Abstract
Plant and animal mitochondrial genomes, although quite distinct in size, structure, expression and evolutionary dynamics both may exhibit the state of heteroplasmy—the presence of more than one type of mitochondrial genome in an organism. This review is focused on heteroplasmy in plants, but we also highlight the most striking similarities and differences between plant and animal heteroplasmy. First we summarize the information on heteroplasmy generation and methods of its detection. Then we describe examples of quantitative changes in heteroplasmic populations of mitochondrial DNA (mtDNA) and consequences of such events. We also summarize the current knowledge about transmission and somatic segregation of heteroplasmy in plants and animals. Finally, factors which influence the stoichiometry of heteroplasmic mtDNA variants are discussed. Despite the apparent differences between the plant and animal heteroplasmy, the observed similarities allow one to conclude that this condition must play an important role in the mitochondrial biology of living organisms.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Heteroplasmy is defined as a state in which more than one mitochondrial genotype occurs in an organism. The ratio of different types of mtDNAs in a heteroplasmic population may be variable, but usually one mitotype is prevalent, while the alternative one(s) are present in a very low proportion. Under such conditions the phenotype of the organism is determined by the predominant mtDNA variant. In animals, heteroplasmy has been related to mitochondrial diseases (Wallace 1994; Zeviani and Antozzi 1997) and aging (Szibor and Holtz 2003). It has also been suggested to play a role in cancer (Chinnery et al. 2002), but evidence for a causative role in this condition is less clear. In the case of plants, this phenomenon has been investigated most often to clarify mitochondrial abnormalities like cytoplasmic male sterility (CMS) (Janska et al. 1998), nonchromosomal stripe mutants in maize (NCS) (Yamato and Newton 1999), the chloroplast mutator mutant in Arabidopsis (CHM) (Martinez-Zapater et al. 1992; Sakamoto et al. 1996), and recently, the mitochondrial mutator system in maize (Kuzmin et al. 2005). Recent studies indicate that heteroplasmy exists also in healthy humans (Kajander et al. 2000) and wild-type plants (Arrieta-Montiel et al. 2001; Taylor et al. 2001). This review is focused on heteroplasmy in plants, but we also highlight the most striking similarities and differences between plant and animal heteroplasmy.
How is heteroplasmy generated?
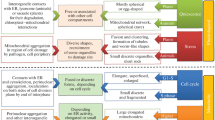
Plant mitochondrial genomes are much larger than the animal ones. The size ranges from 208 kb in Brassica hirta to 2,500 kb in the watermelon, compared to 16–20 kb in animals (Palmer and Herbon 1987; Ward et al. 1981; Boore 1999). Plant mtDNA encodes somewhat more genes but this is not the reason of the significant size difference, which is mainly caused by noncoding sequences that (without introns) make up 82% of Arabidopsis thaliana mtDNA (Unseld et al. 1997) but only 7% of the animal mitochondrial genome (Anderson et al. 1981). The genetic information stored in animal mitochondria is organized as a genome-sized double-stranded circular DNA molecule (Boore 1999). Although a genome-size circular linkage map can also be constructed for most plants, results of electron microscopy and studies using electrophoresis suggest that the genetic information of a plant mitochondrial genome is subdivided among the circular, linear and complex molecules of different sizes (Backert et al. 1997). It is commonly assumed that recombination between large repeated sequences is the most important force responsible for maintaining such multipartite structure as a dynamic entity (Fig. 1a). There is a strong belief that to fulfill their integrative role, the recombinations mediated by large repeats are frequent and easily reversible during plant life. Apart from the main genome whose parts are maintained in a dynamic equilibrium by large repeated sequences, plant mitochondria contain recombinant molecules, so called sublimons, whose number is very low compared to the main mitochondrial genome. The sublimons are products of rare and irreversible recombinations mediated by short repeated sequences (Fig. 1a). Short repeats are common in plant mitochondrial genomes (Notsu et al. 2002; Sugiyama et al. 2005; Kubo et al. 2000; Clifton et al. 2004). It has been proposed that they originate from insertion of reverse-transcribed copies of untranslated RNA (Gualberto et al. 1988) or from the recombinational activity of oligonucleotide motifs (Woloszynska et al. 2001). As a consequence of recombinations via both large and short repeats two types of mtDNA of different quantitative representation coexist in one organism—the predominant mitotype which creates the main genome, and the sublimons at a substoichiometric level.
Recombination and transmission of plant mtDNA. a Subgenomic molecules (solid lines) of the main genome are created by frequent and reversible recombinations mediated by large repeated sequences (L). Substoichiometric molecules (sublimons; dashed lines) are generated via rare and irreversible recombinations across short repeats (S). b Transmission of mtDNA molecules in plants is determined by a hypothetical single replicative unit, which resides in a meristem tissue. This “transmitted form” contains the complete nucleotide information, which in differentiated cells may be located in subgenomic (solid lines) and substoichiometric (dashed lines) molecules
Recombination
The recombination-mediated generation of heteroplasmy is well documented in plants. Rare recombination events across very short repeated sequences (6–36 bp) are the cause of all known deletion nonchromosomal stripe (NCS) mutations of maize (Yamato and Newton 1999). Usually only those NCS plants that contain at least some proportion of the normal mitochondrial genome may survive. Cases of heteroplasmy produced via recombination across short repeats have been detected in maize (Small et al. 1987), Solanaceae (Vitart et al. 1992; Kanazawa et al. 1994; Gutierres et al. 1997), wheat (Hartmann et al. 1994), Arabidopsis (Sakamoto et al. 1996) and other Brassicaceae (Bellaoui et al. 1998), and bean (Woloszynska et al. 2001).
In contrast to the plants, heteroplasmy of animal mtDNA driven by recombination has been only rarely described. Until recently, it has generally been assumed that animal mitochondrial DNA does not undergo recombination. Although this issue is still controversial, it has been suggested that some mitochondrial rearrangements found at a very low level in healthy human tissues result from recombination (Kajander et al. 2000). This conclusion was based on the observation that rearrangement break points occurred at short directly repeated sequences, located chiefly near the ends of the large non-coding control region. Following the terminology applied to plant mitochondrial DNA, the recombination-derived molecules present at very low levels in human mitochondria were also named sublimons.
Small-scale mutations
Beside recombination, new mitochondrial genotypes could be produced by replication errors, defective and inefficient repair or reaction of mtDNA with reactive oxygen metabolites. As a result, point mutations as well as deletions or duplications could be created. Mutations of these types often observed in human heteroplasmy are believed to be mainly produced by a non-recombinational changes attributed to the activity of mtDNA metabolism enzymes and the action of reactive oxygen species (Khrapko et al. 1997). It has been believed for a long time that plant mitochondrial genomes evolve rapidly in structure but slowly in sequence. Consequently, point mutations were assumed to contribute only marginally to heteroplasmy in plants. However, this point of view can no longer be upheld in view of the recent findings of the elevated substitution rates in some plant lineages (Plantago, Cho et al. 2004; Pelargonium, Palmer at al. 2000). According to Cho et al. (2004) the mutation rate may occasionally increase or slow down in particular periods of evolution. Those reports imply that sequence variation due to mutation may be a source of heteroplasmy at least in particular plant lineages. Indeed, a very high level of heteroplasmy associated with point mutations within noncoding sequences was detected in olive tree cultivars (Olea europaea L.) (Garcia-Diaz et al. 2003). The coexistence of a minimum three and maximum five variants differing by point mutations within a single plant was demonstrated. The point mutations causing heteroplasmy are not restricted to noncoding regions which may be expected to accumulate substitutions relatively easily. An analysis of a 2.3 kb mitochondrial region containing several active genes allowed the identification of seven variants in the monocot genera Aegilops and Triticum (Hattori et al. 2002). These sequences were maintained as heteroplasmic populations composed of 2–4 variants within an individual plant. Although tandem repeats are present in the plant mtDNA, there is not a single study reporting heteroplasmy caused by variation in the tandem repeat number (Barr et al. 2005).
Paternal leakage
Mitochondrial genomes are mainly inherited maternally, with some exceptions showing paternal transmission or biparental inheritance (Birky 2001; Ladoukakis and Zouros 2001). Recent findings have shown that in plants and animals, which normally transmit their mtDNA in the female line only, heteroplasmy can be produced by occasional paternal leakage. Aksyonova et al. (2005) observed the transmission of paternal wheat mtDNA in barley-wheat hybrids followed by a selective replication of the paternal mtDNA during repeated backcrosses with the wheat parent. The paternal leakage may lead to further rearrangements, since the mtDNAs passed down from males and females could recombine (Ladoukakis and Zouros 2001). In plants the first evidence for heteroplasmic coexistence of paternal and maternal mtDNA sequences concerned the orf25 region in triticale, a crop obtained by crossing wheat and rye (Laser et al. 1997). However, the origin of the paternal-like sequence in triticale was unclear, because it was also detected at a low level in maternal wheat mitochondria. Conclusive evidence demonstrating an involvement of paternal transmission of mtDNA in generation of heteroplasmy was reported in a nucleus-cytoplasm (NC) hybrid of timopheevi wheat (Kitagawa et al. 2002). In addition to the maternal-identical sequences, the NC hybrids contained paternal-identical and paternal-derived sequences. Moreover, in this case the absence of paternal sequences in maternal progenitor lines was shown.
In animals, transmission of paternal mtDNA has been proven to occur incidentally in humans (Schwartz and Vissing 2002), birds (Kvist et al. 2003) and Drosophila (Kondo et al. 1990), but especially in the interspecific crosses (Shitara et al. 1998). On the other hand, paternal transmission was reported to occur constitutively in mussels in the mode of “doubly uniparental inheritance” (Skibinski et al. 1994; Zouros et al. 1994).
Heteroplasmy in plants and animals seems to be generated via similar events: recombinations, small-scale mutations and transmission of paternal mtDNA. However, the relative input of these mechanisms as the source of heteroplasmy is different for the two groups of organisms. Mutations resulting from the replication errors and oxidative-damage-caused point mutations, deletions, and variation in tandem repeats are the most common explanation for the occurrence of heteroplasmy in animals, while their exact role in the generation of heteroplasmy in plant mitochondria is unclear. In contrast, recombination-mediated generation of heteroplasmy is well documented in plants but remains controversial in animals. Transmission of paternal mtDNA is much more frequently reported for animals than for plants.
How is heteroplasmy detected?
Successful detection of heteroplasmy depends on the type of change in the mitochondrial genome, the ratio of the heteroplasmic variants and the sensitivity of the method applied. The strategies used are mainly based on a Southern hybridization, PCR or DNA sequencing.
Heteroplasmy in plants usually results from large-scale rearrangements mediated by recombination. This was also the case in maize where heteroplasmy was reported for the first time in plants (Small et al. 1987). Four atpA types were found in a range of maize cytoplasms, some of these rearrangements were present at a low stoichiometry. To distinguish between the predominant and substoichiometric atpA variants, Southern hybridization with different times of autoradiogram exposure was utilized. Detection of low-abundance fragments required prolonged exposure. Many subsequent studies on plant heteroplasmy copied the strategy of “overexposed” autoradiograms to visualize mtDNA fragments at an unusually low copy number (Arrieta-Montiel et al. 2001; Gu et al. 1993; Janska et al. 1998; Vitart et al. 1992; Woloszynska et al. 2001). However, some authors were able to show the presence of a “low-abundance” fragment from observation of “normally” exposed autoradiograms (Bellaoui et al. 1998; Kanazawa et al. 1994; Shirzadegan et al. 1991). Alternatively, some sublimons could be visualized by conventional PCR, but detection of others required PCR followed by Southern hybridization (Woloszynska et al. 2001).
In plants substoichiometric mtDNA molecules distinguished from the predominant genome only by point mutations have been rarely reported. In such cases heteroplasmy is detected by PCR amplification of mtDNA fragments encompassing the point mutation followed by sequencing of random PCR clones. Stoichiometry of the mutated variants is then estimated basing on the frequency of clones representing mutated molecules (Hattori et al. 2002). On the other hand, competitive PCR can be used to distinguish and quantify sequence variants. A specific version of orf25 representing only 0.1% of the total orf25 gene sequences in wheat mitochondria was analyzed in this way (Laser et al. 1997).
Currently, some authors exploit the high sensitivity and accuracy of real-time PCR for quantitative detection of heteroplasmy. This method allowed establishing that sublimons containing cytoplasmic male sterility-associated sequence are less abundant than one copy per 100–200 cells in fertile common bean (Arrieta-Montiel et al. 2001).
Even a wider spectrum of techniques has been employed to detect and/or quantify heteroplasmy in humans, especially the heteroplasmy resulting from point mutations. Direct mtDNA sequencing is still used in some laboratories for clinical diagnostics. However, it lacks the sensitivity to detect mtDNA molecules present at less than 20% of the total population (Wong and Boles 2005). Among the variants of sequencing methods, pyrosequencing has been shown to provide high accuracy and exceptional sensitivity allowing detection of 1% heteroplasmy (White et al. 2005).
In the case of known point mutations, the most commonly used methods are PCR/RFLP and PCR/ASO (allele-specific oligonucleotide). In the PCR/RFLP analysis the mutation-containing fragment is amplified and digested with a restriction enzyme that cleaves differentially the wild and the mutated sequence. After electrophoresis the normal and mutant DNA fragments are visualized with ethidium bromide and quantified by densitometric scanning. PCR/ASO involves two steps: PCR, in which multiple products encompassing potential mutations are obtained, and hybridization of PCR products to radioactively labeled probes containing the point mutation in question. This method provides sensitivity even ten-fold higher than that of the PCR/RFLP (down to 1% of heteroplasmy) (Wong and Boles 2005).
Most of the large-scale mtDNA mutations, e.g. deletions and duplications, can be easily detected by Southern blot analysis (reviewed by Moraes et al. 2003 and Wong and Boles 2005). Commonly used are also PCR-based assays, either long-extension PCR (LX-PCR) employing primers capable of amplifying the entire genome or PCR involving sets of primers amplifying only smaller portions of the mitochondrial DNA. In an attempt to quantify human sublimons Kajander et al. (2000) applied multiplex PCR using primers across the deletion junction and primers for a nuclear gene as an internal standard. The authors have estimated that sublimons occur in the range from 190 copies to less than 0.1 copies per cell in healthy human tissues.
Quantitative changes within heteroplasmy
An interesting feature of heteroplasmy is its dynamic nature. Available reports evidence that quantitative changes in the proportion of heteroplasmic variants may occur in both plants and animals. Often such changes are very rapid, leading to the mitochondrial genomic shifting observed as the appearance of a new predominant mitotype in consecutive generations.
Rapid genomic shifts within plant mitochondrial genomes have been observed in plant tissue cultures (Vitart et al. 1992; Kanazawa et al. 1994; Bartoszewski et al. 2004) and under natural growth conditions (Janska et al. 1998). Genomic shifts were reported most often for cultivated plants but also in wild populations of plants (Arrieta-Montiel et al. 2001).
A comprehensive description of genomic shifting occurring during cell culture and plant regeneration was given by Kanazawa et al. (1994). The authors detected significant reversible stoichiometric changes of two tobacco mtDNA fragments containing the atp6 gene. One of these fragments represented only 0.1% of total atp6 gene copies in green tissue, but during the tissue dedifferentiation its fraction increased to almost 75% in calli. During plant regeneration, in green callus, the proportion of the respective DNA fragment decreased to about 50%, while in regenerated plants this fragment comprised only 14% of all the atp6 copies.
The best-documented cases of genomic shifts which can alter the plant phenotype are related to the occurrence of cytoplasmic male sterility (CMS) and spontaneous reversion to fertility in common bean (Janska and Mackenzie 1993; Janska et al. 1998). It was proposed that the emergence of a cytoplasmic male-sterile line involved selective amplification of sublimons present in mitochondria of the fertile progenitor line. Some of these sublimons contain the sterility-inducing pvs sequence. The amplification of the sublimons was accompanied by a decrease in the copy number of the progenitor predominant mitotype. In principle, this genomic shifting was so substantial that the sublimons overcame the predominat mitotype and vice versa, the predominant molecules became sublimons. Furthermore, the spontaneous reversion to fertility of CMS plants was also associated with genomic shifting, however, this time the shift was limited to the part of the predominant mitotype containing the pvs sequence (Fig. 2). It should be emphasized that in bean the switches from fertile to the sterile phenotype and vice versa were coupled with, respectively, amplification and copy number reduction of genes located within the pvs sequence. This means that the genomic shift in bean is a reversible event and can be viewed as a mechanism allowing activation or silencing of gene expression via gene copy number regulation.
Model of quantitative changes occurring as a consequence of genomic shift in Phaseolus vulgaris mitochondrial genome (Janska et al. 1998). Generation of cytoplasmic male sterility in bean is associated with amplification of molecules that are maintained in the fertile line at a substoichiometric level (dashed lines). One of the amplified molecules carries the sterility-inducing sequence (pvs; black square). The amplification is accompanied by simultaneous copy number reduction of the molecule representing the main genome of the fertile progenitor (solid lines). Spontaneous reversion to fertility in bean is also due to a genomic shift, in this case, however, limited to selective reduction of the molecule carrying the pvs sequence
Cases of rapid quantitative shifts have also been observed in animal mitochondria. The first example of such a rapid shift between genotypes differing at a single nucleotide was described in Holstein cows (Hauswirth and Laipis 1982). It occurred within two generations, creating an offspring that was homoplasmic for a mitotype already present in the parental mitochondria at substoichiometric amounts. To date many more cases of a rapid and complete switch to another homoplasmic genotype between mother and offspring have been reported in animals. To our knowledge, the only example of a paternally oriented shift from heteroplasmy to homoplasmy concerned barley–wheat hybdrids backcrossed with wheat parents (Aksyonova et al. 2005).
Transmission and somatic segregation of heteroplasmy
Studies in humans show that the genomic shifts observed between generations do not always result in a complete switch to homoplasmy. It is now clear that the proportion of mutated mitochondrial DNA (the mitochondrial DNA mutation load) may vary widely among siblings, between tissues and during the life span of animals and humans (Sekiguchi et al. 2003; Jenuth et al. 1997). It is thought that the variation in the mitochondrial mutation load results from events operating during germ line and tissue development (Chinnery 2002). Understanding of these events has been greatly advanced by the generation of heteroplasmic mice by cytoplasm transfer experiments (Jenuth et al. 1996). These studies have reinforced the hypothesis of mitochondrial genetic bottleneck occurring during the development of the female germline (Fig. 1). The “bottleneck” refers to reduction of mtDNA copies during early oogenesis, accompanied by their random, often unequal, distribution to primary oocytes and subsequent amplification of the mtDNAs in mature oocytes (Jenuth et al. 1996; Lightowlers et al. 1997). A rapid switch from heteroplasmy to homoplasmy, which is occasionally observed, indicates that the mtDNA copy number in the primary oocyte (the size of the inheritance unit) may be as low as one molecule (Bendall et al. 1997).
The proportions in heteroplasmy may change also during later stages of development as a consequence of non-random cell- (Sekiguchi et al. 2003) or tissue-specific selection (Jenuth et al. 1997) (Fig. 3). It has been suggested that selection is independent from mitochondrial function or cellular proliferation and most probably occurs at the level of the mitochondrial genome (Battersby and Shoubridge 2001; Chinnery 2002).
Accumulation of mutant mtDNA in humans is not only related to mitochondrial diseases, but it has also been observed during carcinogenesis and very often in the course of normal aging (Polyak et al. 1998; Szibor and Holtz 2003; Trifunovic et al. 2004; Wallace 1999). According to a hypothesis proposed by Chinnery et al. (2002), the increase of mutant mtDNA level in aging, cancer and disease occurs due to similar mechanisms, based on the three basic properties of mtDNA: polyploidy, relaxed (independent from the cell cycle) replication, and the presence of a mechanism that regulates the normal cellular mtDNA copy number. The latter may induce proliferation of mtDNA to maintain the respiration capacity of mitochondria affected due to a mtDNA mutation. However, replication of the mutant mtDNA results in an overall increase in the mutation load leading to a further decline of respiration capacity.
In contrast, there is only limited information about the distribution of mitochondrial heteroplasmy among siblings, between tissues and during the life span of plants. The only report suggesting the occurrence of a bottleneck in plant mitochondria comes from studies on olive trees (Garcia-Diaz et al. 2003). The fact that it is one of the most ancient vegetatively propagated plant species makes the olive tree an excellent model to study mtDNA maintenance. Asexually propagated plants exhibit a substantially higher level of heteroplasmy compared to sexually reproduced olive trees. The authors claim that the significant reduction of heteroplasmy in sexually reproducing plants could be explained as a result of genetic drift during the female gamete formation. There are no other reports of a plant mitochondrial bottleneck. If this phenomenon occurs in plants at all, we assume that the number of mitochondrial genomes transmitted by the maternal line to the progeny should be higher than one, since the majority of plant mitochondrial genomes examined are heteroplasmic.
Analysis of bean heteroplasmy and its evolutionary changes brought some important facts about transmission of heteroplasmic molecules (Arrieta-Montiel et al. 2001). Arrieta-Montiel and co-workers examined the presence of the pvs sequence in the mitochondrial genome of 106 wild and 36 cultivated Phaseolus lines. The pvs sequence was present in all of the examined plants, and in 84% of them it was observed at a substoichiometric level. It seems unlikely that the presence of the sublimon molecule carrying pvs in so many bean lines is due to recurrent de novo generation of this molecule by recombination in the course of evolution. It seems rather that this substoichiometric molecule, once created, was subsequently stably transmitted. These conclusions are supported by observations that in some plant species sublimons are not accompanied by a putative parental molecule that would serve as substrate for the recombination event (Janska et al. 1998; Kanazawa et al. 1994). In light of these assumptions it has been proposed that plants must posses a means of ensuring the transmission of a complete mitochondrial genome to subsequent generations, although the mechanism of the transmission and somatic segregation are very poorly understood. Since the plant mitochondrial genome has a multipartite structure, it has been proposed that plants posses a so-called “transmitted form” of the mitochondrial genome containing the complete genetic information of the mitochondria including sublimons. The exact structure of the transmitted form remains unknown. Nevertheless, it was proposed that it is organized into a single replication unit and resides within the meristem (plant mtDNA replication zone) to assure transmission of the complete genetic complement to daughter cells (Arrieta-Montiel et al. 2001; Fig. 1b.). Results obtained by Suzuki et al. (1996) indicate that the organization of mtDNA in root meristem of rice differs from that in differentiated cells (Fig. 1b). According to these results, replication and distribution of mtDNA during cell differentiation may induce changes in the stoichiometry of mtDNA molecules. This was confirmed by results according to which the pvs sequence present as a sublimon in leaf tissue of a bean revertant line was markedly more abundant in the root meristem (Arrieta-Montiel et al. 2001).
Perhaps the best examples of quantitative changes in heteroplasmic mtDNA distribution in plants are the nonchromosomal stripe (NCS) mutants of maize (Yamato and Newton 1999). They carry a deletion within mitochondrial genes and show characteristic phenotypes with green or yellow leaf stripes. The green sectors are heteroplasmic for the mutant and normal forms of mitochondrial genes, while the yellow sectors are homoplasmic containg only the mutated forms. Each NCS mutation is stable in the sense that the deletion can be passed unaltered to the progeny. However, plants derived from the same parents can be affected to various degrees (Wintz 1994).
Heteroplasmy and nuclear copies of mtDNA
It is well known that fragments of mitochondrial DNA are integrated into the nuclear genomes of many organisms including numerous animal and plant species (Bensasson et al. 2001; Timmis et al. 2004). In animals these sequences are named Numts for nuclear mitochondrial pseudogenes. There are no reports on recent mtDNA transfer into metazoan nuclei but in plants this process is still ongoing. Current studies indicate that escape of the genetic material from organelles to the nucleus occurs much more frequently than generally believed (Timmis et al. 2004). In light of this finding, the claim that low abundance DNA fragments in a cell always represent mitochondrial sequences, may be overoptimistic. Nuclear sequences of mitochondrial origin may also be detected using the techniques used to study heteroplasmy; consequently, the experimental design of many heteroplasmy studies has often been criticized, especially when they were based on PCR amplification (Budowle et al. 2002; Brandstatter and Parson 2003). Such assays may lead to false conclusions, as in the case of Numts amplified from patients with Alzheimer disease and mistaken for heteroplasmic mtDNA mutations causing this disease (Wallace et al. 1997). On the other hand, it is equally hard to believe that the DNA fragments of low stoichiometry observed in numerous analyses always represent mitochondrial segments transferred to the nucleus. The fact that genomic shifting could be reversible over a relatively short evolutionary time (Janska et al. 1998; Woloszynska et al. 2001) argues for a view that such rapid quantitative shifts require the pre-existence of low abundance fragments in mitochondria. Therefore, we think that heteroplasmy is a common condition of animal and plant mitochondria but its reliable detection and further analysis require some precautions. It is advisable to enrich the PCR template in mtDNA by purifying mitochondria instead of total DNA or by using tissues containing high amounts of mtDNA relative to nuclear DNA (Bensasson et al. 2001). Another way to verify the identity of suspected Numts can be RT-PCR since these sequences are usually not transcribed, unlike their mitochondrial counterparts.
Is heteroplasmy under nuclear control?
Currently, little is known about the mechanisms involved in the genomic shifting observed in heteroplasmic plant mitochondrial genomes. A lack of precise information about the structure of plant mitochondrial genomes slows down the progress in understanding this phenomenon. Theoretically, three types of events could result in genomic shifting in plants: changes in the replication rate of selected molecules, changes in the state of recombinational equilibrium, and changes in segregation of a heteroplasmic population of mtDNAs; or any combination of them. While it is still uncertain which of these processes leads to mitochondrial genomic shifting in plants, there is no doubt that it is under nuclear control. A few examples of nuclear genes modulating heteroplasmy are described below.
In the common bean the genomic shift leading to reversion to fertility may occur spontaneously or may be induced by the nuclear gene Fr. When introduced into a sterile plant, the Fr gene results in copy number suppression of the pvs sequence (Arrieta-Montiel et al. 2001). The exact mechanism by which Fr drives the pvs copy number decrease has not been explained. However, the Fr-induced phenomenon is indistinguishable from the spontaneous reversion to fertility described here in chapter four (Janska et al. 1998). Thus, it can be assumed that the similarly to the spontaneous reversion also the Fr action involves differential replication of mtDNA molecules.
It has been suggested that the CHM gene in Arabidopsis, preventing the amplification of a specific mitochondrial genome configuration that is associated with the appearance of green-white leaf variegation, acts in a manner similar to Fr (Martinez-Zapater et al. 1992; Sakamoto et al. 1996). The CHM gene has been shown to encode a protein homologous to MutS protein of Escherichia coli that is involved in mismatch repair and DNA recombination (Abdelnoor et al. 2003). Consequently, the locus was named AtMSH1 and the encoded protein MSH1. Surprisingly, chm mutants showed no evidence of increased point mutations. Since mismatch repair components are usually involved in suppression of nonhomologous recombination, it was proposed that the function of the AtMSH1-encoded protein was to control recombination. Mutations disrupting this activity could lead to an increased recombination and finally to a genomic shift. This scenario is possible only if the main genome contains both parental mtDNA configurations which recombine and generate the sequence undergoing the genomic shift. However, this model of MSH1 action does not apply to plant systems where the shifted DNA configuration can not be produced via de novo recombination (Janska et al. 1998). Thus, according to an alternative model, the role of MSH1 would involve control of replication. This scenario assumes that shifted mtDNA molecules are generated via earlier recombinations. The chimeric sites created by the recombinations might serve to trigger site-specific replication stalling by the MSH1 protein (Abdelnoor et al 2003). Similarly to MSH1, also other candidates for nuclear factors controlling heteroplasmy in Arabidopsis—RecA and single-stranded DNA-binding proteins—are involved in DNA recombination and repair (Shedge, unpublished data; Harmon and Kowalczykowski 1998; Steffen and Bryant 2001; Edmondson et al. 2005).
Another example of nuclear control over heteroplasmy in plants is the Psm locus of cucumber. Since the mitochondrial genome of this plant species is paternally inherited, Psm is the first nuclear locus to control sorting of paternally transmitted sublimons (Havey et al. 2004). The most common Psm allele (Psm+) provides sorting that favors the mitochondrial genome variant conditioning a mosaic phenotype. The Psm- locus results in a preferable sorting of sublimons conferring normal phenotype.
In contrast to the above examples of heteroplasmy control driven by single nuclear genes (Fr, AtMSH1, Psm), the mitochondrial mutator system in maize involves complex interactions among nuclear alleles and the mitochondrial genome (Kuzmin et al. 2005). The system was identified in the P2 line of maize characterized by mtDNA destabilization resulting in poor plant growth and pale streaks on leaves. The Fr, Psm and AtMSH1genes, when introduced into a plant or disrupted by mutation, result in reproducible genomic shift. This is not the case for the P2 mutator system. Plants of the P2 line, or plants obtained by introducing P2 nuclear alleles via the male parent, show highly variable mtDNA patterns differing both quantitatively and qualitatively among sibling individuals and between parents and progeny (Kuzmin et al. 2005). In individual P2 siblings the copy number of a particular mitochondrial sequence may increase or decrease differentially, shifting in either direction. The authors propose that the combined outcome of destabilized replication and transmission is variability of mtDNA profiles observed among sibling and between parents and progeny. An important conclusion concerning the P2 mutator system is that the replication of mtDNA molecules and their transmission are controlled by different nuclear alleles.
The complex nature of the nuclear control over heteroplasmy is further supported by studies on the reorganization of the mitochondrial genome of wheat induced by in vitro conditions (Hartmann et al. 2000). It was found that a single rearrangement in mtDNA could be governed by many genes scattered in the nuclear genome. The involvement of a nuclear-cytoplasmic compatibility system in the control of mtDNA heteroplasmy was recently reported for backcrossed barley–wheat hybrids (Aksyonova et al. 2005). It was observed that replacement of the barley nuclear genome with the paternal wheat nuclear genome correlates with the paternally oriented shift of mtDNA composition and the restoration of fertility and plant vigour.
A complexity of the nuclear control has also been observed in animals (Farge et al. 2002; Battersby et al. 2003). A mutant strain of Drosophila subobscura is heteroplasmic with about 80% of the mitochondrial DNA molecules containing a large-scale deletion. Successive backcrosses between mutant females and wild-type males result in a biphasic change in the level of heteroplasmy (Farge et al. 2002). In the early crosses the proportion of deleted molecules decreases relatively quickly, while in subsequent crosses this decrease is much slower. The return to the mutant nuclear genome is followed by a progressive increase in heteroplasmy until stabilization at a level characteristic for the given mutant strain. This dynamics of quantitative changes in mtDNA indicates that several different genes must be involved in heteroplasmy control in D. subobscura. The authors suggest that the mutant nuclear genes controlling the level of heteroplasmy are directly involved in the production of the deleted mtDNA molecules (Farge et al. 2002). On the other hand, Battersby et al. (2003) identified three nuclear loci in the mouse genome that control heteroplasmy via governing tissue-specific and age-dependent segregation of two mtDNA variants. The transmission of both variants in the germ-line is random but one mitotype increases in the liver and kidney with age, whereas the second variant is prevalent in the blood and spleen.
Concluding remarks
Heteroplasmy is generated via the same mechanisms in plants and animals but while in plants the most powerful force is recombination, in animals heteroplasmy mainly results from replication errors, defective and inefficient repair or reaction of mtDNA with reactive oxygen metabolites. Similarly, transmission of paternal mtDNA as a source of heteroplasmy is much more frequently reported for animal than for plant genomes. However, it should be emphasized that the bias of data concerning heteroplasmy origin in plants and animals does not necessarily reflect the true situation. It may simply result from the obvious prevalence for research regarding human mitochondria. In both groups of organisms heteroplasmy was originally recognized as associated with pathological states, but currently it is assumed to occur also in normal individuals. Rapid genomic shifts have been described in plants and animals; however, the switch to homoplasmy seems to be common only in animals while in plants it usually results in a different level of heteroplasmy. Again, this may not result from a real difference but rather from the various methods and experimental procedures applied to both groups of organisms. In the case of plants usually pooled seedlings or leaves from many individuals are sampled, while for animals particular tissues of a single organism are tested. Moreover, as we already mentioned, DNA sequencing that has been widely used to detect animal heteroplasmy is not as sensitive as the PCR or Southern assays with prolonged autoradiography usually applied in plant research. Another common feature of animal and plant heteroplasmy is that this state is controlled by nuclear genes and this control is a complex phenomenon involving many genes. Finally, we would like to emphasize that the universal occurrence of heteroplasmy in plants and animals suggests that it has an important, although still obscure role in mtDNA genetics.
References
Abdelnoor RV, Yule R, Elo A, Christensen AC, Meyer-Gauen G, Mackenzie SA. (2003) Substoichiometric shifting in the plant mitochondrial genome is influenced by a gene homologous to MutS. Proc Natl Acad Sci USA 100:5968–5973
Aksyonova E, Sinyavskaya M, Danilenko N, Pershina L, Nakamura C, Davydenko O (2005) Heteroplasmy and paternally oriented shift of the organellar DNA composition in barley-wheat hybrids during backcrosses with wheat parents. Genome 48:761–769
Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, Schreier PH, Smith AJ, Staden R, Young IG (1981) Sequence and organization of the human mitochondrial genome. Nature 290:457–465
Arrieta-Montiel M, Lyznik A, Woloszynska M, Janska H, Tohme J, Mackenzie S (2001) Tracing evolutionary and developmental implications of mitochondrial stoichiometric shifting in the common bean. Genetics 158:851–864
Backert S, Nielsen BL, Börner T (1997) The mystery of the rings: structure and replication of mitochondrial genomes from higher plants. Trends Plant Sci 2:477–483
Barr CM, Neiman M, Taylor DR (2005) Inheritance and recombination of mitochondrial genomes in plants, fungi and animals. New Phytol 168:39–50
Bartoszewski G, Malepszy S, Havey MJ (2004) Mosaic (MSC) cucumbers regenerated from independent cell cultures possesses different mitochondrial rearrangements. Curr Genet 45:45–53
Battersby BJ, Shoubridge EA (2001) Selection of a mtDNA sequence variant in hepatocytes of heteroplasmic mice is not due to differences in respiratory chain function or efficiency of replication. Hum Mol Genet 10:2469–2479
Battersby BJ, Loredo-Osti JC, Shoubridge EA (2003) Nuclear genetic control of mitochondrial DNA segregation. Nat Genet 33:183–186
Bellaoui M, Martin-Canadell A, Pelletier G, Budar F (1998) Low-copy-number molecules are produced by recombination, actively maintained and can be amplified in the mitochondrial genome of Brassicaceae: relationship to reversion of the male sterile phenotype in some cybrids. Mol Gen Genet 257:177–185
Bendall KE, Macaulay VA, Sykes BC (1997) Variable levels of a heteroplasmic point mutation in individual hair roots. Am J Hum Genet 61:1303–1308
Bensasson D, Zhang D, Hartl DL, Hewitt GM (2001) Mitochondrial pseudogenes: evolution’s misplaced witnesses. Trends Ecol Evol 16:314–321
Birky CW Jr (2001) The inheritance of genes in mitochondria and chloroplasts: laws, mechanisms, and models. Annu Rev Genet 35:125–148
Boore JL (1999) Animal mitochondrial genomes. Nucleic Acids Res 27:1767–1780
Brandstatter A, Parson W (2003) Mitochondrial DNA heteroplasmy or artifacts—a matter of the amplification strategy? Int J Legal Med 117:180–184
Budowle B, Allard MW, Wilson MR (2002) Critique of interpretation of high levels of heteroplasmy in the human mitochondrial DNA hypervariable region I from hair. Forensic Sci Int 126:30–33
Chinnery PF (2002) Modulating heteroplasmy. Trends Genet 18:173–176
Chinnery PF, Samuels DC, Elson J, Turnbull DM (2002) Accumulation of mitochondrial DNA mutations in ageing, cancer, and mitochondrial disease: is there a common mechanism? Lancet 360:1323–1325
Cho Y, Mower JP, Qiu YL, Palmer JD (2004) Mitochondrial substitution rates are extraordinarily elevated and variable in a genus of flowering plants. Proc Natl Acad Sci USA 101:17741–17746
Clifton SW, Minx P, Fauron CM, Gibson M, Allen JO, Sun H, Thompson M, Barbazuk WB, Kanuganti S, Tayloe C, Meyer L, Wilson RK, Newton KJ (2004) Sequence and comparative analysis of the maize NB mitochondrial genome. Plant Physiol 136:3486–3503
Edmondson AC, Song D, Alvarez LA, Wall MK, Almond D, McClellan DA, Maxwell A, Nielsen BL (2005) Characterization of a mitochondrially targeted single-stranded DNA-binding protein in Arabidopsis thaliana. Mol Genet Genomics 273:115–122
Farge G, Touraille S, Le Goff S, Petit N, Renoux M, Morel F, Alziari S (2002) The nuclear genome is involved in heteroplasmy control in a mitochondrial mutant strain of Drosophila subobscura. Eur J Biochem 269:998–1005
Garcia-Diaz A, Oya R, Sanchez A, Luque F (2003) Effect of prolonged vegetative reproduction of olive tree cultivars (Olea europaea L.) in mitochondrial homoplasmy and heteroplasmy. Genome 46:377–381
Gu J, Miles D, Newton KJ (1993) Analysis of leaf sectors in the NCS6 mitochondrial mutant of maize. Plant Cell 5:963–971
Gualberto JM, Wintz H, Weil JH, Grienenberger JM (1988) The genes coding for subunit 3 of NADH dehydrogenase and for ribosomal protein S12 are present in the wheat and maize mitochondrial genomes and are co-transcribed. Mol Gen Genet 215:118–127
Gutierres S, Lelandais C, De Paepe R, Vedel F, Chetrit P (1997) A mitochondrial sub-stoichiometric orf87-nad3-nad1 exonA co-transcription unit present in Solanaceae was amplified in the genus Nicotiana. Curr Genet 31:55–62
Harmon FG, Kowalczykowski SC (1998) RecQ helicase, in concert with RecA and SSB proteins, initiates and disrupts DNA recombination. Genes Dev 12:1134–1144
Hartmann C, Recipon H, Jubier MF, Valon C, Delcher-Besin E, Henry Y, De Buyser J, Lejeune B, Rode A (1994) Mitochondrial DNA variability detected in a single wheat regenerant involves a rare recombination event across a short repeat. Curr Genet 25:456–464
Hartmann C, Henry Y, Tregear J, Rode A (2000) Nuclear control of mitochondrial genome reorganization characterized using cultured cells of ditelosomic and nullisomic-tetrasomic wheat lines. Curr Genet 38:156–162
Hattori N, Kitagawa K, Takumi S, Nakamura C (2002) Mitochondrial DNA heteroplasmy in wheat, Aegilops and their nucleus-cytoplasm hybrids. Genetics 160:1619–1630
Hauswirth WW, Laipis PJ (1982) Mitochondrial DNA polymorphism in a maternal lineage of Holstein cows. Proc Natl Acad Sci USA 79:4686–4690
Havey MJ, Park YH, Bartoszewski G (2004) The Psm locus controls paternal sorting of the cucumber mitochondrial genome. J Hered 95:492–497
Janska H, Mackenzie SA (1993) Unusual mitochondrial genome organization in cytoplasmic male sterile common bean and the nature of cytoplasmic reversion to fertility. Genetics 135:869–879
Janska H, Sarria R, Woloszynska M, Arrieta-Montiel M, Mackenzie SA (1998) Stoichiometric shifts in the common bean mitochondrial genome leading to male sterility and spontaneous reversion to fertility. Plant Cell 10:1163–1180
Jenuth JP, Peterson AC, Fu K, Shoubridge EA (1996) Random genetic drift in the female germline explains the rapid segregation of mammalian mitochondrial DNA. Nat Genet 14:146–151
Jenuth JP, Peterson AC, Shoubridge EA (1997) Tissue-specific selection for different mtDNA genotypes in heteroplasmic mice. Nat Genet 16:93–95
Kajander OA, Rovio AT, Majamaa K, Poulton J, Spelbrink JN, Holt IJ, Karhunen PJ, Jacobs HT (2000) Human mtDNA sublimons resemble rearranged mitochondrial genomes found in pathological states. Hum Mol Genet 22:2821–2835
Kanazawa A, Tsutsumi N, Hirai A (1994) Reversible changes in the composition of the population of mtDNAs during dedifferentiation and regeneration in tobacco. Genetics 138:865–870
Khrapko K, Coller HA, Andre PC, Li XC, Hanekamp JS, Thilly WG (1997) Mitochondrial mutational spectra in human cells and tissues. Proc Natl Acad Sci USA 94:13798–13803
Kitagawa K, Takumi S, Nakamura C (2002) Evidence of paternal transmission of mitochondrial DNA in a nucleus-cytoplasm hybrid of timopheevi wheat. Genes Genet Syst 77:243–250
Kondo R, Satta Y, Matsuura ET, Ishiwa H, Takahata N, Chigusa SI (1990) Incomplete maternal transmission of mitochondrial DNA in Drosophila. Genetics 126:657–663
Kubo T, Nishizawa S, Sugawara A, Itchoda N, Estiati A, Mikami T (2000) The complete nucleotide sequence of the mitochondrial genome of sugar beet (Beta vulgaris L.) reveals a novel gene for tRNA(Cys)(GCA). Nucleic Acids Res 28:2571–2576
Kuzmin EV, Duvick DN, Newton KJ (2005) A mitochondrial mutator system in maize. Plant Physiol 137:779–789
Kvist L, Martens J, Nazarenko AA, Orell M (2003) Paternal leakage of mitochondrial DNA in the great tit (Parus major). Mol Biol Evol 20:243–247
Ladoukakis ED, Zouros E (2001) Direct evidence for homologous recombination in mussel (Mytilus galloprovincialis) mitochondrial DNA. Mol Biol Evol 18:1168–1175
Laser B, Mohr S, Odenbach W, Oettler G, Kuck U (1997) Parental and novel copies of the mitochondrial orf25 gene in the hybrid crop-plant triticale: predominant transcriptional expression of the maternal gene copy. Curr Genet 32:337–347
Lightowlers RN, Chinnery PF, Turnbull DM, Howell N (1997) Mammalian mitochondrial genetics: heredity, heteroplasmy and disease. Trends Genet 13:450–455
Martinez-Zapater JM, Gil P, Capel J, Somerville CR (1992) Mutations at the Arabidopsis CHM locus promote rearrangements of the mitochondrial genome. Plant Cell 4:889–899
Moraes CT, Atencio DP, Oca-Cossio J, Diaz F (2003) Techniques and pitfalls in the detection of pathogenic mitochondrial DNA mutations. J Mol Diagn 5:197–208
Notsu Y, Masood S, Nishikawa T, Kubo N, Akiduki G, Nakazono M, Hirai A, Kadowaki K (2002) The complete sequence of the rice (Oryza sativa L.) mitochondrial genome: frequent DNA sequence acquisition and loss during the evolution of flowering plants. Mol Genet Genomics 268:434–445
Palmer JD, Herbon LA (1987) Unicircular structure of the Brassica hirta mitochondrial genome. Curr Genet 11:565–570
Palmer JD, Adams KL, Cho Y, Parkinson CL, Qiu YL, Song K (2000) Dynamic evolution of plant mitochondrial genomes: mobile genes and introns and highly variable mutation rates. Proc Natl Acad Sci U S A 97:6960–6966
Polyak K, Li Y, Zhu H, Lengauer C, Willson JK, Markowitz SD, Trush MA, Kinzler KW, Vogelstein B (1998) Somatic mutations of the mitochondrial genome in human colorectal tumours. Nat Genet 20:291–293
Sakamoto W, Kondo H, Murata M, Motoyoshi F (1996) Altered mitochondrial gene expression in a maternal distorted leaf mutant of Arabidopsis induced by chloroplast mutator. Plant Cell 8:1377–1390
Schwartz M, Vissing J (2002) Paternal inheritance of mitochondria DNA. N Engl J Med 347:576–580
Sekiguchi K, Kasai K, Levin BC (2003) Inter- and intragenerational transmission of a human mitochondrial DNA heteroplasmy among 13 maternally-related individuals and differences between and within tissues in two family members. Mitochondrion 2:401–414
Shirzadegan M, Palmer JD, Christey M, Earle ED (1991) Patterns of mitochondrial DNA instability in Brassica campestris cultured cells. Plant Mol Biol 16:21–37
Shitara H, Hayashi JI, Takahama S, Kaneda H, Yonekawa H (1998) Maternal inheritance of mouse mtDNA in interspecific hybrids: segregation of the leaked paternal mtDNA followed by the prevention of subsequent paternal leakage. Genetics 148:851–857
Skibinski DOF, Gallagher C, Beynon CM (1994) Sex-limited mitochondrial DNA transmission in the marine mussel Mytilus edulis. Genetics 138:801–809
Small ID, Isaac PG, Leaver CJ (1987) Stoichiometric differences in DNA molecules containing the atpA gene suggest mechanisms for the generation of mitochondrial genome diversity in maize. EMBO J 6:865–869
Steffen SE, Bryant FR (2001) Purification and characterization of the single-stranded DNA binding protein from Streptococcus pneumoniae. Arch Biochem Biophys 388:165–170
Sugiyama Y, Watase Y, Nagase M, Makita N, Yagura S, Hirai A, Sugiura M (2005) The complete nucleotide sequence and multipartite organization of the tobacco mitochondrial genome: comparative analysis of mitochondrial genomes in higher plants. Mol Genet Genomics 272:603–615
Suzuki T, Kawano S, Sakai A, Hirai A, Kuroiwa T (1996) Variability of mitochondrial subgenomic molecules in the meristematic cells of higher plants. Genes Genet Syst 71:329–333
Szibor M, Holtz J (2003) Mitochondrial ageing. Basic Res Cardiol 98:210–218
Taylor DR, Olson MS, McCauley DE (2001) A quantitative genetic analysis of nuclear-cytoplasmic male sterility in structured populations of Silene vulgaris. Genetics 158:833–841
Timmis JN, Ayliffe MA, Huang CY, Martin W (2004) Endosymbiotic gene transfer: organelle genomes forge eukaryotic chromosomes. Nat Rev Genet 5:123–135
Trifunovic A, Wredenberg A, Falkenberg M, Spelbrink JN, Rovio AT, Bruder CE, Bohlooly-Y M, Gidlof S, Oldfors A, Wibom R, Tornell J, Jacobs HT, Larsson NG (2004) Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature 429:417–423
Unseld M, Marienfeld JR, Brandt P, Brennicke A (1997) The mitochondrial genome of Arabidopsis thaliana contains 57 genes in 366,924 nucleotides. Nat Genet 15:57–61
Vitart V, De Paepe R, Mathieu C, Chetrit P, Vedel F (1992) Amplification of substoichiometric recombinant mitochondrial DNA sequences in a nuclear, male sterile mutant regenerated from protoplast culture in Nicotiana sylvestris. Mol Gen Genet 233:193–200
Wallace DC (1994) Mitochondrial DNA sequence variation in human evolution and disease. Proc Natl Acad Sci USA 91:8739–8746
Wallace DC (1999) Mitochondrial diseases in man and mouse. Science 283:1482–1488
Wallace DC, Stugard C, Murdock D, Schurr T, Brown MD (1997) Ancient mtDNA sequences in the human nuclear genome: a potential source of errors in identifying pathogenic mutations. Proc Natl Acad Sci USA 94:14900–14905
Ward BL, Anderson RS, Bendich AJ (1981) The mitochondrial genome is large and variable in a family of plants (cucurbitaceae). Cell 25:793–803
White HE, Durston VJ, Seller A, Fratter C, Harvey JF, Cross NC (2005) Accurate detection and quantitation of heteroplasmic mitochondrial point mutations by pyrosequencing. Genet Test 9:190–199
Wintz H (1994) Analysis of heteroplasmy in cytoplasmic mutant of maize. Plant Physiol Biochem 32:649–653
Woloszynska M, Kieleczawa J, Ornatowska M, Wozniak M, Janska H (2001) The origin and maintenance of the small repeat in the bean mitochondrial genome. Mol Genet Genomics 265:865–872
Wong LJ, Boles RG (2005) Mitochondrial DNA analysis in clinical laboratory diagnostics. Clin Chim Acta 354:1–20
Yamato KT, Newton KJ (1999) Heteroplasmy and homoplasmy for maize mitochondrial mutants: a rare homoplasmic nad4 deletion mutant plant. J Hered 90:369–373
Zeviani M, Antozzi C (1997) Mitochondrial disorders. Mol Hum Reprod 3:133–148
Zouros E, Oberhauser Ball A, Saavedra C, Freeman KR (1994) An unusual type of mitochondrial DNA inheritance in the blue mussel Mytilus. Proc Natl Acad Sci USA 91:7463–7467
Acknowledgments
This work was supported by the State Committee for Scientific Research (Grant No. 3 P06A 007 25).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by R. Bock
Rights and permissions
About this article
Cite this article
Kmiec, B., Woloszynska, M. & Janska, H. Heteroplasmy as a common state of mitochondrial genetic information in plants and animals. Curr Genet 50, 149–159 (2006). https://doi.org/10.1007/s00294-006-0082-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00294-006-0082-1