum T22, T. asperellum T25, T. harzianum T34, T. longibrachiatum T52, and T. virens T59 XbaI-digested. Hybridization was carried out at 65°C using the TvDim1 gene as probe. Molecular sizes (Kb) are indicated
ESTs showing a high degree of identity with Dim1 proteins were only observed in another two cDNA libraries, obtained under simulated conditions of mycoparasitism and stress (Rey et al. 2004): one from T. stromaticum (92% identity) and one from T. aggressivum (91%). We also detected a single homologous gene in the available on-line genomes of T. virens strain Gv29-8 (an identity percentage of 99.3% with the TvDim1 gene), Hypocrea jecorina (anamorph T. reesei) (89%), and T. atroviride (91%) (http://www.genome.jgi-psf.org/).
Expression and TRX activity analysis in T. virens T59
Expression analysis was carried out after culturing T. virens T59 under conditions related to abiotic stresses, using glucose starvation (0% glucose) as control condition (Fig. 3). TvDim1 gene expression was only detected for 1 mM H2O2 at 8 and 24 h and for 0.5 mM Paraquat 8 h, being observed the highest transcript levels in 24 h cultures of 1 mM H2O2. No TvDim1 gene expression was detected in the rest of conditions analyzed: 10% NaCl at 4, 8, and 24 h, 1 mM H2O2 at 4 h, and for 0.5 mM Paraquat at 4 and 24 h.
qRT-PCR analysis of the TvDim1 gene. The experiment was carried out from mycelia of T. virens T59 grown in MM (Penttilä et al. 1987), without glucose, in the presence of 1 mM H2O2 (H), 0.5 mM Paraquat® (Afrasa S.A., Valencia, Spain) (P), 10% NaCl (N), and glucose starvation (g). Cultures were maintained at 25°C on a rotary shaker at 150 rpm for 4, 8, or 24 h. T. harzianum T34 β-tubulin was used as an internal reference gene
Total TRX activity was quantified after the T59 strain had been grown in the presence of 1 mM H2O2 for 8 h, obtaining a value of 6.12 μmol/min mg−1.
Heterologous expression of TvDim1 in T. harzianum T34
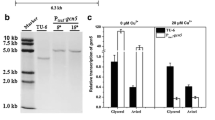
To characterize the T. virens T59 TvDim1 gene functionally, pJLTvDim1 and pSILTvDim1 plasmids were constructed and transformed and cotransformed, respectively, in T. harzianum T34. Ten overexpressing and six silenced transformants showing phleomycin resistance were checked by PCR. A 527-bp fragment, corresponding to the TvDim1 expression cassette, was amplified in nine overexpressing transformants, using the primer pair pKi-614 and 614-3. A 596-bp fragment was amplified with the primers pta-p and 614-3 in the six silenced transformants analyzed. Four TvDim1-overexpressing and four TvDim1-silenced transformants were checked by Southern blot to determine the copy number of the cassette in the T. harzianum T34 genome, using XhoI-digested genomic DNA and the TvDim1 gene as a probe (Fig. 4a, b). Three TvDim1-overexpressing transformants, TvDim1-1, TvDim1-2, and TvDim1-3, and three TvDim1-silenced transformants, pSILT-42, pSILT-45, and pSILT-62, representing different integration patterns for both types of transformants, were selected for further characterization.
Southern blot analysis of wild-type (T34) and transformant strains. Genomic DNAs were XhoI-digested and the TvDim1 ORF was used as probe. EcoRI–HindIII-digested λ DNA (Roche) was used as a marker and molecular sizes (bp) are indicated (line M). a T. harzianum T34 (line 1), TvDim1-1 (line 2), TvDim1-2 (line 3), TvDim1-3 (line 4), and TvDim1-4 (line 5). b T. harzianum T34 (line 1), pSILT-22 (line 2), pSILT-42 (line 3), pSILT-45 (line 4), and pSILT-62 (line 5)
The TvDim1 expression level was investigated in mycelia from T. harzianum T34 and previously selected transformants, three overexpressing and three silenced strains, by qRT-PCR (Fig. 5) and Northern blot analysis (data not shown), respectively. Trichoderma RNAs were obtained from mycelia grown in MM, without glucose, supplemented with 1 mM H2O2 for 8 h. The transformants TvDim1-1, TvDim1-2, and TvDim1-3 showed at least five times higher TvDim1 transcript level than that from the endogenous ThDim1 gene in the wild-type strain T. harzianum T34 under identical growth conditions. Lower TvDim1 transcript levels were observed in the RNA from strain pSIL62, but several hybridization signals were observed in RNAs from the transformants pSIL42 and pSIL45.
qRT-PCR quantification of TvDim1 transcript in the heterologous overexpressed transformants T. harzianum TvDim1-1, TvDim1-2, and TvDim1-3. Values correspond to relative measures against the ThDim1 transcript in T. harzianum T34 wild type (endogenous gene). The experiment was carried out with mycelia grown for 8 h in MM (Penttilä et al. 1987), without glucose, and supplemented with 1 mM H2O2. T. harzianum T34 β-tubulin was used as an internal reference gene
Transformant characterization assays
Since the highest TvDim1 transcript levels in T. virens T59 were observed after growth under oxidative stress, TRX activity was determined in extracts from T. harzianum T34 and the six selected transformant strains after growth in MM supplemented with 1 mM H2O2 for 8 h (Table 1). Higher specific activity levels were observed in the three TvDim1-overexpressing transformants than in T34, with statistically significant differences (P value < 0.05) between the T34 and TvDim1-2, and between the T34 and TvDim1-3 strains. Lower thioredoxin activity levels were observed in the three TvDim1-silenced transformants than in the T34 strain, the three cases showing statistically significant differences (P < 0.05).
Next we investigated the growth of T. harzianum T34 and of the six selected transformants under oxidative conditions, using MM without glucose (control), or MM without glucose but supplemented with 1 or 3 mM H2O2 (Fig. 6a, b). Higher colony diameters were observed only for overexpressing transformant strains than for T34 after growth in the presence of 1 or 3 mM H2O2. As shown in Fig. 6a, these significant solid-medium growth differences were higher in the transformants TvDim1-2 and TvDim1-3 when they were grown on MM with 3 mM H2O2. As shown in Fig. 6b, growth diameters were slightly higher in the wild-type strain than in the silenced transformants on MM plates (control). However, the differences observed in the mycelial diameters of the silenced transformants were only statistically significant (P < 0.05) with respect to the T34 strain after growth in the presence of 1 or 3 mM H2O2 and they were higher on 3 mM plates, where no growth of silenced transformants was observed.
Mycelial growth diameters of T. harzianum T34 and overexpressing and silenced transformant strains measured on MM, without glucose, (MM) (control) or MM, without glucose, supplemented with 1 (MM + 1) or 3 mM H2O2 (MM + 3), after incubation at 28°C for 1 week. a Mycelial growth diameters of T. harzianum T34, TvDim1-1, TvDim1-2, and TvDim1-3. b Mycelial growth diameters of T. harzianum T34, pSILT-42, pSILT-45, and pSILT-62
Similar results were obtained in a liquid growth assay with strain T34 and the TvDim1-1, pSILT-42, and pSILT-62 transformants. A higher quantity of biomass (dry weight) was measured in the overexpressing transformants than in T34, while both silenced transformants showed a lower amount of biomass than T34 after growth for 48 h in MM, with 1% glucose, supplemented with 1 mM H2O2 (Table 2).
No growth differences were observed between the T. harzianum T34 and the TF2 strain, which was transformed with empty pJL43b1 plasmid and was used as a transformation control in all experiments (data not shown).
Plate confrontation experiments between T34, TvDim1-1 or pSILT-62, and the pathogens F. oxysporum f. sp. lycopersici, R. solani, C. acutatum, and B. cinerea were carried out at 25°C, and the plates were observed after 8 days of incubation (data not shown). In all cases, after this incubation time the four pathogens completely covered the surface of 90 mm-diameter PDA or MEA plates used as control. In the dual cultures, Trichoderma strains overgrew the colonies of R. solani and B. cinerea, and they surrounded the colonies of F. oxysporum and C. acutatum. All Trichoderma strains were able to inhibit the growth of the four pathogens tested, reducing the colony diameter of the pathogens to not more than 25 mm, and no significant differences were observed between the inhibition effect of T34 and that of the transformants.
Discussion
Trichoderma is a fungus of high biotechnological value (Monte 2001). Within the framework of the EU-funded TrichoEST project (Rey et al. 2004), a functional genomics project carried out on 10 strains belonging to eight Trichoderma species, only three ESTs showing high identity with Dim1 proteins were identified out of a total of 13,814 unigenes (Vizcaíno et al. 2006, 2007). To date, these proteins have not been reported in filamentous fungi. Here, we explored the role of the TvDim1 gene in Trichoderma.
An EST showing high identity with the evolutionarily conserved Dim1 proteins, belonging to the TRX fold superfamily (Zhang et al. 1999), was identified in library L20 from T. virens T59, a biocontrol strain. EST 614, containing the partial sequence of the TvDim1 gene, was aligned with other Dim1 genes, and the pair of primers designed in that alignment was used to isolate the full-length genomic DNA clone encoding the TvDim1 protein. Southern analysis revealed that the TvDim1 gene was present as a single copy in T. virens T59, and a homologous gene was also present in the other five strains investigated, belonging to four different species. A single TvDim1 homologous gene was also identified in the available online genomes of T. atroviride, T. reesei, and T. virens strain Gv29-8, the latter showing 99.3% of similarity with the TvDim1 gene. In light of these data, it seems reasonable to assume that one Dim1 gene would be present in all Trichoderma genomes. This result is in agreement with the single Dim1 gene detected after exploring genomes from other organisms, such as S. cerevisiae (http://www.yeastgenome.org/), C. elegans (http://www.sanger.ac.uk/Projects/C_elegans/) or A. thaliana (http://www.arabidopsis.org/).
The ORF of TvDim1 encodes a protein of 143 amino acids, and analysis of the sequence revealed a conserved domain with a typical TRX-fold from amino acids 5–137. Upon comparing the TvDim1 sequence with that of Dim1 or hypothetical Dim1 proteins, we observed high homology throughout the entire length of 143 amino acids, Dim1 shows an identity higher than 85% in all cases. This is according to the closer clades observed for Dim1 genes in the NJ tree (Fig. 1). Evolutionarily conserved Dim1 proteins have been described throughout eukaryotic organisms in analyses of mammals, plants, yeasts, and nematode species (Simeoni and Divita 2007).
The highest levels of TvDim1 expression in T. virens T59 were observed after growth under oxidative stress, such as in the presence of H2O2 (Fig. 3). We also observed that TvDim1 transcript levels were increased in the presence of heavy metals—i.e., 100 μM Cd++—in the medium (data not shown). These expression patterns seem to be related to TRX genes. In this sense, H2O2 treatment increased the transcription of the KlTRR1 gene, coding for a TRX reductase, but not of the KlGLR1 gene, encoding a glutathione reductase, in the yeast Kluyveromyces lactis (Tarrío et al. 2004). An enhanced TrxR mRNA level in cells from the yeast S. pombe was detected in the presence of mercuric chloride (Hong et al. 2004). These results suggest that the T. virens T59 TvDim1 gene is one of the stress response-related genes, and they are in agreement with previous work suggesting the involvement of TRXs in defense mechanisms against heavy metals in algae (Lemaire et al. 1999).
We analyzed TvDim1 expression after growing T59 under abiotic stress conditions and gene expression was detected only in the presence of H2O2 and the herbicide Paraquat, indicating its relation with oxidative stress.
The biological function of Dim1 proteins has been related to both pre-mRNA splicing (Reuter et al. 1999) and to the control of cell cycle progression (Berry and Gould 1997), but its biochemical function is not clear. First, TvDim1 showed the TRX-fold but did not display the classic redox active CxxC motif. However, TRX activity was detected in intracellular protein extracts from 8 h-cultures grown in the presence of 1 mM H2O2, in which the highest expression levels were also detected. In view of these results, TvDim1 could act as a single active cysteine protein in redox processes; alternatively, the cysteine from the DxxC motif could form a disulfide bond with other C-terminal cysteines. Regarding this, previous results are contradictory. Thus, in some cases the CxxC motif seemed to be essential for redox-related activity (Holmgren 1979; Martin 1995; Stefankova et al. 2005) while other works have reported a redox function in proteins that lacked one of the two cysteines in this motif (Fomenko and Gladyshev 2003; Jung et al. 2005; Lyles and Gilbert 1994; Reuter et al. 1999).
Trichoderma harzianum T34 is a strain commonly used in basic research studies. To investigate the function of TvDim1 in vivo, heterologous expressions in the strain T34 were performed and TvDim1-overexpressing and TvDim1-silenced transformants were obtained. Since Southern analysis revealed that T. harzianum T34 had a homologous gene to TvDim1, but no ESTs with Dim1 homology were detected in the 3,478 unigenes of strain T34 identified, we used the TRX-1 and 614-3 primer pair to isolate the gene from T. harzianum T34 genomic DNA. The ThDim1 gene showed 94% similarity with TvDim1, and ThDim1 shared 100% identity with the TvDim1 protein. However, H2O2 treatment increased the transcription of ThDim1 less than that of TvDim1. This result indicates that both genes were subjected to different forms of regulation. We have also previously reported that the hsp23 gene, encoding a small heat-shock protein, showed 83% identity between strains T34 and T59 but displayed different expression patterns in both strains (Montero-Barrientos et al. 2007). A different expression pattern could explain why no ESTs with Dim1 homology were present out of the total of 3,478 unigenes obtained from T. harzianum T34 (Vizcaíno et al. 2006).
To characterize the transformants, we analyzed TvDim1 gene expression and TRX activity in strain T34 as well as in three overexpressing and three silenced strains after culturing them in the presence of 1 mM H2O2 (Fig. 5, Table 1). These transformants were chosen because in Southern analyses they showed different integration patterns of cassette transformation (Fig. 4a, b). In a relative quantification by qRT-PCR under identical growth conditions, the overexpressing transformants showed at least 5 times higher TvDim1 transcript level referred to that from the ThDim1 in the wild-type strain T. harzianum T34 (endogenous gene). These transformants also showed higher TRX activity than strain T34. The highest expression and activity levels were observed in strain TvDim1-2, an overexpressing transformant with more insertions of the transformation cassette. Similar TvDim1 expression levels were detected in strains TvDim1-1 and TvDim1-3 although the latter exhibited a higher TRX activity level even though both transformant strains had a similar integration pattern of pJLTvDim1, the plasmid used for transformation. In this sense, it has also been reported that no correlation exists between the number of inserted copies of the chit33, hsp23, and hsp70 genes and the expression levels observed in Trichoderma transformant strains (Limón et al. 1999; Montero-Barrientos et al. 2007, 2008). The silenced transformant strains showed lower TRX activity than that observed in T. harzianum T34 corresponding the lowest level to strain pSILT-62, transformant analyzed with the least inserted copies of plasmid pSILTvDim1 (Table 1). Considering the hybridization profiles obtained from the three silenced transformants analyzed, it is difficult to propose a direct correlation between TRX activity and expression levels. Since the method used to silence the TvDim1 gene expression in T. harzianum T34 was based on the generation of an intron containing self-complementary “hairpin” RNAs (ihpRNA) or perfect duplex hairpin RNAs (Dykxhoorn et al. 2003), the strong and abundant Northern signals could be due to degraded transcripts.
To analyze the contribution of the TvDim1 gene to abiotic stress resistance in T. harzianum T34, we compared the growth of strain T34 and of three overexpressing and three silenced transformant strains with or without H2O2 in the medium. The higher growth observed in the TvDim1-overexpressing transformants and the lower growth pattern seen in the TvDim1-silenced transformants than in the wild-type strain only in presence of this stress, together with the enhanced differences when the amount of H2O2 added to the medium was increased, are in agreement with the redox activity observed in the insulin assays. Statistically significant biomass quantities, higher in the TvDim1-1 transformant and lower in the silenced pSILT-42 and pSILT-62 transformants than in strain T34, were obtained after growing the strains in liquid medium containing 1 mM H2O2 for 48 h. These results point to the ability of TvDim1 to confer resistance to oxidative stress and are in agreement with others previously reported for the Prx6 human peroxiredoxin protein, which has only one active cysteine but maintains its capacity to reduce H2O2 (Pedrajas et al. 2000).
To date, Dim1 proteins have been related to the control of cell cycle progression, which seems to be a consequence of a fault in the pre-mRNA splicing of the proteins responsible for entry into mitosis (Simeoni and Divita 2007). In this sense, we observed a lower growth of TvDim1-silenced transformants than that of strain T34 in MM (a basal medium). Dim1 proteins are related to the cell cycle, since Dim1-deficient mutants in S. pombe (Berry and Gould 1997) and in C. elegans (Zhang et al. 2000) are lethal, and this could account for the reduced growth of TvDim1-silenced transformants under basal conditions. In contrast, no growth differences were detected between T34 and the overexpressing transformants in the basal medium. Considering that the Dim1 protein acts as part of a complex, in these transformants only the level of Dim1, but not that of other associated proteins, was increased and hence it could be expected that their growth would not be affected after growth in basal conditions.
Since T. harzianum is considered as a good biocontrol agent, we also analyzed the TvDim1 overexpression and silencing effect in two T. harzianum T34 transformants (TvDim1-1 and pSILT-62) in a direct confrontation assay against four phytopathogenic fungi. The behavior of both transformant strains was similar to that of strain T34 against the four pathogens assayed. In light of these data, it may be proposed that the TvDim1 gene would not be related to biocontrol activity.
Our results indicate that TvDim1 is a stress-related TRX gene and confirm the involvement of TRXs in defense mechanisms against oxidative stresses.
References
Alfano G, Lewis Ivey ML, Cakir C, Bos JIB, Miller SA, Madden LV, Kamoun S, Hoitink HAJ (2007) Systemic modulation of gene expression in tomato by Trichoderma hamatum 382. Phytopathology 97:429–437
Benítez T, Rincón AM, Limón MC, Codón AC (2004) Biocontrol mechanisms of Trichoderma strains. Int Microbiol 7:249–260
Berry LD, Gould KL (1997) Encodes a functionally conserved polypeptide essential for mitosis. J Cell Biol 137:1337–1354
Bradford M (1976) A rapid and sensitive method for quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 62:248–254
Brunner K, Omann M, Pucher ME, Delic M, Lehner SM, Domnanich P, Kratochwill K, Druzhinina I, Denk D, Zeilinger S (2008) Trichoderma G protein-coupled receptors: functional characterisation of a cAMP receptor-like protein from Trichoderma atroviride. Curr Genet 54:283–299
Cardoza RE, Vizcaíno JA, Hermosa R, Monte E, Gutiérrez S (2006) A comparison of the phenotypic and genetic stability of recombinant Trichoderma spp. generated by protoplast—and Agrobacterium—mediated transformation. J Microbiol 44:383–395
Carnahan RH, Feoktistova A, Ren L, Niessen S, Yates JR, Gould KL (2005) Dim1p is required for efficient splicing and export of mRNA encoding Lid1p, a component of the fission yeast anaphase-promoting complex. Eukaryot Cell 4:577–587
Djonovic S, Pozo MJ, Dangott LJ, Howell CR, Kenerley CM (2006) Sm1, a proteinaceous elicitor secreted by the biocontrol fungus Trichoderma virens induces plant defense responses and systemic resistance. Mol Plant Microbe Interact 19:838–853
Djonovic S, Vargas WA, Kolomiets MV, Horndeski M, Wiest A, Kenerley CM (2007) A proteinaceous elicitor Sm1 from the beneficial fungus Trichoderma virens is required for induced systemic resistance in maize. Plant Physiol 145:875–889
Dykxhoorn DM, Novina CD, Sharp PA (2003) Killing the messenger: short RNAs that silence gene expression. Mol Cell Biol 4:457–467
Esquivel-Naranjo EU, Herrera-Estrella A (2007) Enhanced responsiveness and sensitivity to blue light by blr-2 overexpression in Trichoderma atroviride. Microbiology 153:3909–3922
Ferrari DM, Nguyen P, Kratzin HD, Söling H (1998) ERp28, a human endoplasmic-reticulum-lumenal protein, is a member of the protein disulfide isomerase family but lacks a CXXC thioredoxin-box motif. Eur J Biochem 255:570–579
Fomenko DE, Gladyshev VN (2003) Identity and functions of CxxC-derived motifs. Biochemistry 42:11214–11225
Grant CM (2001) Role of the glutathione/glutaredoxin and thioredoxin systems in yeast growth and response to stress conditions. Mol Microbiol 39:533–541
Gutiérrez S, Velasco J, Marcos AT, Fernández FJ, Fierro F, Barredo JL, Díez B, Martín JF (1997) Expression of the cefG gene is limiting for cephalosporin biosynthesis in Acremonium chrysogenum as shown by promoter replacement studies. Appl Microbiol Biotechnol 48:606–614
Harman GE, Howell CR, Viterbo A, Chet I, Lorito M (2004) Trichoderma species-opportunistic, avirulent plant symbionts. Nat Rev Microbiol 2:43–56
Holmgren A (1979) Thioredoxin catalyzes the reduction of insulin disulfides by dithiothreitol and dihydrolipoamide. J Biol Chem 254:9627–9632
Hong SM, Lim HW, Kim IH, Kim K, Park EH, Lim CJ (2004) Stress-dependent regulation of the gene encoding thioredoxin reductase from the fission yeast. FEMS Microbiol Lett 234:379–385
Jung J, Yee A, Wu B, Arrowsmith CH, Lee W (2005) Solution structure of YKR049C, a putative redox protein from Saccharomyces cerevisiae. J Biochem Mol Biol 38:550–554
Kumar S, Tamura K, Jakobsen IB, Nei M (2001) MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17:1244–1245
Lemaire S, Keryer E, Stein M, Schepens I, Issakidis-Bourguet E, Gérard-Hirne C, Miginiac-Maslow M, Jacquot JP (1999) Heavy-metal regulation of thiredoxin gene expression in Chlamydomonas reinhardtii. Plant Physiol 120:773–778
Limón MC, Pintor-Toro JA, Benítez T (1999) Increased antifungal activity of Trichoderma harzianum transformants that overexpress a 33-kDa chitinase. Phytopathology 89:254–261
Lorito M (1998) Chitinolytic enzymes and their genes. In: Kubicek CP, Harman GE (eds) Trichoderma and gliocladium. Taylor & Francis, London, pp 73–99
Lyles MM, Gilbert HF (1994) Mutations in the thioredoxin sites of protein disulfide isomerase reveal functional nonequivalence of the N- and C-terminal domains. J Biol Chem 269:30946–30952
Mach RL, Schindler M, Kubicek CP (1994) Transformation of Trichoderma reesei based on hygromycin B resistance using homologous expression signals. Curr Genet 25:567–570
Malagnac F, Klapholz B, Silar P (2007) PaTrx1 and PaTrx3, two cytosolic thioredoxins of the filamentous ascomycete Podospora anserina involved in sexual development and cell degeneration. Eukaryot Cell 6:2323–2331
Martin JL (1995) Thioredoxin-a fold for all reasons. Structure 3:245–250
Monte E (2001) Understanding Trichoderma: between agricultural biotechnology and microbial ecology. Int Microbiol 4:1–4
Montero-Barrientos M, Cardoza RE, Gutiérrez S, Monte E, Hermosa R (2007) The heterologous overexpression of hsp23, a small heat-shock protein gene from Trichoderma virens, confers thermotolerance to T harzianum. Curr Genet 52:45–53
Montero-Barrientos M, Hermosa R, Nicolás C, Cardoza RE, Gutiérrez S, Monte E (2008) Overexpression of a Trichoderma HSP70 gene increases fungal resistance to heat and other abiotic stresses. Fungal Genet Biol 45:1506–1513
Pedrajas JR, Miranda-Vizuete A, Javanmardy N, Gustafsson J, Spyrou G (2000) Mitochondria of Saccharomyces cerevisiae contain one-conserved cysteine type peroxiredoxin with thioredoxin peroxidase activity. J Biol Chem 275:16296–16301
Penttilä M, Nevalainen H, Ratto M, Salminen E, Knowles J (1987) A versatile transformation system for the cellulolytic filamentous fungus Trichoderma reesei. Gene 61:155–164
Raeder U, Broda P (1985) Rapid preparation of DNA from filamentous fungi. Lett Appl Microbiol 1:17–20
Reuter K, Nottrott S, Fabrizio P, Lüehrmann R, Ficner R (1999) Identification, characterization and crystal structure analysis of the human spliceosomal U5 snRNP specific 15 kD protein. J Mol Biol 294:515–525
Rey M, Llobell A, Monte E, Scala F, Lorito M (2004) Genomics of trichoderma. In: Kachatourians GG (ed) Fungal genomics applied mycology and biotechnology, vol 4. Elsevier Science, Amsterdam, pp 225–248
Rocha-Ramírez V, Omero C, Chet I, Horwitz BA, Herrera-Estrella A (2002) Trichoderma atroviride G-protein—subunit gene tga1 is involved in mycoparasitic coiling and conidiation. Eukaryot Cell 1:594–605
Rubio MB, Hermosa R, Reino JL, Collado IG, Monte E (2009) Thctf1 transcription factor of Trichoderma harzianum is involved in 6-pentyl-2H-pyran-2-one production and antifungal activity. Fungal Genet Biol 46:17–27
Sambrook J, Russel DW (2001) Molecular cloning: a laboratory manual, 3rd edn. Cold Spring Harbor Laboratory Press, New York
Shoresh M, Harman GE (2008) The molecular basis of shoot responses of maize seedlings to Trichoderma harzianum T22 inoculation of the root: a proteomic approach. Plant Physiol 147:2147–2167
Simeoni F, Divita G (2007) The Dim protein family: from structure to splicing. Cell Mol Life Sci 64:2079–2089
Sivan A, Chet I (1989) The possible role of competition between Trichoderma harzianum and Fusarium oxysporum on rhizosphere colonization. Phytopathology 79:198–203
Sousa S (2004) Mejora del sistema de expresión en Trichoderma harzianum CECT 2413 para la producción de proteínas de interés biotecnológico. Ph. D. thesis. University of Sevilla, Spain
Stefankova P, Kollárová M, Barák I (2005) Thioredoxin-structural and functional complexity. Gen Physiol Biophys 24:3–11
Tarrío N, Díaz-Prado S, Cerdán ME, González-Siso MI (2004) Isolation and characterization of two nuclear genes encoding glutathione and thioredoxin reductases from the yeast Kluyveromyces lactis. Biochem Biophys Acta 1678:170–175
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Vinale F, Marra R, Scala F, Ghisalberti EL, Lorito M, Sivasithamparam K (2006) Major secondary metabolites produced by two commercial Trichoderma strains active against different phytopathogens. Lett Appl Microbiol 43:143–148
Viterbo A, Chet I (2006) TasHyd1, a new hydrophobin gene from the biocontrol agent Trichoderma asperellum, is involved in plant root colonization. Mol Plant Pathol 7:249–258
Vizcaíno JA, González FJ, Suárez B, Redondo J, Heinrich J, Delgado-Jarana J, Hermosa R, Gutiérrez S, Monte E, Llobell A, Rey M (2006) Generation, annotation and analysis of ESTs from Trichoderma harzianum CECT 2413. BMC Genomics 7:193–207
Vizcaíno JA, Redondo J, Suárez MB, Cardoza RE, Hermosa MR, González FJ, Rey M, Monte E (2007) Generation, annotation, and analysis of ESTs from four different Trichoderma strains grown under conditions related to biocontrol. Appl Microbiol Biotechnol 75:853–862
Zhang Y, Gould KL, Dunbrack RL, Cheng H, Roder H, Golemis EA (1999) The evolutionarily conserved Dim1 protein defines a novel branch of the thioredoxin fold superfamily. Physiol Genomics 2:85–92
Zhang Y, Lindblom T, Chang A, Sudol M, Sluder AE, Golemis EA (2000) Evidence that Dim1 associates with proteins involved in pre-mRNA splicing, and delineation of residues essential for Dim1 interactions with hnRNP F and Npw38/PQBP-1. Gene 257:33–43
Acknowledgments
Research project funding was from the Junta de Castilla y León (GR67) and the Ministry of Education and Science (AGL2008-0512/AGR) is gratefully acknowledged.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by K. Kuchler.
Rights and permissions
About this article
Cite this article
Morán-Diez, M.E., Cardoza, R.E., Gutiérrez, S. et al. TvDim1 of Trichoderma virens is involved in redox-processes and confers resistance to oxidative stresses. Curr Genet 56, 63–73 (2010). https://doi.org/10.1007/s00294-009-0280-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00294-009-0280-8