Abstract
Aspergillus fumigatus is the most important airborne fungal pathogen causing life-threatening infections in immunosuppressed patients. One of the important questions concerning A. fumigatus is the identification of pathogenicity determinants. To obtain a comprehensive overview about the proteins produced at different physiological conditions that are related to the infectious process a proteomic approach has been applied. Here, 2-D gel electrophoresis for filamentous fungi was optimised concerning removal of interfering compounds, protein extraction and separation methods. A trichloroacetic acid-based precipitation method of proteins with their subsequent solubilisation by the use of a combination of CHAPS with a second sulfobetaine detergent gave the best results. The optimised protocol was evaluated by the analysis of the proteomes of A. fumigatus grown on two different carbon sources, i.e., glucose and ethanol. Carbon catabolite repression has not been studied in detail at the protein level in A. fumigatus yet. In addition, growth on ethanol leads to activation of the glyoxylate cycle which was shown to be essential for pathogenesis in bacteria and fungi. In A. fumigatus, differential patterns of enzymes of the gluconeogenesis, glyoxylate cycle and ethanol degradation pathway during growth on glucose and ethanol were observed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In the last decades, A. fumigatus has become the most important airborne fungal pathogen of humans. Diseases caused by A. fumigatus can be divided into three categories: allergic reactions and colonisation with restricted invasiveness are observed in immunocompetent individuals while systemic infections with high mortality rates occur in immunocompromised patients (Ellis 1999; Brakhage and Langfelder 2002; Brakhage 2005; Brakhage and Liebmann 2005). Due to the improvement in transplant medicine and the therapy of haematological malignancies the number of invasive aspergillosis has increased. Specific diagnostics are still limited, as are the possibilities of therapeutic intervention, leading to a high mortality rate of 30–98% for invasive aspergillosis [reviewed in Latgé (1999)]. The infectious agent of A. fumigatus is conidia, which are inhaled during routine daily activities (Braude 1986).
The genome of the A. fumigatus isolate Af293 has been fully sequenced (http://www.tigr.org) and the annotation has been almost completed. The genome consists of eight chromosomes with a total size of 29.2 Mb, of which 9,922 protein-encoding sequences were identified (Pain et al. 2004; Niermann et al. 2005). With the genome data available the regulation of genes and proteins of A. fumigatus on a global scale can be analysed, also under conditions that are related to infection. 2-D gel electrophoresis introduced by O’Farrel (1975) is still the most established method for studying properties of proteins in parallel (proteomics). Not many proteomic data are available for filamentous fungi, and no extensive study from any Aspergillus proteome has been published yet (Bruneau et al. 2001; Nandakumar and Marten 2002; Nandakumar et al. 2003). Preparation of protein samples for 2-D gel electrophoresis is a critical step and essential for reproducible results. This is also influenced by the presence of a cell wall which makes up the majority of the cell mass in filamentous fungi. The cell wall is exceptionally robust (Latgé et al. 2005; Shimizu and Wariishi 2005). Thus, cell lysis is an important step in sample preparation for 2-D gel electrophoresis. An often used method to remove contaminants which can interfere with the 2-D gel electrophoresis separation is the classical trichloroacetic acid (TCA)/acetone precipitation. This method has often been applied to the preparation of extracts from plants (Damerval et al. 1986; Carpentier et al. 2005) and fungi (Wildgruber et al. 2002; Nandakumar et al. 2003). However, until now this method has not been compared with methods employing commercial kits for protein precipitation. Another critical step during 2-D gel electrophoresis is the solubility of proteins during isoelectric focusing. Many proteins tend to aggregate or precipitate at their isoelectric point. Increased solubility was obtained by the use of thiourea (Rabilloud 1998) and new zwitterionic detergents (Rabilloud 1996; Rabilloud et al. 1997; Molloy et al. 1999; Luche et al. 2003).
Here, a comparison of several protein precipitation methods and lysis buffers helped to optimise a 2-D gel electrophoresis protocol for the human pathogenic fungus A. fumigatus. Furthermore, the optimised protocol was applied to analyse the proteomes when A. fumigatus was grown with glucose or ethanol as the carbon source. In saprophytic filamentous fungi, the expression of catabolic pathways involved in the utilisation of carbon compounds strongly depends on available nutrients in the environment. Energetically favourable carbon sources are used preferentially to less readily metabolisable carbon sources. The synthesis of the enzymes for the utilisation of the latter is reduced and the corresponding genes are repressed. This mechanism known as carbon catabolite repression has been intensively studied in the filamentous fungus A. nidulans (see review Flipphi and Felenbok 2004; Kelly 2004). Until now, little is known about carbon catabolite repression and ethanol metabolism in A. fumigatus. Homologous genes coding for the transcriptional regulators involved in A. nidulans, such as CreA, FacB and AlcR are present in the genome of A. fumigatus, showing 78, 65 and 43% amino acid identity to the proteins in A. nidulans, respectively. Therefore, it is likely that similar regulation patterns of carbon catabolite repression exist in both fungi A. nidulans and A. fumigatus. Moreover, previously the glyoxylate cycle was shown to be essential for pathogenesis of bacteria and fungi, i.e., Mycobacterium tuberculosis (McKinney et al. 2000) and Candida albicans (Lorenz and Fink 2001 ), respectively. Therefore, it is of considerable interest to analyse the proteome of A. fumigatus grown under glyoxylate cycle-activating conditions, e.g., when the fungus was grown with ethanol as the sole carbon source.
Here, a 2-D gel electrophoresis protocol for A. fumigatus was established that resolved more than 1,000 proteins on a large format gel with a broad pH range. By comparison of the protein pattern of A. fumigatus grown on glucose versus ethanol the method was verified and a first proteome map was obtained.
Material and methods
Strains and culture conditions
The Aspergillus fumigatus wild-type strain ATCC 46645 was used for this study. A. fumigatus was cultivated at 37°C in Aspergillus minimal medium (AMM) as previously described (Weidner et al. 1998). The medium contained 50 mM glucose or 50 mM ethanol as the sole carbon source. One hundred millilitres of medium was inoculated to give a spore concentration of 3×106 conidia/ml. The glucose-grown cultures were incubated on a rotary shaker for 15 h with 200 rpm, the ethanol-grown cultures for 30 h. The glucose concentration of the medium was monitored enzymatically using glucose oxidase, peroxidase and 2,2′-azino-bis(3-ethylbenzthiazoline-6-sulfonic acid) (ABTS) as described in Brock and Buckel (2004). The ethanol concentration was determined using ethanol dehydrogenase (Beuther 1988) at the end of cultivation. After harvesting, the mycelium was rinsed with tap water, pressed to remove any liquid and the dried mycelium was frozen in liquid nitrogen. Protein extracts were prepared as described below.
Sample preparation
Extraction protocol
Frozen mycelium was ground in a precooled mortar with a pestle in the presence of liquid nitrogen. About 50 mg of homogenate were directly dissolved in lysis buffer (described below). Alternatively, the homogenate was purified by TCA precipitation or by the application of the “Clean Up Kit” (GE Healthcare Bio-Sciences, Freiburg, Germany) with or without prior extraction of the proteins with an SDS lysis buffer.
TCA/acetone precipitation
TCA/acetone precipitation was carried out as described by Damerval et al. (1986) with some modifications. Briefly, 50 mg of homogenate were precipitated over night with 500 μl acetone/13.3% (w/v) TCA/0.093% (v/v) 2-mercaptoethanol. After centrifugation for 15 min at 20,000g at 4°C the supernatant was removed and the pellet was rinsed twice in ice-cold acetone containing 0.07% (v/v) 2-mercaptoethanol. The suspension was centrifuged again and the pellet was air-dried for 5 min at room temperature, resuspended in 300 μl lysis buffer [7 M urea, 2 M thiourea, 4% (w/v) CHAPS, 0.8% (v/v) ampholytes (Serva), 20 mM Tris and 20 mM DTT] and incubated for 1 h at room temperature. To improve the solubilisation of the proteins the samples were sonicated for 10 min in an ultrasonic bath. After centrifugation at 20,000g for 30 min at 16°C, the supernatant was collected.
“Clean Up Kit”
Fifty milligrams of homogenate were resuspended in 300 μl solubilising solution consisting of 2% (w/v) SDS, 40 mM Tris base and 60 mM DTT. The extract was heated at 95°C for 5 min and insoluble debris was removed by centrifugation for 10 min at 20,000g. The supernatant was transferred to another microcentrifuge tube and three volumes of precipitant of the “Clean Up Kit” were added. The following steps were performed according to the protocol B of the manufacturer’s manual (GE Healthcare Bio-Sciences). The air-dried pellet was resuspended in 300 μl lysis buffer [7 M urea, 2 M thiourea, 4% (w/v) CHAPS, 0.8% (v/v) ampholytes (Serva), 20 mM Tris and 20 mM DTT].
In an alternative approach, 50 mg of homogenate was directly resuspended in 300 μl precipitant without prior SDS solubilisation. The subsequent steps were performed as described above.
Evaluation of different lysis buffers
Five different lysis buffers for protein extraction were compared. The ground mycelium was purified by TCA precipitation with the “Clean Up Kit” (GE Healthcare Bio-Sciences). Fifty milligrams of homogenate were directly resuspended in 300 μl precipitant solution of the “Clean Up Kit”. After washing with acetone the dried pellet was suspended in 300 μl of the following lysis buffers: buffer I [9 M urea, 4% (w/v) CHAPS, 0.8% (v/v) ampholytes, 20 mM DTT, 20 mM Tris], buffer II [7 M urea, 2 M thiourea, 4% (w/v) CHAPS, 0.8% (v/v) ampholytes, 20 mM DTT, 20 mM Tris], buffer III [7 M urea, 2 M thiourea, 2% (w/v) CHAPS, 1% (w/v) Zwittergent 3–10, 0.8% (v/v) ampholytes, 20 mM DTT, 20 mM Tris], buffer IV [7 M urea, 2 M thiourea, 1% (w/v) ASB 14, 0.8% (v/v) ampholytes, 20 mM DTT, 20 mM Tris] and buffer V (equal mixture of buffer III and buffer IV).
Protein quantification
The protein concentration was determined according to the method of Bradford (1976) using BIO-RAD protein assay (BIO-RAD Lab., Hartfordshire, USA).
2-D gel electrophoresis
The protein sample was applied via anodic cup loading to 2-D gel electrophoresis. The amount of 50 μg protein was loaded for comparison of precipitation methods, the amount of 250 μg protein for comparison of the different lysis buffers, whereas 300 μg protein was applied for comparison of ethanol and glucose-grown mycelia. The strips of 7 and 24 cm covering a non-linear pH range of pH 3–11 or 7–11 (GE Healthcare Bio-Sciences) were rehydrated over night in 150 or 450 μl rehydration buffer, respectively. The rehydration buffer contained the following: 7 M urea, 2 M thiourea, 2% (w/v) CHAPS and/or 1% of another zwitterionic detergent in accordance with the lysis buffer, 0.002% (w/v) bromophenol blue, 0.5% (v/v) IPG buffer, 1.2% (v/v) De-Streak Reagent (Olsson et al. 2002) (GE Healthcare Bio-Sciences). Isoelectric focusing was carried out using the IPGphor II with a manifold ceramic tray (GE Healthcare Bio-Sciences) at 20°C with a current of 50 μA/strip. For 7 cm strips the following protocol was applied: 5 h at 300 V (gradient), 4 h at 600 V (gradient), 3 h at 1,000 V, 2 h at 5,000 V and 10,000 V h at 5,000 V. Isoelectric focusing of 24 cm strips was carried out according to the following protocol: 3 h at 300 V (gradient), 3 h at 600 V (gradient), 4 h at 1,000 V (gradient), 3 h at 8,000 V (gradient) and 24,000 V h at 8,000 V (step). The amount of 500 μg protein was loaded for the separation of basic protein (pH 7–11). The focusing time was extended to 4 h at 600 V and the last step at 8,000 V to 32,000 V h. Prior to the start of separation by the second dimension the strips were equilibrated for 15 min in 10 ml equilibration buffer containing 1% (w/v) DTT and subsequently for 15 min in 10 ml equilibration buffer containing 2.5% (w/v) iodoacetamide. The separation in the second dimension was realised on a Minigel-Twin System for the 7 cm strips (Biometra, Göttingen, Germany) or on the Ettan DALT System (GE Healthcare Bio-Sciences) with lab cast 1.0 mm SDS polyacrylamide gels [12.5% (w/v)] containing Rhinohide as a gel strengthener (Molecular Probes, Leiden, NL): 20 mA/gel for minigels and 30 min 30 W, 4 h 100 W for six Ettan Dalt gels. Protein standard Mark 12 was purchased from Invitrogen (Karlsruhe, Germany).
Protein visualisation, image analysis and quantification
Proteins were visualised by colloidal CBB staining (Neuhoff et al. 1988). The gels were fixed for 2 h in a fixing solution [40% (v/v) methanol, 7% (v/v) glacial acetic acid], washed twice with ultrafiltrated water for 20 min and stained over night in a staining solution [10% (v/v) methanol, 0.1% (w/v) CBB G-250, 1.6% (v/v) o-phosphoric acid, and 10% (w/v) ammonium sulfate]. Gels were neutralised (0.1 M Tris-base titrated to pH 6.5 with o-phosphoric acid) for 5 min and washed in 25% (v/v) methanol for 1 h. Stained gels were scanned and calibrated with Labscan software 5 (GE Healthcare Bio-Sciences). Images were analysed using Image Master Platinum (Melanie 5). The spots were quantified using the % volume criterion and the ratio and gap parameter. Only spots with a ratio value greater than two were regarded as significant. Three replicates of three gels for each growth condition were obtained. For matching the detected spots a synthetic master gel was created by matching eight representative gels with each other by keeping only triplicate spots.
MS identification
Protein spots were excised manually. Tryptic digest of proteins, MALDI-TOF and MALDI TOF/TOF were carried out at TOPLAB, Martinsried, Germany. Alternatively, protein spots were digested according to the protocol of Bruker (Bruker Daltonics, Bremen, Germany) that was adapted from Shevchenko et al. (1996). MALDI TOF/TOF spectra were generated with an Ultraflex I device (Bruker Daltonics). Peptide mass fingerprint and peptide fragmentation spectra of each sample were submitted for identification using MASCOT interface (MASCOT 2.1.0, Matrix Science, London, UK) or ProFound (Genomic Solutions, USA). Hits were considered significant according to the MASCOT score (P=0.05).
Results
Optimisation of protein cleanup methods
Filamentous fungi possess a robust and rigid cell wall (Latgé et al. 2005). Therefore, cell lysis is a critical step in preparation of protein extracts. As shown in Fig. 1, direct lysis of ground mycelia resulted in irreproducible gels with streaks, strong background and faint spots. In total, 516 spots were detected. TCA/acetone precipitation of proteins is a well-established technique for the purification of protein extracts, helping in the elimination of carbohydrates. The TCA/acetone precipitation from A. fumigatus crude extracts resulted in 690 detectable spots on a mini-gel (Fig. 1). Compared with the protein samples that had been directly dissolved, the gels loaded with TCA/acetone precipitated protein showed clear spots with less background. A few spots displayed vertical streaking. The “Clean Up Kit” from GE Healthcare is a modified TCA precipitation method for quantitative precipitation of proteins while excluding interfering substances. It was specially designed for the preparation of samples for 2-D gel electrophoresis. Direct treatment of the sample with the “2-D Clean Up Kit” resulted in gels of similar quality compared to the TCA/acetone precipitated samples. Fewer spots were detected (573), which, however, showed less vertical streaking. Compared to the TCA/acetone precipitation method, the protein yield was more than two times higher (5.6 mg/ml protein versus 2.6 mg/ml protein). Extraction of proteins with an SDS buffer containing DTT and Tris at 95°C prior to the application of the “Clean Up Kit” did not produce superior results. Although a definite protein pellet was produced, only fewer spots (532) were visible and the gel pictures looked more diffuse (Fig. 1).
Extraction of mycelia of A. fumigatus grown in liquid cultures with glucose as the carbon source. a Direct resuspension of mycelia in lysis buffer; b purification of protein samples with “Clean Up Kit”; c SDS lysis of mycelia and subsequent purification with “Clean Up Kit”; d purification of protein samples by TCA/acetone precipitation
Evaluation of several lysis buffers
Two-dimensional gel electrophoresis is hampered by the low solubility of proteins in the first dimension, especially when they migrate close to their isoelectric points. This leads to protein streaking and protein loss. In particular, hydrophobic proteins are difficult to study by 2-D gel electrophoresis. These problems were in part overcome by the use of new zwitterionic detergents and of thiourea which led to an increase of resolution and capacity of 2-D gels.
Here, five different lysis buffers for the solubilisation of proteins from the filamentous fungus A. fumigatus were tested. Buffer 1 was composed of urea and the zwitterionic detergent CHAPS, ampholytes, Tris buffer and DTT. Thiourea was additionally added to buffer 2 while buffer 3 contained the zwitterionic sulfobetaine Zwittergent 3–10 and CHAPS. The zwitterionic detergent ASB 14 was added to buffer 4. Finally, buffer 5 consisted of an equal mixture (v/v) of buffer 4 and 5. The use of buffers 1 and 4 resulted in gels with the lowest number of detectable spots, i.e., 756±81 spots and 714±88 spots, respectively (Fig. 2). Buffer 1 led to the production of gels with horizontal streaking, whereas gels loaded with protein samples obtained by the use of buffer 4 showed clear spots (Fig. 2). However, when lysis buffer 4 was applied, it was difficult to reproduce the quality of gels. The application of buffers 2, 3 and 5 resulted in gels of almost the same quality and with an approximate number of spots of 828±13 for buffer 2, 861±51 for buffer 3 and 827±64 for buffer 5. With buffer 5, more horizontal streaking was observed. Compared with lysis buffer 2, additional spots were detected when buffers 3 or 5 were used (see Fig. 2).
Test of different lysis buffers for the solubilisation of A. fumigatus proteins. a Lysis buffer 1 (9 M urea, 4% CHAPS, 0.8% ampholytes, 20 mM DTT, 20 mM Tris); b lysis buffer 2 (7 M urea, 2 M thiourea, 4% CHAPS, 0.8% ampholytes, 20 mM DTT, 20 mM Tris); c lysis buffer 3 (7 M urea, 2 M thiourea, 2% CHAPS, 1% Zwittergent 3–10, 0.8% ampholytes, 20 mM DTT, 20 mM Tris); d lysis buffer 4 (7 M urea, 2 M thiourea, 1% ASB 14, 0.8% ampholytes, 20 mM DTT, 20 mM Tris); e lysis buffer 5 (equal mixture of buffer III and buffer IV)
Proteomes of A. fumigatus grown on glucose or ethanol
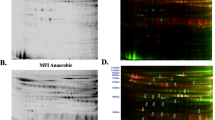
The combination of protein precipitation with “Clean Up kit” and subsequent solubilisation with lysis buffer 3 containing CHAPS and Zwittergent 3–10 showed the best results. To evaluate the optimised method of proteome analysis, the global response of A. fumigatus at the proteomic level by cultivating the fungus on 50 mM ethanol or 50 mM glucose was analysed. The experiments were carried out in triplicate and 18 gels were analysed. After spot detection, the gels were matched to each other with the help of the landmark function of the Image Master Platinum software (see Materials and Methods). When 300 μg protein was loaded, on average 1,235±159 spots per gel were detected (Fig. 3a). 980 spots were detected which matched with a spot in the reference gel and were assigned to a group number (see below). For the analysis of changes in the protein pattern, mean quantitative values and SDs were calculated. Spots representing the same protein on different gels were defined as groups. Only spot groups with a ratio higher than 2 were considered as significant change. The ratio value comprises the lower limit of one class interval and the upper limit of the other class interval. In Table 1, the fold changes of the raw central tendency and the gap value [maximum difference between the range of the current class (either glucose or ethanol) and the range of one of the other classes (either glucose or ethanol)] are displayed. Negative values indicate overlapping, positive values non-overlapping class ranges.
a 2-D gel electrophoresis of proteins extracted from A. fumigatus grown in AMM with glucose (A) or ethanol (B) as the sole carbon source. The orientation of the IEF gel is indicated. Mr standards are indicated on the right. The numbers refer to proteins induced during growth on glucose or ethanol (see Table 1). Black arrow upregulation of the protein, white arrow downregulation of the protein. b shows a section of gel A and B of Fig. 3a indicated by the black frame
Thirty-seven spots were identified by MALDI-TOF/TOF showing consistent differences in intensity between growth on glucose and ethanol (Table 1 and Fig. 3b). A sequence coverage between 21 and 87% was obtained (Table 1) during MS analysis. Some proteins appeared in gels as more than one spot with the same apparent molecular mass, but with different pI values and abundance, presumably due to post-translational modifications or isoenzyme variation. Proteins for which several variants were identified, included aconitate hydratase, isocitrate lyase, alcohol dehydrogenase (ADH), aldehyde dehydrogenase, acetyl-CoA synthetase and phosphoenolpyruvate carboxykinase. Thirty-seven spots were sorted into six different categories (Table 1). Among the enzymes that are involved in glycolysis, pyruvate dehydrogenase changed most significantly in quantity. During growth on glucose it showed a 3.1-fold increase. During growth on ethanol some enzymes of the citrate cycle were upregulated. Aconitate hydratase, which is involved in the tricarboxylic acid cycle, revealed a 2.8-fold increase during growth on ethanol. Two different malate dehydrogenases (MDHs) exhibited a higher abundance during growth on ethanol, i.e., MDH with the locus tag Afu6g05210 increased 3.1-fold, whereas the other MDH (locus tag Afu7g05740) did not increase significantly (1.7-fold). Both enzymes possess a conserved domain characteristic of glycosomal-mitochondrial MDH, but only Afu7g05740 exhibited an N-terminal signal peptide for import into the mitochondria. The upregulated MDH is therefore presumably localised in the glyoxysome or the cytosol. Enzymes of the glyoxylate cycle and the ethanol utilisation pathway showed the most significant change, e.g., isocitrate lyase increased 415-fold. Three different spots with different isoelectric points represented this key enzyme of the glyoxylate shunt (Fig. 3b). Spot No. 5 exhibited the biggest spot with a proportion of 86%. Two additional, but minor spots, which were identified as isocitrate lyase, were only detected in gels from ethanol-grown cultures (spot 6 and 7). The abundance of ADH (103- and 17-fold), aldehyde dehydrogenase (352-, 12- and 15-fold) and acetyl-CoA synthetase (33- and 8-fold) was strongly increased during growth on ethanol (Table 1). In all cases, spots of the same molecular mass but with different isoelectric points were detected. Alcohol and aldehyde dehydrogenase together represented a total of 3.8 vol% of all spots.
When A. fumigatus was grown with ethanol as the sole carbon source, the amount of the key enzyme of gluconeogenesis, phosphoenolpyruvate carboxykinase, increased 67-fold, a variant of this enzyme with a different pI, 29-fold. Under the same conditions, malic enzyme increased 10.5-fold and fructose-1,6-bisphosphatase 5-fold. The second key enzyme of the glyoxylate cycle, malate synthase, was only detected in gels covering the pH range from 7 to 11. No significant amounts of malate synthase were detectable in glucose-grown mycelium (Fig. 4).
Section of a 2-D gel electrophoresis gel separating basic polypeptides (pI 7–11) from A. fumigatus grown with glucose (A) or ethanol (B) as the sole carbon source. The numbers refer to proteins induced during growth on glucose or ethanol (see Table 1). Black arrow upregulation of the protein, white arrow downregulation of the protein. Spot number 8 and 9 represented the malate synthase, which could not be detected when a broad pH range from 3 to 11 was applied for separation
During growth on ethanol the abundance of two aspartate aminotransferases increased. The aspartate aminotransferase with the locus tag Afu4g10410 represented presumably the mitochondrial form with a 29 amino acid long signal peptide (6.2-fold increase) and the enzyme with locus tag Afu2g09650 the cytosolic form, because it did not show an obvious signal peptide. The protein Afu2g09650 was only detected in gels run with protein from ethanol-grown mycelia. A putative carnitine acetyl transferase showed an increased level during growth on ethanol of 7-fold. This carnitine acetyl transferase with the locus tag Afu2g12530 contains a signal peptide of 41 amino acids with a 99.4% probability for export to mitochondria calculated by Mitoprot (Claros and Vincens 1996). Additionally, several other proteins with different physiological functions changed their abundance significantly during growth of A. fumigatus on glucose and ethanol (Table 1; Fig. 3a, b). A flavohemoprotein (4.1-fold), an acetyltransferase and several other proteins showed a higher abundance during growth on glucose. Also, enzymes of the primary metabolism were identified which did not show a significant change. These enzymes are listed in Table 2.
Discussion
Here, we optimised a 2-D gel electrophoresis protocol for A. fumigatus which is based on the purification of protein samples with “Clean Up Kit” in combination with a lysis buffer containing CHAPS and Zwittergent 3–10. Lysis buffer 3 was further evaluated in experiments resulting in the first proteome maps of A. fumigatus grown on different carbon sources, i.e., on a glycolytic (glucose) and a gluconeogenic substrate (ethanol). Ethanol-grown mycelia of A. fumigatus showed a strong up-regulation of enzymes required for the glyoxylate cycle, gluconeogenesis and ethanol oxidation (see Table 1, Fig. 3b). Isocitrate lyase was the most up-regulated enzyme with a 415-fold increase. Post-translational modification of isocitrate lyase or very similar isoforms may be the reason for the occurrence of three different spots with different isoelectric points, but only a single isocitrate lyase sequence is present in the genome of A. fumigatus (De Lucas et al. 1994). The second enzyme of the glyoxylate cycle, malate synthase, was only detected in gels with a pH range from 7 to 11. The application of such a narrow pH range improved the spatial resolution of proteins, resulting in the finding that spot No. 52 in a gel covering a pH range from 3 to 11 represented two spots, namely spot 52 and 8 (Fig. 4).
All key enzymes of gluconeogenesis, especially PEP carboxykinase and fructose-1,6-bisphosphatase, were upregulated during growth of A. fumigatus on ethanol. In addition, malic enzyme showed a higher abundance during growth on ethanol. Consistently, in A. nidulans malic enzyme which converts malate to pyruvate was shown to have higher activity during growth on ethanol (Kelly and Hynes 1981). Besides the PEP carboxykinase, this enzyme is essential for growth on C2 compounds (Hondmann and Visser 1994).
As expected, the ethanol utilisation enzymes were strongly upregulated in A. fumigatus during growth on ethanol. In A. nidulans, they are under the control of the AlcR protein. Moreover, in A. nidulans growth on ethanol resulted in high levels of both alcohol and aldehyde dehydrogenase (up to 1% of total protein) (Felenbok et al. 2001). As shown here, similarly, in A. fumigatus both enzymes represented a total of 3.8 vol% of all spots. Interestingly, the genome of A. fumigatus contains more than 30 putative ADHs, but only the ADH with the locus tag Afu7g010110 represents the ethanol-oxidising enzyme. This ADH also shows high similarity to ADH I from A. nidulans, the enzyme required for the utilisation of ethanol as sole carbon source (Gwynne et al. 1987). Furthermore, carnitine acetyltransferase was found to be inducible by acetate in A. nidulans (Stemple et al. 1998) and, in S. cerevisiae, by ethanol (Claus et al. 1982). This protein is required for the shuttle of acetyl groups between intracellular compartments. The detected carnitine acetyl transferase Afu2g12530 presumably represents the protein localised in both mitochondria and peroxisomes. The second annotated, but not detected carnitine acetyl transferase Afu1g12340 possesses no obvious signal peptide and shows a high degree of similarity to the cytosolic carnitine acetyl transferase FacC of A. nidulans (Stemple et al. 1998). Furthermore, aconitate hydratase and MDH revealed an increase in quantity during growth on ethanol. This can be explained by the necessary replenishment of the glyoxylate cycle by the activity of citrate synthase and aconitase as shown for A. nidulans (Sims et al. 2004). Furthermore, two MDHs were detected. The up-regulated MDH identified here is presumably located either in glyoxysomes or the cytoplasm. In contrast to S. cerevisiae (Steffan and Mc Alister-Henn 1992), there is no evidence for three MDH isoenzymes (mitochondrial, glyoxysomal, cytosolic) in A. fumigatus. Although the genome of A. fumigatus encodes two additionally annotated MDHs with the locus tags Afu2g13800 and Afu7g02420, these ORFs were presumably misinterpreted as MDHs because convincing similarity to MDHs is missing.
Gluconeogenesis requires metabolite transport between mitochondria, the glyoxysomes and the cytosol. During growth on C2 compounds, oxaloacetate has to be transported from the mitochondria to the glyoxysomes via the MDH route or, alternatively, aspartate aminotransferase route (Bakker et al. 2001). The latter would explain the higher abundance of aspartate transaminase during growth on ethanol.
During growth on glucose less proteins showed an increase in abundance. Not surprisingly, enzymes of the glycolysis, such as pyruvate dehydrogenase were upregulated. Presumably due to the higher growth rate on glucose, transaldolase and transketolase of the pentose phosphate cycle (Hondmann and Visser 1994), phosphoglucomutase involved in cell wall biosynthesis and trehalose production (Hoffmann et al. 2000), and the glycerol-synthesising glycerol dehydrogenase were upregulated (Schuurink et al. 1990).
In summary, most of the key enzymes for gluconeogenic growth were detected in ethanol-grown mycelia. The obtained results helped to improve functional assignments to the annotated ORFs of the A. fumigatus genome. Future experiments will reveal the role of the key enzymes for the metabolism of C2 carbon compounds during infection. It is interesting to note that in C. albicans, the genes encoding isocitrate lyase, carnitine acetyl transferase and other genes involved in β-oxidation were induced after phagocytosis by macrophages. The encoded proteins were shown to play an important role in microbial pathogenesis (Lorenz and Fink 2001; Prigneau et al. 2003; Prigneau et al. 2004).
References
Bakker BM, Overkamp KM, van Maris AJA, Kötter P, Luttik MAH, van Dijken JP, Pronk JT (2001) Stoichiometry and compartmentation of NADH metabolism in Saccharomyces cerevisiae. FEMS Microbiol Rev 25:15–37
Beuther H-O (1988) Ethanol. In: Bergmeyer HU (eds) Methods of enzymatic analysis, vol VI. VCH, Cambridge, pp 598–606
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 7:248–254
Brakhage AA (2005) Systemic fungal infections caused by Aspergillus species: epidemiology, infection process and virulence determinants. Curr Drug Targets 6:875–886
Brakhage AA, Langfelder K (2002) Menacing mold: the molecular biology of Aspergillus fumigatus. Annu Rev Microbiol 56:433–455
Brakhage AA, Liebmann B (2005) Aspergillus fumigatus conidial pigment and cAMP signal transduction: significance for virulence. Med Mycol 43:S75-S82
Braude AI (1986) The aspergilli. In: Braude AI, Davis CE, Fierer J (eds) Infectious diseases and medical microbiology. W.B. Saunders, Philadelphia, pp 592–597
Brock M, Buckel W (2004) On the mechanism of action of the antifungal agent propionate. Eur J Biochem 271:3227–3241
Bruneau J-M, Magnin T, Tagat E, Legrand R, Bernard M, Diaquin M, Fudali C, Latgé J-P (2001) Proteome analysis of Aspergillus fumigatus identifies glycosylphosphotidylinositol-anchored proteins associated to the cell wall biosynthesis. Electrophoresis 22:2812–2823
Carpentier SC, Witters E, Laukens K, Deckers P, Swennen R, Panis B (2005) Preparation of protein extracts from recalcitrant plant tissues: an evaluation of different methods for two-dimensional gel electrophoresis analysis. Proteomics 5:2497–2507
Claus R, Käppeli O, Fiechter A (1982) Influence of different physiological states on carnitine-acetyl-transferase activity in Saccharomyces cerevisiae. Experientia 38:1377
Claros MG, Vincens P (1996) Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur J Biochem 241:779–786
Damerval C, Devienne D, Zivy M, Thiellement H (1986) Technical improvements in two-dimensional electrophoresis increase the level of genetic variation detected in wheat-seedling proteins. Electrophoresis 7:52–54
De Lucas JR, Valenciano S, Laborda F, Turner G (1994) Glucose-induced inactivation of isocitrate lyase in Aspergillus nidulans. Arch Microbiol 162:409–413
Ellis M (1999) Therapy of Aspergillus fumigatus-related diseases. In: Brakhage AA, Jahn B, Schmidt A (eds) Aspergillus fumigatus: biology, clinical aspects and molecular approaches to pathogenicity. Contributions to microbiology, vol 2. Karger, Basel, pp 105–129
Felenbok B, Flipphi M, Nikolaev I (2001) Ethanol catabolism in Aspergillus nidulans: a model system for studying gene regulation. Prog Nucleic Acid Res Mol Biol 69:149–204
Flipphi M, Felenbok B (2004) The onset of carbon catabolic repression and interplay between specific induction and carbon catabolite repression in Aspergillus nidulans. In: Brambl R, Marzluf GA (eds) The mycota III—biochemistry and molecular biology, Springer, Berlin Heidelberg New York, pp 403–420
Gwynne DI, Buxton FP, Sibley S, Davies RW, Lockington RA, Scazzocchio C, Sealy-Lewis HM (1987) Comparison of the cis-acting control regions of two coordinately controlled genes involved in ethanol utilization in Aspergillus nidulans. Gene 51:205–216
Hoffmann B, LaPaglia SK, Kübler R, Andermann M, Eckert SE, Braus GH (2000) Developmental and metabolic regulation of the phosphoglucomutase-encoding gene, pgmB, of Aspergillus nidulans. Mol Gen Genet 262:1001–1011
Hondmann DHA, Visser J (1994) Carbon metabolism. Prog Ind Microbiol 29:61–139
Kelly JM (2004) The regulation of carbon metabolism in filamentous fungi. In: Brambl R, Marzluf GA (eds) The mycota III—biochemistry and molecular biology. Springer, Berlin Heidelberg New York, pp 386–401
Kelly JM, Hynes MJ (1981) The regulation of phosphoenolpyruvate carboxykinase and the NADP-linked malic enzyme in Aspergillus nidulans. J Gen Microbiol 123:371–375
Latgé J-P (1999) Aspergillus fumigatus and aspergillosis. Clin Microbiol Rev 12:310–350
Latgé J-P, Mouyna F, Tekaia F, Beauvais A, Debeaupuis JP, Nierman W (2005) Specific molecular features in the organization and biosynthesis of the cell wall of Aspergillus fumigatus. Med Mycol 43:S15–S22
Lorenz MC, Fink GR (2001) The glyoxylate cycle is required for fungal virulence. Nature 412:83–86
Luche S, Santoni V, Rabilloud T (2003) Evaluation of nonionic and zwitterionic detergents as membrane protein solubilizers in two-dimensional electrophoresis. Proteomics 3:249–253
Mc Kinney JD, Honer zu Bentrup K, Munoz-Elias EJ, Miczak A, Chen B, Chan WT, Swenson D, Sacchettini JC, Jacobs Jr WR, Russell DG (2000) Persistence of Mycobacterium tuberculosis in macrophages and mice requires the glyoxylate shunt enzyme isocitrate lyase. Nature 406:735–738
Molloy MP, Herbert BR, Williams KL, Gooley AA (1999) Extraction of Escherichia coli proteins with organic solvents prior to two-dimensional electrophoresis. Electrophoresis 20:701–704
Nandakumar MP, Marten MR (2002) Comparison of lysis methods and preparation protocols for one- and two-dimensional electrophoresis of Aspergillus oryzae intracellular proteins. Electrophoresis 23:2216–2222
Nandakumar MP, Shen J, Raman B, Marten MR (2003) Solubilization of TCA precipitated microbial proteins via NaOH for two-dimensional gel electrophoresis. J Proteome Res 2:89–93
Neuhoff V, Arold N, Taube D, Ehrhardt W (1988) Improved staining of proteins in polyacrylamide gels including isoelectric focusing gels with clear background at nanogram sensitivity using Coomassie Brilliant Blue G-250 and R-250. Electrophoresis 9:255–262
Niermann WC, May G, Kim HS, Anderson MJ, Chen D, Denning DW (2005) What the Aspergillus genomes have told us. Med Mycol 43:S3–S5
O’Farrel PH (1975) High resolution two-dimensional electrophoresis of proteins. J Biol Chem 250:4007–4021
Olsson I, Larsson K, Palmgren R, Bjellqvist B (2002) Organic disulfides as a means to generate streak-free two-dimensional maps with narrow range basic immobilized pH gradient strips as first dimension. Proteomics 2:1630–1632
Pain A, Woodward J, Quail MA et al (2004) Insight into the genome of Aspergillus fumigatus: analysis of a 922 kb region encompassing the nitrate assimilation gene cluster. Fung Genet Biol 41:443–453
Prigneau O, Porta A, Poudrier JA, Colonna-Romano S, Noel T, Maresca B (2003) Genes involved in β-oxidation, energy metabolism and glyoxylate cycle are induced by Candida albicans during macrophage infection. Yeast 20:723–730
Prigneau O, Porta A, Maresca B (2004) Candida albicans CTN gene family is induced during macrophage infection: homology, disruption and phenotypic analysis of CTN3 gene. Fung Genet Biol 41:783–793
Rabilloud T (1996) Solubilization of proteins for electrophoretic analyses. Electrophoresis 17:813–829
Rabilloud T (1998) Use of thiourea to increase the solubility of membrane proteins in two-dimensional electrophoresis. Electrophoresis 19:758–760
Rabilloud T, Adessi C, Giraudel A, Lunardi J (1997) Improvement of the solubilization of proteins in two-dimensional electrophoresis with immobilized pH gradients. Electrophoresis 18:307–316
Schuurink R, Busink R, Hondmann DHA, Witteveen CFB, Visser J (1990) Purification and properties of NADP+-dependent glycerol dehydrogenases from Aspergillus nidulans and A. niger. J Gen Microbiol 136:1043–1050
Shevchenko A, Wilm M, Vorm O, Mann M (1996) Mass spectrometric sequencing of proteins from silver stained polyacrylamide gels. Anal Chem 68:850–858
Shimizu M, Wariishi H (2005) Development of a sample preparation method for fungal proteomics. FEMS Microbiol Lett 247:17–22
Sims AH, Robson GD, Hoyle DC, Oliver SG, Turner G, Prade RA, Russell HH, Dunn-Coleman NS, Gent ME (2004) Use of expressed sequence tag analysis and cDNA microarrays of the filamentous fungus Aspergillus nidulans. Fung Genet Biol 41:199–212
Steffan JS, Mc-Alister-Henn L (1992) Isolation and characterization of the yeast gene encoding the MDH3 isozyme of malate dehydrogenase. J Biol Chem 267:24708–24715
Stemple CJ, Davis MA, Hynes MJ (1998) The facC gene of Aspergillus nidulans encodes an acetate-inducible carnitine acetyltransferase. J Bacteriol 180:6242–6251
Weidner G, d’Enfert C, Koch A, Mol PC, Brakhage AA (1998) Development of a homologous transformation system for the human pathogenic fungus Aspergillus fumigatus based on the pyrG gene encoding orotidine 5′-monophosphate decarboxylase. Curr Genet 33:378–385
Wildgruber R, Reil G, Drews O, Parlar H, Görg A (2002) Web-based two-dimensional database of Saccharomyces cerevisiae proteins using immobilized pH gradients from pH 6 to pH 12 and matrix-assisted laser desorption/ionization-time of flight mass spectrometry. Proteomics 2:727–732
Acknowledgements
We are grateful to Chi Wong and Geoffrey Turner of MBB at Sheffield University for introducing us in proteome analysis. Silke Steinbach is acknowledged for excellent technical assistance and Matthias Brock for critically reading of the manuscript. This research was supported by the Priority Program 1160 of the Deutsche Forschungsgemeinschaft.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by G. Braus
Rights and permissions
About this article
Cite this article
Kniemeyer, O., Lessing, F., Scheibner, O. et al. Optimisation of a 2-D gel electrophoresis protocol for the human-pathogenic fungus Aspergillus fumigatus . Curr Genet 49, 178–189 (2006). https://doi.org/10.1007/s00294-005-0047-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00294-005-0047-9