Abstract
Gibberella zeae causes Fusarium head blight of cereal crops, and sexual spores of the fungus play an important role as primary inocula. We isolated a restriction enzyme-mediated integration (REMI) transformant, ZH431, of G. zeae with defects in perithecia formation and virulence. Integration of the REMI vector resulted in disruption of GzCHS7 gene, which encodes a putative class VII chitin synthase with high similarity to Fusarium oxysporum ChsVb. A second chitin synthase, GzCHS5, is adjacently located in a head-to-head configuration with GzCHS7, and its deduced protein sequence showed similarity with a class V chitin synthase in F. oxysporum. Neither ΔGzChs5 nor ΔGzChs7 mutants produced perithecia or caused disease on barley heads. Microscopic observation revealed that both mutants formed balloon-shaped hyphae and intrahyphal hyphae and that cell wall rigidity of the mutants was weaker than that of the wild-type strain. Transcription profiles of GzCHS5 and GzCHS7 were not altered in ΔGzChs7 and ΔGzChs5, respectively, suggesting that transcription regulations of the genes are independent of each other. Our results demonstrate that GzCHS5 and GzCHS7 are indispensable for perithecia formation and pathogenicity as well as normal septa formation and hyphal growth in G. zeae.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Gibberella zeae (anamorph: Fusarium graminearum) is a filamentous ascomycete fungus that causes Fusarium head blight (FHB) of cereal crops and Fusarium ear and stalk rot of maize in many regions of the world (Leslie and Summerell 2006). The fungus produces mycotoxins such as trichothecene and zearalenone that pose serious threats to human and animal health (Desjardins 2006).
Sexual reproduction in G. zeae is an important factor for survival under field conditions and helps maintain a genotypically diverse population structure. Sexual spores (ascospores) of G. zeae overwinter within a fruiting body (perithecium) on plant debris, and may serve as the primary inoculum for initiating disease epidemics (Sutton 1982; Trail et al. 2002). The osmotic pressure generated by concentrating potassium ions and chloride ions within the asci triggers forcible discharge of ascospores and initiates the transfer of ascospores from plant debris to cereal flowers (Trail et al. 2005). Thus, studies of sexual developments in G. zeae have gained attraction because certain genes responsible for sexual reproduction in G. zeae are closely related to disease development (Han et al. 2007; Hou et al. 2002; Lee et al. 2009; Shim et al. 2006; Urban et al. 2003; Yu et al. 2008).
Fungal cell walls are the outermost layer of fungal cells against adverse environmental conditions. Chitin, a β-1,4-linked polysaccharide of N-acetylglucosamine, is a key structural component of the fungal cell wall (Bartnicki-Garcia 1968). Chitin also cross-links other cell wall components and plays an important role in mycelial growth and polarity maintenance in filamentous fungi (Takeshita et al. 2002).
Fungal chitin synthases have been divided into at least seven classes based on the structural properties (Mandel et al. 2006). Myosin motor-like chitin synthases, which are unique to filamentous fungi, are important for the maintenance of cell wall integrity and host infection (Liu et al. 2004; Madrid et al. 2003; Weber et al. 2006). These class V and class VII chitin synthases are characterized by the presence of two conserved domains: an N-terminal myosin motor-like domain (MMD) and a C-terminal chitin synthase domain (Martín-Urdíroz et al. 2008; Nino-Vega et al. 2004; Takeshita et al. 2006). The characteristics differentiating these two classes of chitin synthases are the length of MMD and the presence of ATP-binding motifs, i.e., P-loop, Switch I, and Switch II motifs (Nino-Vega et al. 2004; Takeshita et al. 2006).
One of our strategies to isolate genes associated with sexual development and virulence in G. zeae is the use of random insertional mutagenesis. We have generated over 20,000 mutants from a representative strain of G. zeae, GZ03643, by restriction enzyme-mediated integration (REMI) strategy (Han et al. 2004). REMI has been used successfully to identify novel genes associated with virulence not only in G. zeae but also in other filamentous fungi (Dufresne et al. 2000; Han et al. 2004; Inoue et al. 2002; Lu et al. 1994). During our REMI screen, we isolated a transformant (ZH431) with aberrant hyphal growth and perithecia formation, and significantly reduced virulence. We determined that REMI vector was inserted into a locus that encodes a chitin synthase (GzCHS7), and subsequently we learned that another chitin synthase gene (GzCHS5) is located adjacent to GzCHS7 in a head-to-head configuration. The objective of this study was to functionally characterize the two chitin synthase genes, GzCHS5 and GzCHS7. We demonstrate that these two chitin synthase genes, one class V and the other class VII, play an important role in sexual reproduction and pathogenesis in G. zeae. Furthermore, we show that GzCHS5 and GzCHS7 are required for normal hyphal tip growth and septum formation.
Materials and methods
Strains, culture condition, and plasmids
Gibberella zeae strain GZ03643, a zearalenone and deoxynivalenol producer, was obtained from Dr Robert L. Bowden (US Department of Agriculture, Manhattan, KS, USA) and was used as the wild-type strain. The mutant strain ZH431 was generated from GZ03643 by using REMI mutagenesis, as previously described (Han et al. 2004). Transgenic G. zeae strain, T39∆M1-3, which carries mat1-1 deletion and thus self-sterile, was used as the female test strain for outcross analysis (Lee et al. 2003). For sexual crosses, carrot agar was used as previously described (Leslie and Summerell 2006). All strains were stored at −80°C in 25% glycerol. For DNA extraction, strains were grown in 30 ml of complete medium (CM) (Leslie and Summerell 2006) for 3 days at 25°C with shaking (150 rpm). For RNA extraction, fungi were grown in 50 ml CM at 25°C with shaking (150 rpm). To determine the effect of osmotic stabilizers, strains were grown on minimal medium supplemented with 1 M sorbitol, 1 M mannitol, 0.7 M NaCl, or 0.7 M KCl as osmotic stabilizer. Recombinant plasmids, pUCH1, and pII99 (Namiki et al. 2001) carrying the gene for resistance to hygromycin B (hygB) and geneticin (gen), respectively, were used as selection needs.
Nucleic acid manipulations, plasmid rescue, PCR primers, and sequencing
Fungal genomic DNA was extracted by using acetyltrimethyl ammonium bromide procedure (Leslie and Summerell 2006). Total RNA from G. zeae mycelia was prepared with TRI reagent (Molecular Research Center Inc., Cincinnati, OH, USA) following the manufacturer’s instructions. Plasmid DNA of recombinant E. coli strain was isolated using a NucleoGen plasmid purification kit (NucleoGen Biotech, Siheung, Korea). Restriction endonuclease digestion, gel electrophoresis, gel blotting, ligation, Southern, and northern hybridization with 32P-labeled probes were performed following standard techniques (Sambrook and Russell 2001). PCR primers (Table 1) were suspended in sterile water to achieve 100 μM final concentration and stored at −20°C. DNA sequencing was performed at National Instrumentation Center for Environmental Management (Seoul National University, Seoul, Korea). The primers described by Han et al. (2004) were used to sequence the rescued plasmid from the mutant strain ZH431, and the sequence obtained was compared against the Fusarium Comparative Database at Broad Institute (http://www.broad.mit.edu/annotation/genome/fusarium_group/MultiHome.html) and the GenBank at National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/) by BLAST algorithm.
Vector constructions and fungal transformation
Gene deletion constructs were generated by double-joint PCR method (Yu et al. 2004). For targeted deletion of GzCHS5 and GzCHS7, DNA fragments of 5′ and 3′ flanking regions of GzCHS5 and GzCHS7 were amplified from the wild-type GZ03643 genomic DNA using primer pairs CHS5-5f/CHS5-5r and CHS5-3f/CHS5-3r for GzCHS5, and CHS7-5f/CHS7-5r and CHS7-3f/CHS7-3r for GzCHS7 (Fig. 1a, b). The primers used in this study were listed in Table 1. Simultaneously, a 1.6-kb fragment of geneticin resistance gene (gen) cassette was amplified from plasmid pII99 using the primers Gen-f and Gen-r. The 5′ and 3′ flanking regions of each gene were mixed with gen amplicon in a 1:2:1 molar ratio (5′ fragment: gen: 3′ fragment) and joined by PCR without any primers. Subsequently, the 6.0- and 3.5-kb PCR products harboring the gen marker fused to the flanking regions of GzCHS5 and GzCHS7 were amplified with primer pairs NCHS5-5/NCHS5-3 and NCHS7-5/NCHS7-3, respectively. For ∆GzChs5/7 double mutant, the 3′ flanking regions of GzCHS5 and GzCHS7 amplified above were fused with gen as described above (Fig. 1c). Nested primers SNCHS5-3 and NCHS7-3 were used to amplify the 3.6-kb amplicon carrying the gen marker fused to the 3′ flanking regions of GzCHS5 and GzCHS7, and the construct was transformed into GZ03643 to generate a double deletion of the GzCHS5 and GzCHS7 genes. We isolated two gene replacement mutant strains of ΔGzChs7, three of ΔGzChs5, and three of ΔGzChs5/7 in this study.
Targeted gene deletion of GzCHS5 (a), GzCHS7 (b), and both GzCHS5 and GzCHS7 (c) in the wild-type G. zeae GZ03643 strain. Restriction enzymes used in this experiment are XbaI (X) and BglII (B). In Southern blots, lane 1 is GZ03643, and lane 2 is the mutant that the target gene was replaced with geneticin-resistant gene (gen). The probe used for each hybridization is indicated by a bar. The sizes of the DNA standards (in kilobases) are indicated on the left of the blot
For overexpression of GzCHS7 gene in the ∆GzChs5/7 double-deletion mutant, the entire GzCHS7 gene including the native terminator was amplified, and the 3′ flanking region of GzCHS5 was amplified with primers CCHS7-3f and CHS7-3r/CHS5-3r and CCHS7-5f, respectively. The strong constitutive promoter of Neurospora crassa isocitrate lyase gene, ICL, was amplified from pIGPAPA (Horwitz et al. 1999) with primers ICL-f and ICL-r. These three amplicons were fused after purification with same nested primer pair used to construct the double-deletion construct by using a double-joint PCR method (Yu et al. 2004), and the final construct was transformed into the ∆GzChs5/7 double-deletion mutant by co-transformation with pUCH1.
Sexual reproduction and virulence assays
Sexual crosses were performed as previously described (Lee et al. 2003). For self-fertilization, GZ03643 strain was inoculated on carrot agar and incubated at 25°C for 7 days (Leslie and Summerell 2006). Aerial mycelia were removed by scrubbing with 1 ml of sterile 2.5% Tween60® solution (v/v) using a glass rod. The plates were incubated for additional 10 days at 25°C under the mixture of fluorescent cool white and black lights with a 12-h photoperiod.
For outcrossing ZH431, a mycelia agar block of T39ΔM1-3 was placed on a carrot agar plate and incubated at 25°C for 7 days. A conidial suspension (1 × 105 conidia ml−1) of ZH431 was applied to the mycelia of T39∆M1-3 strain that was established on carrot agar as described above. The plates were incubated for an additional 10–14 days under the same conditions as the self-fertilized plates.
Fungal strains were incubated in CMC liquid medium (Capellini and Peterson 1965) at 25°C with shaking (150 rpm) for 5 days. Macroconidia of each isolate were grown in CMC and then harvested and re-suspended in sterile water (1 × 106 spores ml−1). The spore suspension was sprayed on barley heads at early anthesis. Inoculated plants were incubated in a growth chamber at 25°C with 100% humidity for 2 days and then transferred to a greenhouse. FHB symptoms were observed 10 days after inoculation.
Microscopy
For scanning electron microscope (SEM), mycelial plugs from PDA plates were immersed at 4°C overnight in modified Karnovsky’s fixation buffer (2% paraformaldehyde and 2% glutaraldehyde in 50 mM sodium cacodylate buffer, pH 7.2) and washed with the same buffer three times each for 10 min. Post-fixation with 1% osmium tetroxide in 0.05 M sodium cacodylate buffer (pH 7.2) was performed at 4°C for 2 h, and the samples were washed twice with distilled water. The fixed samples were dehydrated in a graded ethanol series (30, 50, 70, 80, 90, and 100%, and three times in 100% each for 10 min) and then transferred to 100% isoamyl acetate. The samples were dried with liquid carbon dioxide and then mounted on metal stubs. Samples were coated with gold and observed with a SEM JSM 5410LV (JEOL Ltd, Tokyo, Japan).
For transmission electron microscopy (TEM), the fixation protocol was the same as described for SEM. After post-fixation, the samples were en bloc stained with 0.5% uranyl acetate at 4°C for overnight, were dehydrated in a graded ethanol series. The samples were treated twice with 100% propylene oxide for 10 min and were embedded in Spurr’s resin and sectioned using ultramicrotome MT-X (RMC, Tucson, AZ, USA). The specimens were stained with 2% uranyl acetate and Reynolds’ lead citrate each for 7 min, and then observed with a TEM JEM-1010 (JEOL Ltd).
For atomic force microscopy (AFM), autoclaved cover glasses (each 18 mm × 18 mm) with growing hyphae on the surface were collected from cultures growing on PDA plates. The samples were fixed to a steel disc (18 mm in diameter) with the hyphae upward using two-sided adhesive tape. The steel disc was magnetically mounted on a piezoscanner (maximum xy-scan range of 5 μm) of an AFM (AutoProbe-CP; Park Scientific Instruments, Sunnyvale, CA, USA). Force modulation microscopy was used to acquire elastic images under ambient conditions by using V-shaped silicon cantilevers with a force constant of 0.26 N/m. The microscope was operated by applying forces of 10.2 nN to the specimen surface with optimized feedback parameters at a scan frequency of 1 Hz. Elastic images over 1 × 1 μm2 scan areas were recorded from four hyphae on the cover glass of each strain. Based on the amplitude of cantilever modulation, the z-axis values of elastic images represent the hardness of the specimen surface (Li et al. 1998). The mean height (voltage in the z-axis) values of each elastic image were derived by using a software package (ProScan version 1.51b; Park Scientific Instruments) and processed to compare the arithmetic mean values of the four mean height values between strains.
Results
Genetic analysis of the REMI mutant ZH431
A total of 3,912 REMI transformants were screened for defects in perithecia formation and FHB virulence. We isolated one transformant, designated ZH431, that failed to produce perithecia on carrot agar. ZH431 and GZ03643 also differed dramatically in other phenotypes including hyphal growth, pigmentation, and virulence. The mutant grew slower and produced less aerial mycelia than did GZ03643 on both PDA and carrot agar. Also, ZH431 produced a yellow–orange pigment absent in GZ03643. ZH431 showed drastically reduced virulence; while the wild-type progenitor caused severe FHB symptoms 7 days after inoculation, ZH431 developed few small necrotic spots on spikelets of barley heads 10 days after inoculation.
To determine if the phenotypic changes in ZH431 were caused by the insertion of REMI vector, we crossed ZH431 with the heterothallic strain, T39∆M1-3, carrying a mat1-1 deletion (Lee et al. 2003). One hundred random ascospores from the outcross segregated into parental phenotypes at 53:47 that fits a 1:1 ratio (χ 2 = 0.36), and all hygromycin-resistant progeny had the ZH431 phenotypes. This result suggested that the mutation in ZH431 occurred at a single locus and that this mutation was tightly linked to or resulted directly from the REMI vector insertion.
To confirm the insertion of REMI vector in ZH431, a Southern analysis was performed. ZH431 genomic DNA was digested with HindIII and BglII, separated by electrophoresis in an agarose gel, and transferred to a nylon membrane. We probed the blot with REMI vector pUCH1 (5.2 kb) and observed a single band, which indicated the integration of a single copy of pUCH1 into the ZH431 genome (data not shown).
Molecular characterization of disrupted loci in ZH431
We hybridized 32P-labeled pUCH1 to the BglII-digested ZH431 genomic DNA blot and identified a single 7.2-kb band, which is expected to contain G. zeae genomic DNA flanking pUCH1. The DNA fragment, designated pZH431B, was rescued and subsequent sequencing revealed that the 5′-flanking region (0.5 kb) and the 3′-flanking region (1.6 kb) correspond to G. zeae gene FGSG_12039.3, which is 5,305-bp in length and encodes a 1,558-amino acid polypeptide interrupted by three putative introns. The vector insertion occurred at a HindIII site located 2,448-bp downstream of the FGSG_12039.3 translational start site. FGSG_12039.3 share a significant similarity (91%) with ChsVb, a F. oxysporum f. sp. lycopersici class VII chitin synthase (GenBank EF673037.1) that is required for septation (Martín-Urdíroz et al. 2008). The N-terminal MMD in FGSG_12039.3 is approximately 220 amino acids in length and lacks ATP-binding motifs, which are conserved in class VII chitin synthases such as F. oxysporum ChsVb and Aspergillus nidulans CsmB (Martín-Urdíroz et al. 2008; Nino-Vega et al. 2004). We identified a second gene 4,350-bp upstream of the FGSG_12039.3 translational start site, located in a head-to-head orientation (Fig. 1). This gene (FGSG_01964.3) encodes a 1,865-aa protein that shares 94% identity with a F. oxysporum class V chitin synthase gene (GenBank AF484941.1) (Madrid et al. 2003); the MMD is about 740 amino acids in length and the ATP-binding motifs are highly conserved. This gene is similar to class V chitin synthases F. oxysporum ChsV and A. nidulans CsmA. Based on this similarity and the structural properties, we designated these two putative G. zeae chitin synthase genes FGSG_12039.3 and FGSG_01964.3 as GzCHS7 and GzCHS5, respectively.
Targeted deletion of GzCHS5 and GzCHS7
To further characterize the functions of GzCHS5 and GzCHS7 in G. zeae, we generated GzCHS5-deletion (∆GzChs5), GzCHS7-deletion (∆GzChs7), and double-deletion (∆GzChs5/7) mutants from wild-type strain GZ03643. A 5.7-kb region containing GzCHS5 and a 5.3-kb region containing GzCHS7 were completely replaced with the 1.6-kb geneticin (gen) marker via double homologous recombination. Southern analyses confirmed these replacements in the GzCHS5 and GzCHS7 loci. XbaI-digested genomic DNA of ΔGzChs5 mutants had a 9.4-kb band instead of the 7.4-kb wild-type band when a 5′ PCR fragment of GzCHS5 was used as a probe (Fig. 1a). BglII-digested genomic DNA of ΔGzChs7 mutants contained a 9.3-kb band when the gen cassette was used as a probe, confirming that the GzCHS7 gene was replaced with the marker (Fig. 1b). BglII-digested genomic DNA of ΔGzChs5/7 strains had a single 4.7-kb hybridizing band instead of the 5.0-kb wild-type band, suggesting that the 15.3-kb region harboring GzCHS5 and GzCHS7 was deleted and replaced with the gen marker (Fig. 1c).
Phenotypes of ΔGzChs5, ΔGzChs7, and ΔGzChs5/7 strains
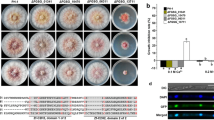
All mutants showed phenotype similar to the REMI mutant ZH431. The mycelia growth rate of deletion mutants, ΔGzChs5, ΔGzChs7, and ΔGzChs5/7, was slower than that of GZ03643 on PDA (Fig. 2a). GZ03643 strain produced aerial mycelia when growing on PDA, but very little aerial mycelia was observed in ΔGzChs5, GzChs7, and ΔGzChs5/7. All mutants failed to produce perithecia on carrot agar and showed drastically reduced virulence on barley heads when compared to the wild type (Fig. 2b). Conidia production was also severely reduced in all mutants. When incubated for 7 days GZ03643 produced 2.3 × 106 conidia ml−1in CMC medium, however, ΔGzChs5, ΔGzChs7, and ΔGzChs5/7 averaged 7.8 × 104, 6.3 × 104, and 6.0 × 104 conidia ml−1, respectively, suggesting that GzCHS5 and GzCHS7 contribute to G. zeae conidia production. To determine whether GzCHS5 and GzCHS7 can mediate osmotic stability, ΔGzChs5, ΔGzChs7, and ΔGzChs5/7 strains were grown on minimal media supplemented with 1.0 M sorbitol, 1.0 M mannitol, 0.7 M NaCl, or 0.7 M KCl as an osmotic stabilizer. Defective mycelial growth observed in ΔGzChs5, ΔGzChs7, and ΔGzChs5/7 strains grown on minimal media was partially restored by the addition of osmotic stabilizers (Fig. 3). In order to examine the effects of mutations on cell wall rigidity, we analyzed the wild type and mutants with an AFM. The cell walls of all three mutants were less rigid than those of the wild-type strain, with the ΔGzChs5 cell wall being more rigid than that of ΔGzChs7 strain (Fig. 4).
Growth of wild type and deletion mutants on PDA (a) and virulence on barley heads (b). Photographs were taken 5 and 10 days after inoculation on PDA and barley heads, respectively. GZ03643, wild-type G. zeae strain; ΔGzChs5, GzCHS5-deleted mutant; ΔGzChs7, GzCHS7-deleted mutant; ΔGzChs5/7, double-deletion mutant of GzCHS5 and GzCHS7
Comparison of cell wall rigidity of hyphae measured by atomic force microscopy. The y-axis represents mean height (voltage in unit) of elastic images from each fungal specimen by force modulation microscopy. Higher voltages indicate increased hardness. Error bar indicates standard deviation of four biological replicates, and the significance among strains is labeled with a and b, which are significantly different according to Tukey’s test (P < 0.05), on the error bar
Microscopic observation of ΔGzChs5, ΔGzChs7, and ΔGzChs5/7 strains
We investigated whether chitin synthase mutations had deleterious effect on G. zeae hyphae and conidia development. Under the light microscope, hyphae of three mutants appeared swollen when grown on PDA. Further analyses by SEM asserted that mutant hyphae were heavily swollen in contrast to the wild-type strain (Fig. 5). Swollen balloon-shaped hyphae, as well as occasional crumpled balloon shapes, also were observed in the mutant hyphae (Fig. 5). TEM revealed typical hyphae enclosed with an electron-translucent cell wall in the wild-type strain (Fig. 6a). While intrahyphal hyphae can be found in the older mycelia of the wild-type strain (Fig. 6b), they were quite often found in the balloon-shaped hyphae of mutants (Fig. 6c, d). Moreover, one hypha often contained more than one intrahyphal hypha in the mutants (Fig. 6d). Swelling was evident in the mutant hyphae except for the septal region where normal hyphal thickness is likely maintained (Fig. 6e). At a higher magnification, we observed the septal pores in the mutant hyphae plugged by a woronin body-like structure (Fig. 6f).
Transmission electron micrographs of hyphae. a Hyphae of the wild-type strain GZ03643. b Intrahyphal hyphae (arrow) in old mycelia of GZ03643. c Intrahyphal hyphae (arrow) in a balloon-shaped hypha of ΔGzChs7. d Three intrahyphal hyphae in mycelia of ΔGzChs7. e Hyphae of ΔGzChs7. f Woronin body-like organelle (arrowhead) plugging a septal pore in ΔGzChs7. (Scale bars in a, c and e represent 3.0 μm, in b and f 1.0 μm, and in d 2.0 μm)
Overexpression of GzCHS7 in ΔGzchs5/7
Our original REMI mutation was in GzCHS7 gene, and we were interested in testing whether the GzCHS7 overexpression can overcome the double mutant phenotype. The GzCHS7-coding region was amplified from GZ03643 strain, fused to a constitutive N. crassa ICL promoter, and subsequently transformed into ∆Gzchs5/7 strain (Fig. 7a). Two out of ten hygromycin-resistant transformants had the construct integrated into the GzCHS7-deletion locus by homologous recombination, which was confirmed by Southern analysis (Fig. 7b). However, none of the transformants carrying the ICLp::GzCHS7 construct could be clearly distinguished morphologically from the ΔGzChs5/7 mutant (Fig. 7c). In Cchs7 strain, the transformant with ICLp::GzCHS7, GzCHS7 was expressed at a higher level than that in the wild type, but GzCHS5 expression was not detected (Fig. 8).
Overexpression of GzCHS7. a Schematic depiction of overexpression construct and homologous recombination strategy to overexpress GzCHS7 in the double mutant ΔGzChs5/7. GzCHS7 ORF was fused to the ICL promoter, and the fused construct was transformed into ΔGzChs5/7 to generate Cchs7 mutants. Restriction enzyme BglII site is indicated with b. The probe used for Southern hybridization is indicated with a bar. b Southern hybridization to confirm the integration of the fused construct. Lane 1 the wild-type G. zeae GZ03643, lanes 2 and 3 independent Cchs7 mutants. The sizes of the DNA standards (in kilobases) are indicated on the left of the blot. c Growth of Cchs7 mutants on PDA. The plate on top is the wild-type GZ03643, the plate on bottom left is Cchs7 mutant, and the plate on bottom right is ΔGzChs5/7
Northern analyses of GzCHS5 and GzCHS7 during hyphal growth
GzCHS5 and GzCHS7 genes are positioned in a head-to-head configuration, which suggests that two genes share a common promoter region. We investigated whether the expression of GzCHS5 or GzCHS7 was altered in ΔGzChs5, ΔGzChs7, and ΔGzChs5/7 strains. The overall expression levels of GzCHS5 and GzCHS7 in ΔGzChs7 and ΔGzChs5, respectively, were similar to those of the corresponding genes in the wild type (Fig. 8). However, while both genes exhibited gradual increase in expression in the wild type, we did notice that GzCHS5 and GzCHS7 expressions peaked on day 4 in the mutant strains.
Discussion
Chitin comprises 10–20% of filamentous fungal dry weight, and the chitin synthases play critical roles in hyphal development and fungal pathogenicity (Madrid et al. 2003; Martín-Urdíroz et al. 2008; Takeshita et al. 2006; Werner et al. 2007). Fungal chitin synthases have been divided into seven classes based on the structural properties (Mandel et al. 2006). In A. nidulans, six different genes designated chsA, chsB, chsC, chsD, csmA, and csmB correspond to classes II, III, I, IV, V, and VI, respectively (Culp et al. 2000; Horiuchi et al. 1999; Motoyama et al. 1994, 1996; Takeshita et al. 2006). In the tomato pathogen F. oxysporum f. sp. lycopersici, five chitin synthases, designated CHS1, CHS2, CHS3, CHSV, and CHSVb, have been isolated and characterized (Martín-Urdíroz et al. 2004, 2008). The F. graminearum genome database (http://www.broad.mit.edu/annotation/genome/) recognizes eight chitin synthases (FGSG_01272.3, FGSG_01949.3, FGSG_01964.3, FGSG_02483.3, FGSG_03418.3, FGSG_10116.3, FGSG_10327.3, and FGSG_12039.3). The predicted amino acid sequence of these genes showed the presence of conserved chitin synthase 2 (CS2) domain. Furthermore, phylogenetic analysis using the CS2 domain indicated that all eight genes can be assigned to one of the chitin synthase classes (data not shown), suggesting that G. zeae chitin synthases are also highly conserved.
Chitin synthases are known to directly associate with hyphal growth and asexual development in filamentous fungi. In A. nidulans chsA, chsC, and chsD play a role in conidiation (Ichinomiya et al. 2005; Motoyama et al. 1996). Deletion of chsB in particular resulted in hyper branching of hyphae, enlarged tips, and disorganized lateral cell walls. Disruption of csmA, the first class V chitin synthase to be identified, resulted in morphological abnormalities such as balloon-like hyphal tips, sparse conidiophores, and abnormal septa formation (Horiuchi et al. 1999). The follow-up study on A. nidulans csmB, a class VII chitin synthase gene, was determined to have a role in hyphal tip and septa formation (Takeshita et al. 2006). Significantly, in several plant pathogenic fungi, class V and VII chitin synthases have important roles in pathogenicity. F. oxysporum CHSV and CHSVb, class V and class VII chitin synthases, respectively, were directly linked to pathogenicity (Madrid et al. 2003; Martín-Urdíroz et al. 2008). In maize anthracnose fungus Collectotrichum graminicola, ChsV is essential for appressorium formation thus directly impacting host penetration. Appressoria of ChsV deletion mutant were severely distorted and the appressorial cell wall appeared to be disintegrated (Werner et al. 2007).
To date, only FgCHS1, a class I chitin synthase gene, has been cloned, but the report was limited to gene sequence analysis and its transcription profile during fungal development (Li et al. 2003). However, our understanding of chitin synthases in G. zeae, particularly the functional roles in development and virulence, is limited. In this study, we characterized the functional roles of GzCHS5 and GzCHS7 by using gene-deletion mutants. Similar to other filamentous fungi, mutants were viable but showed a number of abnormal hyphal morphologies. Both ΔGzChs5 and ΔGzChs7 mutants had swollen hyphal tips and reduced hyphal growth without apparent aerial hyphae. TEM revealed that intrahyphal hyphae were formed frequently in the mutants. Similar intrahyphal hyphae were observed in A. nidulans csmA and csmB mutants, suggesting that septum formation and hyphal development were uncoordinated in these mutants (Horiuchi et al. 1999; Takeshita et al. 2006). In some cases, we observed a woronin body-like organelle plugging a septum pore in the swollen mycelia of mutants (Fig. 6f). Woronin bodies are dense-core vesicles that are localized in the close vicinity of septal pores, and when hyphae are damaged, these organelles plug the septal pores rapidly to prevent cytoplasmic content leakage (Asiegbu et al. 2004). While further characterization is needed to unambiguously determine whether these organelles are indeed woronin bodies, we can hypothesize that a class V or class VII chitin synthase deficiency lead to the formation of deviant woronin body or that abnormal phenotypes caused by gene mutations induce the movement of woronin body to septal pores. These observations also suggest that ΔGzChs5 and ΔGzChs7 mutants are under stress, perhaps osmotic, due to the weakened or irregular hyphal tip development, and therefore exhibit aberrant phenotypes such as intrahyphal hyphae and woronin body-plugged septal pores.
When we compared the ΔGzChs5 and ΔGzChs7 mutant phenotypes with other filamentous fungi, we recognized one unique feature in G. zeae that was not observed in other fungi. Previous studies indicated fungal chitin synthases are essential for cell wall synthesis and maintenance, conidiation, hyphal tip growth, and septum formation. In G. zeae, mutations in GzCHS5 and GzCHS7 impacted the ability to produce perithecia. The function of those genes is directly related to the production of female fruiting bodies, perithecia, since the mutants completely lost their self-fertility while maintaining their male fertility. Perithecia consist of several layers of cells (Trail and Common 2000), and in particular this structure may require higher chitin content than do vegetative growth or asexual development. Therefore, it is reasonable to believe that the defect in fertility in the mutants is due to the defect of chitin in perithecia formation rather than defects in fertilization or post-fertilization.
The class V and VII chitin synthases in G. zeae are important for proper hyphal development and perhaps specialization. In A. nidulans, CsmA mutants form abnormal conidiophores and balloon hyphae. Significantly, while chitin synthase genes are redundant in filamentous fungi, functional role of genes from these two classes are not complementary in G. zeae. Similar to what we have observed in this study, it is likely that expression of certain class of chitin synthase may not be influenced when either or both GzCHS5 and GzCHS7 are deleted in G. zeae.
We showed that mutation in GzCHS5 and GzCHS7 caused G. zeae to be avirulent. In order to establish successful infection, fungal pathogens must overcome highly effective, constitutive physical and chemical host barriers, and employ a range of different infection strategies. A number of important steps in the infection process are common to fungal pathogens, including adhesion to the surface of the plant, penetration of the plant surface, and acquisition of nutrients from the plant cells (Hardham 2001). After successful penetration, hyphae of pathogenic fungi colonize host cells mechanically by expansion of the growing hyphal tips. Therefore, it is reasonable to hypothesize that opportune fungal cell wall biogenesis and hyphae development are critical for virulence. However, testing this hypothesis has been difficult in G. zeae due to the lack of mutant strains and other tools. Our study may provide a new insight into the complex mechanisms associated with early host–G. zeae interactions. In addition, chitin synthases characterized in this study may provide opportunities to develop a new chemical for controlling FHB.
References
Asiegbu FO, Choi W, Jeong JS, Dean RA (2004) Cloning, sequencing and functional analysis of Magnaporthe grisea MVP1 gene, a hex-1 homolog encoding a putative ‘woronin body’ protein. FEMS Microbiol Lett 230:85–90
Bartnicki-Garcia S (1968) Cell wall chemistry, morphogenesis, and taxonomy of fungi. Annu Rev Microbiol 22:87–108
Capellini RA, Peterson JL (1965) Macroconidium formation in submerged cultures by a nonsporulating strain of Gibberella zeae. Mycologia 57:962–966
Culp DW, Dodge CL, Miao Y, Li L, Sag-Ozkal D, Borgia PT (2000) The chsA gene from Aspergillus nidulans is necessary for maximal condition. FEMS Microbiol Lett 182:349–353
Desjardins AE (2006) Fusarium mycotoxins chemistry. Genetics, and biology. American Phytopathological Society, St Paul
Dufresne M, Perfect S, Pellier AL, Bailey JA, Langin T (2000) A Gal4-like protein is involved in the switch between biotrophic and necrotrophic phases of the infection process of Colletotrichum lindemuthianum on common bean. Plant Cell 12:1597–1590
Han YK, Lee T, Han KH, Yun SH, Lee YW (2004) Functional analysis of the homoserine O-acetyltransferase gene and its identification as a selectable marker in Gibberella zeae. Curr Genet 46:205–212
Han YK, Kim MD, Lee SH, Yun SH, Lee YW (2007) A novel F-box protein involved in sexual development and pathogenesis in Gibberella zeae. Mol Microbiol 63:768–779
Hardham AR (2001) Cell biology of fungal infection of plants. In: Gow NAR, Howard RJ (eds) The mycota VIII. Springer, Berlin, pp 91–123
Horiuchi H, Fujiwara M, Yamashita S, Ohta A, Takagi M (1999) Proliferation of intrahyphal hyphae caused by disruption of csmA, which encodes a class V chitin synthase with a myosin motor-like domain in Aspergillus nidulans. J Bacteriol 181:3721–3729
Horwitz BA, Sharon A, Lu SW, Ritter V, Sandrock TM, Yoder OC, Turgeon BG (1999) A G protein α subunit from Cochliobolus heterostrophus involved in mating and appressorium formation. Fungal Genet Biol 26:19–32
Hou Z, Xue C, Peng Y, Katan T, Kistler HC, Xu JR (2002) A mitogen-activated protein kinase gene (MGV1) in Fusarium graminearum is required for female fertility, heterokaryon formation, and plant infection. Mol Plant Microbe Interact 15:1119–1127
Ichinomiya M, Yamada E, Yamashita S, Ohta A, Horiuchi H (2005) Class I and class II chitin synthases are involved in septum formation in the filamentous fungus Aspergillus nidulans. Eukaryot Cell 4:1125–1136
Inoue I, Namiki F, Tsuge T (2002) Plant colonization by the vascular wilt fungus Fusarium oxysporum requires FOW1, a gene encoding a mitochondrial protein. Plant Cell 14:1869–1883
Lee J, Lee T, Lee YW, Yun SH, Turgeon BG (2003) Shifting fungal reproductive mode by manipulation of mating type genes: obligatory heterothallism of Gibberella zeae. Mol Microbiol 50:145–152
Lee SH, Lee J, Lee S, Park EH, Kim KW, Kim MD, Yun SH, Lee YW (2009) GzSNF1 is required for normal sexual and asexual development in the ascomycete Gibberella zeae. Eukaryot Cell 8:116–127
Leslie JF, Summerell BA (2006) The Fusarium lab manual. Blackwell, Ames
Li FB, Thompson GE, Newman RC (1998) Force modulation atomic force microscopy: background, development and application to electrodeposited cerium oxide films. Appl Surf Sci 126:21–33
Li HP, Fu CY, Peschen D, Ling XY, Fischer R, Liao YC (2003) Cloning and characterization of a gene coding for a class I chitin synthase from Fusarium graminearum. Can J Plant Pathol 25:240–248
Liu HB, Kauffman S, Becker JM, Szaniszlo PJ (2004) Wangiella (Exophiala) dermatitidis WdChs5p, a class V chitin synthase, is essential for sustained cell growth at temperature of infection. Eukaryot Cell 3:40–51
Lu S, Lyngholm L, Yang G, Bronson C, Yoder OD, Turgeon BG (1994) Tagged mutations at the Tox1 locus of Cochliobolous heterostrophus by restriction enzyme-mediated integration. Proc Natl Acad Sci USA 91:12649–12653
Madrid MP, Di Peitro A, Roncero MIG (2003) Class V chitin synthase determines pathogenesis in the vascular wilt fungus Fusarium oxysporum and mediates resistance to plant defense compounds. Mol Microbiol 47:257–266
Mandel MA, Galgiani JN, Kroken S, Orbach MJ (2006) Coccidioides posadasii contains single chitin synthase genes corresponding to classes I to VII. Fungal Genet Biol 43:775–788
Martín-Urdíroz M, Madrid MP, Roncero MIG (2004) Role of chitin synthase genes in Fusarium oxysporum. Microbiology 150:3175–3187
Martín-Urdíroz M, Roncero MIG, Gonzalez-Reyes JA, Ruiz-Roldan C (2008) ChsVb, a class VII Chitin synthase involved in septation, is critical for pathogenicity in Fusarium oxysporum. Eukaryot Cell 7:112–121
Motoyama T, Kojima N, Horiuchi H, Otha A, Takagi M (1994) Isolation of a chitin synthase gene chsV of Aspergillus nidulans. Biosci Biotechnol Biochem 58:2254–2257
Motoyama T, Fujiwara M, Kojima N, Horiuchi H, Otha A, Gakagi M (1996) The Aspergillus nidulans genes chsA and chsD encode chitin synthases which have redundant functions in conidia formation. Mol Gen Genet 251:520–528
Namiki F, Matsunaga M, Okuda M, Inoue I, Nishi K, Fujita Y, Tsuge T (2001) Mutation of an arginine biosynthesis gene causes reduced pathogenicity in Fusarium oxysporum f. sp. melonis. Mol Plant Microbe Interact 14:580–584
Nino-Vega GA, Carrero L, San-Blas G (2004) Isolation of the CHS4 gene of Paracoccidioides brasiliensis and its accommodation in a new class of chitin synthases. Med Mycol 42:51–57
Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Shim WB, Sagaram US, Choi YE, Wilkinson HH, Lee YW (2006) FSR1 is essential for virulence and female fertility in Fusarium verticillioides and F. graminearum. Mol Plant Microbe Interact 19:725–733
Sutton JC (1982) Epidemiology of wheat head blight and maize ear rot caused by Fusarium graminearum. Can J Plant Pathol 4:195–209
Takeshita N, Yamashita S, Ohta A, Horiuchi H (2002) csmA, a gene encoding a class V chitin synthase with a myosin motor-like domain of Aspergillus nidulans, is translated as a single polypeptide and regulated in response to osmotic conditions. Biochem Biophys Res Commun 298:103–109
Takeshita N, Yamashita S, Ohta A, Horiuchi H (2006) Aspergillus nidulans class V and VI chitin synthases CsmA and CsmB, each with a myosin motor-like domain, perform compensatory functions that are essential for hyphal tip growth. Mol Microbiol 59:1380–1394
Trail F, Common R (2000) Perithecial development by Gibberella zeae: a light microscopy study. Mycologia 92:130–138
Trail F, Xu H, Lorganger R, Gadoury D (2002) Physiological and environmental aspects of ascospore discharge in Gibberella zeae. Mycologia 94:181–189
Trail F, Gaffoor I, Vogel S (2005) Ejection mechanics and trajectory of the ascospores of Gibberella zeae (anamorph Fusarium graminearum). Fungal Genet Biol 42:528–533
Urban M, Mott E, Farley T, Hammond-Kosack K (2003) The Fusarium graminearum MAP1 gene is essential for pathogenicity and development of perithecia. Mol Plant Pathol 4:347–359
Weber I, Assmann D, Thines E, Steinberg G (2006) Polar localizing class V myosin chitin synthases are essential during early plant infection in the plant pathogenic fungus Ustilago maydis. Plant Cell 18:225–242
Werner S, Sugui JA, Steinberg G, Deising HB (2007) A chitin synthase with a myosin-like motor domain is essential for hyphal growth, appressorium differentiation, and pathogenicity of the maize anthracnose fungus Collectotrichum graminicola. Mol Plant Microbe Interact 12:1555–1567
Yu JH, Hamari Z, Han KH, Seo JA, Reyes-Dominguez Y, Scazzocchio C (2004) Double-joint PCR: a PCR-based molecular tool for gene manipulations in filamentous fungi. Fungal Genet Biol 41:973–981
Yu HY, Seo JA, Kim JE, Han KH, Shim WB, Yun SH, Lee YW (2008) Functional analyses of heterotrimeric G protein Gα and Gβ subunits in Gibberella zeae. Microbiology 154:392–401
Acknowledgments
This work was supported by a grant CG1411 from the Crop Functional Genomics Center of the twenty-first century Frontier Research Program funded by the Korean Ministry of Education, Science and Technology, and by the Korea Science and Engineering Foundation (KOSEF) grant by the Korea government (R11-2008-062-01001-0).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by S. Hohmann.
Rights and permissions
About this article
Cite this article
Kim, JE., Lee, HJ., Lee, J. et al. Gibberella zeae chitin synthase genes, GzCHS5 and GzCHS7, are required for hyphal growth, perithecia formation, and pathogenicity. Curr Genet 55, 449–459 (2009). https://doi.org/10.1007/s00294-009-0258-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00294-009-0258-6