Abstract
Comparative genomics allows comparison of different proteins that execute presumably identical functions in different organisms. In contrast to paralogues, orthologues per definition perform the same function and interact with the same partners and, consequently, should display conservation in all these properties. We have employed 20 fungal genomes to analyse key components of the high osmolarity glycerol signalling pathway of Saccharomyces cerevisiae. Among the proteins scrutinised are a complete phosphotransfer module, a MAP kinase, two scaffold proteins, one of which is also a MAPKK, and two transcription factors. Sequence alignments, domain structure and size analysis, combined with the rich information available in the literature, allowed us to probe previous structural and functional studies and to generate hypotheses for future experimental studies. Although certain domains are too highly conserved across fungal species for meaningful comparative studies, others, like interaction domains, can be studied in closely related species. Moreover, putative functionally relevant sites for protein modifications can be identified in such comparative studies. We provide several relevant examples and present a number of previously un(der)characterised domains of potential functional significance in osmosensing and signal transduction. We propose that any functional protein analysis in fungi should make use of the unique resource that fungal genome sequences offer.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The availability of genome sequences from closely related species offers opportunities to study the evolution of genome organisation (Langkjaer et al. 2003; Dietrich et al. 2004; Dujon et al. 2004; Fabre et al. 2004; Kellis et al. 2004; Koszul et al. 2004), to identify regulatory sequences (Cliften et al. 2003) or to facilitate genome annotation (Brachat et al. 2003; Dujon et al. 2004; Lafontaine et al. 2004). In addition, such genome sequences are invaluable tools for the functional analysis of proteins and cellular pathways. Because we are limited in our ability to decode the information of primary sequences in terms of protein structure and function, the interpretation of protein sequences is still largely based on comparison. Genome sequences from yeasts and fungi offer the opportunity to investigate functional protein domains and to identify residues of functional importance. Hence, comparative genomics is a tool to probe previous functional and structural analysis studies, to aid the interpretation of the effects of specific mutations and to generate hypotheses for functional analysis.
Comparing sequences of orthologous proteins from different organisms to aid functional studies is done under the implicit assumption that protein function is exactly the same in the set of the organisms studied. In the accompanying paper (Krantz et al. 2006) we have investigated the osmosensing HOG (high osmolarity glycerol) signalling pathway from Saccharomyces cerevisiae and tried to identify orthologues for 40 proteins in 19 different species. For components that are part of the core of the system, this was readily possible. For a number of proteins, however, either sequence conservation was too poor to identify reliable orthologues in all organisms studied, or they simply do not exist. This suggests that certain features of the pathway are not identical across fungi and indeed experimental studies on this popular signalling network in different species indicate that signal perception, the connection of certain pathway modules as well as response mechanisms are different (see e.g. (Hull and Heitman 2002; Alonso-Monge et al. 2003; Smith et al. 2004; Furukawa et al. 2005)). Hence, certain protein properties may differ between fungi and this needs to be taken into account when comparing fungal protein sequences for functional studies.
Different taxonomic groups of fungal species with different lifestyle and evolutionary relationship are subject to genome sequencing, making the growing set of sequences particularly valuable. For instance, the genomes of five very closely related sensu strictu plus two senso lato Saccharomyces yeasts have been sequenced and these species have similar lifestyles and hence likely have highly similar properties also at the proteome level (Cliften et al. 2003; Kellis et al. 2003). Furthermore, eight other yeast genomes are available from organisms that have different life styles and belong to different taxonomic subgroups (Wood et al. 2002; Dietrich et al. 2004; Dujon et al. 2004; Jones et al. 2004; Kellis et al. 2004). The molecular properties of their proteins may have diverged to different extent. In addition, genome sequences from five filamentous fungi have been included in this analysis, four of which belong to the ascomycetes (published only N. crassa (Borkovich et al. 2004)) and one (Ustilago maydis) to the basidiomycetes. Further functional divergence can be expected in proteins from these organisms. While functional divergence cannot be expected to affect principal biochemical properties such as protein kinase activity or DNA binding, it likely concerns features such as interaction with or modification by other proteins.
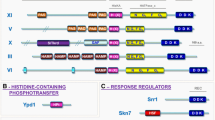
The HOG pathway from S. cerevisiae (Fig. 1) consists of two branches that seem to sense osmotic changes in different ways (de Nadal et al. 2002; Hohmann 2002; O’Rourke et al. 2002; Saito and Tatebayashi 2004; Sheikh-Hamad and Gustin 2004; Westfall et al. 2004). The Sln1-branch consists of the Sln1-Ypd1-Ssk1 phosphorelay system, where Sln1 is a sensor histidine kinase, Ypd1 a phosphotransfer protein and Ssk1 a response regulator (Posas et al. 1996; Santos and Shiozaki 2001; Wolanin et al. 2002; Catlett et al. 2003). Hyperosmotic shock deactivates Sln1, leading to enhanced levels of dephospho-Ssk1, which is an activator of the MAPKKKs Ssk2 and Ssk22 (Posas et al. 1996). In this study we have included the entire three-protein phosphorelay system, since its detailed function is of great interest. Ssk2 and Ssk22 activate the MAPKK Pbs2 by phosphorylation, which in turn activates the MAPK Hog1 by phosphorylation (Maeda et al. 1994). We included Hog1 in this study as an example of a highly conserved protein. The Sho1-branch contains two scaffold proteins that are crucial for signalling and specificity: the plasma membrane-localised Sho1 (Raitt et al. 2000; Seet and Pawson 2004) as well as the MAPKK Pbs2 (Posas and Saito 1997; O’Rourke and Herskowitz 1998). Sho1 recruits Pbs2 to the cell surface during signalling (Raitt et al. 2000; Reiser et al. 2000). Both Sho1 and Pbs2 can bind the MAPKKK Ste11 (Zarrinpar et al. 2004). Since both Sho1 and Pbs2 are crucially important in signalling and have a range of known functional features, both were included in this study. Ste11 is activated by phosphorylation, which is mediated by the Ste20 kinase and probably Cla4 (Raitt et al. 2000; Reiser et al. 2000) and requires Ste50 (Ramezani-Rad 2003). Ste20 activation in turn depends on the membrane-bound G-protein Cdc42 (Elion 2000; Ramezani-Rad 2003). Activated Hog1 has numerous targets in the cell, such as the transcription factors Msn2/Msn4, Hot1, Sko1 and Smp1 (Rep et al. 1999a, 2000, 2001; Alepuz et al. 2001, 2003; Proft et al. 2001; Proft and Struhl 2002; de Nadal et al. 2003). We included Hot1 and Sko1 in this analysis as an example of relatively poorly conserved “peripheral” proteins in signalling (Krantz et al. 2006).
The HOG pathway. Osmotic stress is perceived via two distinct input branches, the Sho1 and Sln1 branch, both of which activate the MAPKK Pbs2 via distinct MAPKKK. Sln1-Ypd1-Ssk1 constitutes a phosphotransfer system, which links to the MAPK module via either of the two MAPKKK Ssk2 and Ssk22. The mechanism of activation via the Sho1 branch is less clear, but requires co-scaffolding by Sho1 and the MAPK Pbs2, as well as the MAPKKK Ste11. Once activated, Pbs2 activates the MAPK Hog1, which, in turn stimulates transcription via a number of transcription factors. Hog1 also activates cytoplasmic targets such as the MAPKAP Rck2
Materials and methods
Multiple alignments and motif prediction
Multiple alignments were performed with Clustal W (http://www.ebi.ac.uk/clustalw/) (Thompson et al. 1994), using default settings, and the quality histograms were derived from Jalview. Domain definitions were retrieved using Pfam (http://www.sanger.ac.uk/Software/Pfam/) (Bateman et al. 2002) and FingerPRINTScan (http://www.ebi.ac.uk/printsscan/) (Attwood et al. 2000). Transmembrane predictions were made at (http://www.cbs.dtu.dk/services/TMHMM-2.0/) (Moller et al. 2001). Searches of protein databases were performed using FASTA (http://www.ebi.ac.uk/fasta) (Pearson 1990). Helix predictions were done according to (Suzuki and Brenner 1995).
Analysis of size
Proteins, and protein domains, from yeasts and filamentous fungi were considered as two separate populations. Each protein size was compared to the average and standard deviation based on the rest of its population. Probability was calculated from the resulting t-distribution, and the threshold of significance was set to 0.001. After elimination of all outliers, yeast and filamentous fungi were compared with heteroscedastic t tests, with a threshold of significance of 0.01.
Results
Sln1
Sln1 is the sensor histidine kinase of the Sln1 branch of the HOG pathway. A number of functional properties are encoded in its protein sequence, some of which may overlap. (1) Localisation in the plasma membrane; (2) Osmosensing; (3) Control of histidine kinase activity; (4) Dimerisation; (5) Binding of ATP; (6) Hydrolysis of ATP and transfer of the phosphate group to a histidine residue; (7) Acceptance of the phosphate group on a histidine residue; (8) Transfer of the phosphate group to an aspartate in the C-terminal receiver domain; (9) Interaction with the Ypd1 phosphotransfer protein; (10) Transfer of the phosphate group to a histidine in Ypd1.
We included 16 sequences in the analysis. The truncated Saccharomyces kluyveri open reading frame was restored using DNA sequence information and the truncated Debaryomyces hansenii open reading frame was included in this analysis as such (Krantz et al. 2006). The well-characterised S. cerevisiae protein has 1,220 amino acids (Fig. 1 and Supplementary Fig. 1).
Membrane insertion of Sln1 is achieved by means of two transmembrane domains. The first transmembrane domain (TMD1) is located close the N-terminus and its deletion renders the protein unresponsive to osmotic changes, i.e. constitutively active (Ostrander and Gorman 1999). TMD1 is preceded by a fully conserved Gln residue (position 22 in S. cerevisiae), which in turn is preceded by one or two basic residues two and six positions upstream of Gln22. A surplus of positively charged residues commonly marks the cytoplasmic face of TMDs (von Heijne 1989; White and von Heijne 2004). The prediction of the exact position of the TMD varies between proteins and hence differs somewhat from the sequence alignment. The transmembrane domain is characterised by a central block of small hydrophobic residues in which two positions (Val27 and Leu35, S. cerevisiae positions) are perfectly conserved. The distal end of TMD1 is predicted for S. cerevisiae Sln1 after Phe46. This position is followed by hydrophilic residues in all proteins.
The second TMD probably starts with the perfectly conserved Ile334. Also TMD2 is preceded by positively charged residues, commonly two, which are commonly one and four residues upstream of the conserved Ile334. In this case, however, they mark the predicted periplasmic face. TMD2 probably ends at Trp356, which is conserved in all yeast Sln1 (except Yarrowia lipolytica). The sequence of TMD2 is well conserved, for instance Gly337 is conserved in 15/16 sequences, Pro352 in 15/16 sequences.
Deletion of the large extracellular domain between TMD1 and TMD2 inactivates Sln1. Since replacing the extracellular domain with a leucine zipper renders the construct constitutively active, it was suggested that the extracellular domain mediates dimerisation (Ostrander and Gorman 1999). The length of this domain varies considerably between species, but is rather consistent within the three different groups of organisms. The Y. lipolytica domain (391 residues) is significantly larger than that of the other yeasts (290±7 residues, P=6.8×10−8). Sln1 from filamentous fungi has a significantly larger extracellular domain of 364±8 amino acids (P=1.16×10−5, t test).
The extracellular domain is overall poorly conserved at the sequence level. However, our comparison reveals certain patterns. Up to Gly99 (there is a Gly in this region in all 16 proteins) the sequences align without gaps; several residues are conserved in the majority of the sequences and there seems to be a consistent pattern of charged, hydrophilic and hydrophobic residues. In particular, there is a conserved pattern of no less than ten hydrophobic residues (Leu53, Leu58, Ile60, Leu64, Ile69, Leu73, Leu76, Leu83, Leu89 and Leu93; Leu, Val or Ile are acceptable in any of these positions), potentially forming a leucine zipper.
Following Gly99, sequences diverge and the alignment contains numerous gaps. However, deletion of residues 138–150 impairs Sln1 function (Reiser et al. 2003) while mutation of the putative N-glycosylation sites Asn138 and Asn142 does not have an effect. Asn138 is not conserved, even not among closely related Saccharomyces species and Asn142 is not conserved beyond Ashbya gossypii, confirming that those residues probably do not have a specific role. Pro158 is conserved but this region aligns differently in different ClustalW runs. Hence, the functional significance and conservation of this region remain uncertain.
A block between Tyr185 and Ile237 aligns rather well. Gly211 and Ala219 are conserved and Tyr212 and Thr214 are conserved in 15/16 sequences. Moreover, several residues are chemically conserved. In particular, this region also contains a pattern of small hydrophobic residues, although not as well pronounced as the putative zipper at the beginning of the extracellular loop: Leu191, Ile193, Val206, Ile210, Ile212 (or Ile213), Ala216, Leu219. This region has, to our knowledge, not been investigated.
Following this domain the Y. lipolytica protein has an insert that makes the extracellular domain longer. Finally, sequences just preceding TMD2 are also well conserved: Gly302 and Trp313 are fully conserved and there is chemical conservation in several other positions as well as a pattern of acidic residues. Using the entire extracellular domain in FASTA searches against all proteins in databases identifies osmosensing histidine kinases but no other significant matches.
The second transmembrane domain is followed by a HAMP (Histidine kinases, Adenylyl cyclases, Methyl-binding proteins, Phosphatases) domain, which is found especially in transmembrane sensor histidine kinases (Aravind and Ponting 1999). It seems to serve as a linker between the transmembrane domain and the histidine kinase domain and may also be important for dimerisation. The sequence 359PIVRLQKAT367 contains 5/9 perfectly conserved amino acids (bold) and three further residues are chemically conserved. HAMP domains are then commonly followed, at some distance, by a second conserved block, which is characterised by a conserved Gly. This is not apparent in Sln1. Typical for the third block of the HAMP domain are the conserved Asp520, Asn 529, Met531 and Leu535. This region of the HAMP domain is predicted to fold into two helices, followed by a coiled-coil region that extends into Helix 1 of the highly conserved histidine kinase acceptor region (Tao et al. 2002). As compared to the bacterial EnvZ two-component osmosensor, Sln1 has an insertion of 25 residues, which exists in all fungal Sln1 orthologues. The coiled-coil is required for kinase activity and presumed to position kinase and acceptor domain of the two subunits of a dimer (Tao et al. 2002). Glu543 and Thr550 were suggested to be important based on mutagenesis studies (Tao et al. 2002). Thr550 is fully conserved while Glu543 tolerates the chemically identical Asp in this position. Interestingly, the size of the HAMP region is well conserved in filamentous fungi (138±2), and significantly shorter than in the yeast species (178±27 (P=3.0×10−3, t test)). This difference is due to the spacer between the two conserved blocks.
Immediately distal to the HAMP domain and the associated coiled-coil follows the histidine kinase phosphoacceptor domain. This H-domain is important both for dimerisation and accepts the phosphate group from the second kinase in the Sln1 dimer. The phosphorylated histidine residue is at position His576. The domain is composed of two helices, as predicted from the bacterial EnvZ, which have almost identical sequences in all fungal Sln1 orthologues. Also sequences following the second helix are well conserved.
The histidine kinase ATPase domain starts with Gly680 and runs until Glu931. The distance to the H domain is perfectly conserved in yeast (48 residues), and shorter than in filamentous fungi (77±2 (P=2×10−4, t test)). The ATPase domain contains four blocks of conserved residues called the N, G1, F and G2 boxes, which form the ATP-binding cleft (Dutta et al. 1999; Dutta and Inouye 2000). The N-box is located around Asn691 followed by a spacer (residue 716–847), which is unique for Sln1s. While its sequence is poorly conserved, the size is similar within filamentous fungi (99±15) and yeasts (154±10), with the exception of Candida albicans (335 residues, P=8.7×10−8). The G1, F and G2 boxes are close together around Gly861 (G1 box, two fully conserved Gly), Phe873 (F-box, two fully conserved Phe) and Gly888 (G2-box, four fully conserved Gly).
The ATPase domain is again followed by a spacer (residues 932–1,088) that does not show obvious sequence conservation. The response regulator receiver domain of Sln1 starts with Lys1089 (always a basic residue in this position) and extends to about position 1,200. The receiver residue is Asp1151. Overall, the domain is highly conserved. In database searches, the Sln1 orthologues cluster together, separate from other fungal histidine kinases, including the closest Sln1 homologues in Schizosaccharomyces pombe and Ustilago maydis, which lack transmembrane domains (Fig. 2). The crystal structure of the Sln1 response regulator domain in complex with Ypd1 has been determined (Xu et al. 2003).
The C-terminus is commonly short, although in some filamentous fungi more than 100 amino acids long, and rich in charged amino acids.
Ypd1
The phosphotransfer proteins (Hpts) are commonly small proteins (167 amino acids in S. cerevisiae) but Sz. pombe Spy1 is predicted to be 295 amino acids due to a unique N-terminal extension that is unrelated to any other protein. Hpts have two functions: (1) interaction with response regulator domains; (2) exchange of phosphate groups with response regulator, specifically between an aspartate and a histidine residue. It appears that Hpts do not encode any particular discrimination between different response regulator domains (Xu et al. 2003; Porter and West 2005), consistent with the observation that fungal genomes only encode one single Hpt but several histidine kinases and response regulator proteins (Catlett et al. 2003).
The crystal structure of Ypd1 has been determined individually (Xu and West 1999) as well as in a complex with the response regulator domain of Sln1 (Xu et al. 2003). In addition, a detailed mutational analysis for the requirements for interaction with the Ssk1 response regulator (Porter et al. 2003) as well as all three yeast response regulators (Ssk1, Sln1 and Skn7) has been performed (Porter and West 2005). These studies provide the framework for interpretation of the multiple alignments.
Twenty proteins were included in the analysis (Supplementary Figure 2). The predicted start of the Saccharomyces castelli and S. kluyveri proteins is most probably a sequencing or annotation error. According to the crystal structure of Ypd1, the protein folds into a bundle of four helices, αA, αB, αC and αD (Fig. 3). Helix C contains the phospho-accepting histidine in position 64. The mutational analysis was performed as alanine scanning and of course other substitutions may confer different effects (Porter et al. 2003; Porter and West 2005). Ile13 and Glu16 seem to be important for interaction with Skn7, Ssk1 and to a lesser extent Sln1. Ile13 seems to tolerate Val and Thr, and Glu16 is replaced by Gln in several organisms. Mutation of Met20, Asp21, Asp23, Asp24, Glu27, Leu31 and Gln38 affect interaction with all response regulators. Met20 is conserved and followed by a block of usually four acidic residues (only two instances where this block is only two or three residues). Then Asp60 seems to play an important role in protein interaction but this residue is only conserved in closely related yeasts. The sequences following the phospho-accepting His64 play roles in interaction with specifically Skn7, and to different extent also with the other response regulators. This entire region is highly conserved. Residues Phe65 (Tyr in one instance), Lys67 (Arg in one instance), Gly68 (conserved), Ser69 (conserved), Ser70 (Ala in two instances), Leu73 (can be Val or Ile), Gln76 (not conserved), Trp80 (not conserved) and Asp83 (can be Asn) seem to be important according to mutational and structural analysis (Porter et al. 2003; Porter and West 2005). The availability of such multiple alignments allows now to extend the mutational analyses and interpretation of the structural data.
Ypd1 conservation. The ratio of conservation for each of the residues providing the interaction surfaces with the response regulators in Sln1, Ssk1 and Skn7. The colour coding of the bars indicates for which interactions their corresponding residues is highly important; black (all), striped (Skn7 and Ssk1), chequered (Skn7 and Sln1), grey (only Skn7) and white (none). Compare to (Porter et al. 2005). The four boxes at the bottom indicate the positions of the helixes
Mutation of Gly74 to Cys has recently been shown to cause pradimicin resistance (Hiramoto et al. 2005). This residue is one of two conserved Glys (the other being Gly 68) that introduce a turn between two helices, where the first helix contains the phosphorylated His64.
Ssk1
Ssk1 contains the following functionalities: (1) interaction with Ypd1; (2) accepting a phosphate group on an aspartate; (3) interaction with Ssk2/Ssk22 when dephosphorylated.
Nineteen proteins were included in the analysis (Supplementary Fig. 3). A second candidate found in U. maydis was excluded, because its response regulator is different from that of the other Ssk1 proteins (Fig. 4). Ssk1 orthologues differ substantially in size between 522 amino acids in Sz. pombe and 1,658 amino acids in U. maydis.
The large (501 amino acids in S. cerevisiae) N-terminal domain is poorly conserved and does not contain known functional domains. There are, however, some elements that show conservation. Ile246 marks the start of a stretch of three hydrophobic residues, followed by the element Asp/Glu249 Pro250 Asp/Glu251, which is conserved in yeasts (not Sz. pombe). Another short conserved patch of hydrophobic amino acids follows Ile385. Ssk1 of the four filamentous ascomycetes, however, display an extensive conserved stretch following Arg340 until about Ser438 (in Maghaporthe grisea). The functional significance of any of those regions remains to be studied.
The only known functional domain is the response regulator receiver domain at Pro502-Leu651. The phospho-receiving aspartate is located at position Asp554. The response regulator domain also overlaps with the domain needed for interaction with Ssk2, which is located between 475 and 670 (Posas and Saito 1998).
When the Ssk1 receiver domain is used in FASTA searches against databases, it identifies with highest score putative Ssk1 orthologues from fungi, followed by response regulators of histidine kinases from bacteria and plants. The response regulator domains from Sln1 and Ssk1 are far more different from each other than they are from orthologues of other organisms (Fig. 4). This adds confidence to the orthologue identification and is consistent with the observation that the response regulator domain of Sln1 fails to interact with Ssk2 (Posas and Saito 1998).
Sho1
Sho1 fulfils the following functions: (1) localisation to the plasmamembrane by means of transmembrane domains; (2) interaction with an unknown protein to localise Sho1 to places of cell expansion; (3) interaction with Pbs2, Fus1, Ste11 and probably other proteins.
Seventeen proteins were used for this analysis. All proteins are characterised by four predicted transmembrane domains and a C-terminal SH3 (Src homology 3) domain. The U. maydis protein appears well conserved in domain structure and size but often differs where all other sequences are identical (Supplementary Fig. 4).
The N-terminal block with four TMDs stretches from Asp31 to Lys161 (S. cerevisiae). The four TMDs are densely packed. The loops are only five (between TMD2 and 3) or eight (between TMD3 and 4) amino acids long. The first loop is somewhat variable in filamentous fungi. Although the prediction of the exact location of the TMDs differs somewhat between different proteins (probably an artefact of the prediction), there is strong conservation of the size of the entire block: 124±4 residues in yeasts and 129±3 in filamentous fungi, including U. maydis). Assuming aligned endpoints, the variation virtually disappears in yeasts, unlike filamentous fungi, which vary due to a variable insert between TMD1 and TMD2 (±1 and ±5, respectively). The start of the TMD1 is characterised by the conserved peptide Asp31 Pro32 Phe33 Ala34 (except U. maydis). TMD2 starts with Phe65 (Tyr in some species, Val in U. maydis) and is characterised by an almost perfectly conserved Trp67 Trp68 dipeptide (Trp Phe in two filamentous fungi, including U. maydis). The start of TMD3 may be marked by Arg93 (Lys or His in some species). TMD4 may start around Ala124 and is characterised by the perfectly conserved (also U. maydis) dipeptide Ala127 Gly128. TMD4 seems to be the best conserved TMD at primary sequence level.
Between the transmembrane section and the SH3 domain at 303–359, there is a linker domain that has been regarded as poorly conserved. However, within groups of organisms there is size conservation: 100±2 residues in filamentous ascomycetes, while U. maydis is significantly longer (141 residues, P=0.00041), and 146±11 residues in yeasts, excluding Y. lipolytica (109), D. hansenii (119) and C. albicans (200). In addition, the sequence is rather well conserved within the yeasts (again excluding Y. lipolytica, D. hansenii and C. albicans) and within the filamentous ascomycetes (Fig. 5). Linker size in yeasts is significantly longer than in filamentous ascomycetes (P=5.7×10−8), t test). Moreover, this linker appears to contain sequences of potentially more specific importance. Leu223 and Glu227 are perfectly conserved and Gly225 and Asn228 are conserved in 16/17 proteins (except U. maydis). This uncharacterised element deserves investigation. The region between residues 170 and 210 has been implicated in the binding of Ste11 (Zarrinpar et al. 2004). However, this region is not conserved outside the Saccharomyces sensu strictu group and is of low complexity. However, it has been pointed out that additional sequences must be involved in Ste11 binding (Zarrinpar et al. 2004).
Multiple alignment of the Sho1 linker. Despite the poor overall alignment with identities as low as 4%, there is a relatively high conservation within groups. Among yeasts, Y. lipolytica stands out (4–15% identity towards other yeasts), as do D. hansenii and C. albicans (32% towards each other, 12–21% and 17–27% versus. other yeasts, respectively). Apart from these, this domain is rather well conserved in yeasts (34–55% outside the sensu strictu group (86–92% inside)). Similarly, the sequences from filamentous fungi are rather well conserved (31–54%), excluding that of U. maydis (7–18%). In contrast, there is very low conservation between these two groups (8–20%)
The SH3 domain, which interacts with Pbs2 and Fus1, shows a high degree of conservation. In this domain the U. maydis protein does not deviate from the others. Using the SH3 domain in database searches reveals with highest scores the fungal Sho1 orthologues studied here, followed by tyrosine kinases from different organisms that have a structural organisation (SH3)-(SH2)-(Tyr-kinase) and play a role in cell shape organisation. The overall sequence identity within this 57 amino acid region is 35% (61% similarity). Pro352, which has been shown to be essential for Fus1 interaction (Nelson et al. 2004), is conserved. The region between Ile302 and Glu360 has been studied in detail by mutational and comparative analysis to identify residues that determine the strength of interaction with Pbs2 (Marles et al. 2004). This study focused on three residues based on the conservation with the FYN tyrosine kinase SH3 domain. Exchange of Tyr309 by Ala reduced binding affinity. This residue is conserved (in Neurospora crassa: Phe). Glu317 is fully conserved. Tyr355, mutation of which strongly affects interaction with Pbs2, is also fully conserved.
Pbs2
Pbs2 is another particularly interesting protein since it serves as a scaffold protein for several components of the HOG pathway, as the MAPKK of the system and as the node where the two upper branches of the pathway converge. Hence, the following functions are encoded in the protein sequence: (1) interaction with the MAPKKK Ste11; (2) interaction with the MAPKKKs Ssk2 and Ssk22; (3) phosphorylation by these kinase within the activation loop; (4) interaction with the Hog1 MAPK; (5) binding of ATP; (6) transfer of the phosphate group to the activation domain of Hog1; (7) interaction with Sho1; (8) interaction with Nbp2 to target the phosphatase Ptc1 to Pbs2.
Seventeen proteins were included in this analysis. The likely Pbs2 orthologue from U. maydis is lacking large portions of the N-terminus, indicative of either an annotation problem or a fundamentally different pathway architecture. Fusarium graminearum and N. crassa both have a second Pbs2 homologue, which, however, also lacks large parts of the N-terminus and hence those were eliminated from this analysis ((Krantz et al. 2006) Supplementary Fig. 5).
The protein kinase domain is the best conserved part and located between residues 360 and 623 in S. cerevisiae Pbs2. It starts with the ATP-binding domain, which is followed by about 40 amino acids insertion in the sequences of the filamentous ascomycetes. The following sequences are highly conserved in all proteins but there are certain sections where filamentous fungi differ from yeasts, such as around position 535 and towards the distal part of the protein kinase domain. The overall sequence identity between residues 360 and 623 is as high as 43%. The residues in the activation domain phosphorylated by the MAPKKKs for activation of Pbs2 (Ser514 and Thr518) are conserved; 27/31 residues in this section are identical across all sequences.
The large N-terminal part of Pbs2 is poorly conserved, the alignment shows many gaps and its size differs substantially. Pbs2 from filamentous ascomycetes tends to be smaller (P=0.002, t test), but there is an overlap of the size distribution between the groups (278±19 as compared to 345±56 in yeasts).
Mutational analysis showed that residues 46–56 are important for signalling through the Sln1 branch and contain a docking site for the Ssk2/Ssk22 MAPKKK (Tatebayashi et al. 2003). Residues Ala52, Arg53, Val54 and Ala56 were found to be important. Ala52 and Ala56 are conserved in all yeasts (except Sz. pombe) and Arg53 and Val54 are chemically conserved. In filamentous fungi and in Sz. pombe this docking site is not conserved. However, Arg53 is part of a stretch of several basic residues that is well conserved among yeasts down to C. glabrata, with the exception of Sz. pombe Wis1. Arg61 is conserved in all proteins, except Wis1. Hence it is possible that for interaction with a MAPKKK these basic residues play a role. The Ste11-binding site has been mapped to residues 56–162 and hence may overlap that for Ssk2/Ssk22, as well as the binding site for Sho1 [see below (Zarrinpar et al. 2004)].
Residues 91–101 (VNKPLPPLPVA), forming a proline-rich element (PXXP, SIM element for SH3-interacting motif), mediate interaction with the SH3 domain of Sho1 and are necessary for signalling through the Sho1 branch (Raitt et al. 2000; Reiser et al. 2000). The core of this element (KPLPPLP) is perfectly conserved down to D. hansenii. Also Sz. pombe (which is lacking a Sho1 orthologue) has a proline-rich element (PPLPRAVP) in this region; its possible interaction partner is not known. Y. lipolytica, M. grisea, N. crassa and F. graminearum appear to have some proline residues in this region, however, the sequence arrangement is different. C. albicans and A. nidulans lack any prolines in this area. It has recently been noted that A. nidulans Sho1 does indeed not signal to Pbs2 (Furukawa et al. 2005).
Residues 283–353, i.e. the sequences immediately proximal of the kinase domain, are required for signalling from Pbs2 to Hog1 (Zarrinpar et al. 2004), probably as a docking site. In fact, sequences distal from Leu316 align without gaps and show significant similarity, with several residues fully or chemically conserved. This region deserves further investigation.
Recently, evidence has been reported that Pbs2 interacts with the adaptor protein Nbp2, which has an SH3 domain (Mapes and Ota 2004). Nbp2 is thought to target the protein phosphatase Ptc1 to Pbs2. Interaction with the SH3 domain of Nbp2 does not appear to use the same proline-rich domain as that of Sho1, but rather a sequence located at residues 187–190 (PRRP). This area contains a second such element (196–199), which does not seem to be required for Nbp2 interaction (Mapes and Ota 2004). The specific element at 187–190 is only conserved in Pbs2 of Saccharomyces sensu strictu yeasts. However, Pbs2 orthologues from most species have a PXXP element within this region. We did not locate such an element in this area of Pbs2 from only D. hansenii and C. albicans. This suggests that indeed the region around position 190 could serve as target for interaction of SH3-containing proteins. It should, however, be noted that all Pbs2 sequences contain several PXXP motifs at various, non-conserved positions.
Following the protein kinase domain S. cerevisiae Pbs2 has a short C-terminus of about 45 amino acids. In filamentous ascomycetes there is a poorly conserved insertion of about 30 amino acids, which is rich in acid residues. This is followed in all proteins by a region rich in basic residues and then a stretch VPALHMGGL, which is well conserved in yeasts (except Sz. pombe). This segment is not functionally characterised but has been implicated in Hog1 localisation in a recent paper (Sharma and Mondal 2005).
Hog1
The MAPK Hog1 has the following functional features: (1) interaction with Pbs2; (2) activation by phosphorylation in the activation loop; (2) shuttling between cytosol and nucleus and hence interaction with several relevant proteins; (3) binding of ATP; (4) ATP hydrolysis and phosphorylation of target proteins; (5) interaction with a range of target proteins (Rck2, Hot1, Sko1, Msn2, Smp1 and probably more); (6) interaction with several protein phosphatases (Ptp2, Ptp3, Ptc1) and dephosphorylation by those.
Hog1 orthologues from all 20 organisms included in the study were compared. The overall identity is no less than 62% (Supplementary Fig. 6). This high sequence identity makes it difficult to distinguish certain functional domains. The only remarkable differences are small insertions in the proteins from F. graminearum, A. nidulans and N. crassa. Still, the proteins from filamentous fungi are significantly smaller than their yeast counterparts (364±10 and 421±31, respectively (P=7.1×10−6, t test)). It appears that the C-terminus of the N. crassa protein may be incorrectly annotated. Yeasts seem to have an alanine-rich C-terminus, which is missing in the filamentous fungi.
A set of mutations has been isolated that cause Hog1 to be active independently of phosphorylation by Pbs2 (Bell et al. 2001; Yaakov et al. 2003). Those mapped to the following positions: Tyr68 (conserved), Glu170 (conserved), Ala314 [also accepts Cys and was mutated to Thr in (Bell et al. 2001; Yaakov et al. 2003)], Phe318 (conserved), Trp 320 (conserved), Phe322 (conserved), Trp332 (conserved) and Asn391 (not conserved).
Sko1
Sko1 is a DNA-binding protein that mediates both repression and activation of expression of a subset of Hog1 target genes (Proft et al. 2001; Rep et al. 2001; Proft and Struhl 2002). Sko1 has multiple functions that must be decoded in its sequence: (1) sequence-specific binding to DNA (2) interaction with Hog1 (3) phosphorylation by Hog1 (4) interaction with the Tup1/Ssn6 corepressor complex (5) phosphorylation by cAMP-dependent protein kinase (PKA) (6) shuttling between the cytosol and the nucleus and hence interaction with relevant proteins. We included 12 yeast proteins in this analysis (Supplementary Fig. 7). Putative orthologues from filamentous fungi and fission yeast are conserved only within the DNA-binding domain and hence may be regulated in different ways.
The DNA-binding domain is of the basic leucine zipper (bZIP) type and located at positions 427–488 in the S. cerevisiae protein. Sixteen residues are identical in this region. The DNA-binding domain is characterised by a short block of acidic residues, followed by a first block of basic amino acids and the almost perfectly conserved sequence FLERNRVAAS, a second block of basic amino acids and the actual leucine zipper. However, none of the leucine residues is conserved, rather the zipper reads I K/Q K I/M E X D/E L/V X F/I Y/L E X E/G Y X D/E L/M X X I/V/M/L X X L/F X X I/V/L. The zipper consists of two parts with regularly spaced small hydrophilic residues.
Sequences required for nuclear localisation of Sko1 have been reported to overlap with those involved in DNA-binding, or at least being close to those (Pascual-Ahuir et al. 2001). Indeed, preceding the bZIP region, most Sko1 orthologues have sequences that could serve as bipartite nuclear localisation signal (two adjacent basic residues, ten arbitrary residues and then three basic residues out of five). At the same time, the basic blocks within the bZIP also fulfil this requirement.
Hog1 phosphorylates Sko1 on Ser108, Thr113 and Ser126 (Proft et al. 2001). Phosphorylation sites are characterised by Ser/Thr Pro. All three phosphorylation sites are conserved, although the first site is moved by one position in D. hansenii and Saccharomyces mikatae. The entire section of about 20 residues is quite well conserved and characterised by a central, almost perfectly conserved GGSKRLPPL element that separates the second and the third phosphorylation site. The significance of this sequence has not been studied so far and it is tempting to speculate that it plays a role in protein interaction.
Protein kinase A phosphorylates Sko1 on Ser380, Ser 393 and Ser399 (Pascual-Ahuir et al. 2001). Sites for PKA phosphorylation are commonly characterised by two basic residues, followed by an arbitrary residue and the phosphorylated Ser/Thr. The block of PKA-dependent phosphorylation sites is located within the region of basic residues preceding the bZIP domain and constituting a possible bipartite nuclear localisation sequence. Since PKA stimulates nuclear accumulation of Sko1 (Pascual-Ahuir et al. 2001) it is possible that phosphorylation activates this localisation signal. However, neither the possible nuclear localisation signal upstream of bZIP nor the PKA-dependent phosphorylation sites are conserved across yeasts. This could indicate that the PKA-dependent regulation of Sko1, whose precise function is not fully understood (Pascual-Ahuir et al. 2001), is a speciality of Saccharomyces yeasts. It is worth noting that Sko1 is the main factor in yeast binding to consensus cAMP-response elements (CREs), which mediate transcriptional PKA responses in eukaryotes, while in S. cerevisiae the effect of PKA on expression from CREs is moderate (Hohmann 2002).
Binding of Hog1 as well as Tup1 requires the first 315 amino acids of the protein, but has not been mapped to any further detail. Apart from the Hog1-dependent phosphorylation sites there are two short regions of high sequence similarity. Right at the N-terminus, following residue 15, the sequence FDLEPNPFEQSF is almost perfectly conserved, in fact also in Sko1 homologues from some filamentous fungi. The sequence does not bear any information as to its function and does not identify any other sequences in the databases. Around position 220 the element KSGLTPNESNIRTGLTP is highly conserved (except K. lactis). Using this sequence in a database search yields only Sko1 homologues from fungi, including some filamentous fungi and in fact also K. lactis (following position 162; has not been captured by ClustalW). To our knowledge these two conserved elements have not been studied so far and may be important for interaction with other proteins.
Hot1
Hot1 is a DNA-binding protein that is required for expression of a subset of Hog1-regulated genes. It has been shown to bind to target promoters and recruit Hog1 to such promoters. Hog1 phosphorylates Hot1. Hence, Hot1 has at least three functions: (1) DNA binding, (2) Hog1 binding, (3) phosphorylation by Hog1.
Hot1 is poorly conserved and we did not identify orthologues beyond yeasts. For this reason, only ten yeast sequences were included in the analysis (Supplementary Fig. 8). Sequence conservation is restricted to the very C-terminus. In this region (residues 620–719), however, 47/100 residues are fully conserved. This region likely represents the DNA-binding domain (this assumption is based also on moderate similarity to the DNA-binding region of Gcr1, see below). The region may fold into four helices of nine residues (helices 1–3) and 15 residues (helix 4), respectively. These could be arranged in a configuration of two helix-turn-helix motifs: the first and second helix as well as the third and fourth helixes are each separated by two conserved glycine residues, which likely introduce turns. The two helix-turn-helix motifs are separated by a stretch rich in basic residues, which extends into the third helix and could make contact with the DNA phosphate backbone. Sequence conservation extends to the very C-terminus, which may form a fifth helix, preceded by another short stretch of basic residues.
Using the putative DNA-binding region in FASTA searches against databases only reveals the Hot1 orthologues used in this study, as well as Msn1 from yeasts. Msn1 is known to have some redundant or overlapping function with Hot1 (Rep et al. 1999b). The C-terminus of Hot1 has some sequence and predicted structural similarity to Gcr1, a regulator of glycolytic and ribosomal protein genes, as well as the uncharacterised protein Ymr111. Hence, these four proteins seem to share a common DNA-binding domain, which, however, is not conserved at the sequence level beyond yeasts. The S. cerevisiae Hot1 DNA-binding domain is more similar to that of Hot1 orthologues than to the other three homologues from S. cerevisiae.
There are five SerPro motifs in S. cerevisiae Hot1, which seem to be phosphorylated by Hog1. A mutant of Hot1 in which all those Ser residues (Ser30, Ser70, Ser153, Ser360 and Ser410) were mutated to Ala was not phosphorylated anymore by Hog1 (Alepuz et al. 2003). Unexpectedly, this unphosphorylatable Hot1 derivative turned out to be fully functional. None of these five SerPro motifs is conserved among yeast Hot1 orthologues, different orthologues have different numbers of such SerPro dipeptides and Hot1 from S. kluyveri is lacking possible Hog1 phosphorylation sites altogether.
The entire 620 amino acids N-terminus is not conserved and we were unable to identify any regions that could be likely candidates for interaction with Hog1.
Discussion
In this study we have undertaken detailed sequence comparisons of fungal orthologues of eight different proteins in the HOG signal transduction pathway. We employed the fungal genome sequences for the identification and characterisation of functional domains that display little sequence conservation, or conservation for only a few critical residues. Such motifs occur frequently by chance, but their significance can be inferred from their degree of ubiquity within orthologous. Examples are residues for protein modification, such as phosphorylation or glycosylation, and domains involved in protein–protein interaction. Emphasis is placed on poorly conserved sequences as comparison of proteins from relatively closely related species tends to be less informative for characterisation of functional residues in highly conserved domains. For instance, protein kinase domains are highly conserved across all eukaryotes; the entire sequence of the MAPK Hog1 sequence showed more than 60% overall sequence identity across the 20 species included in this study. To pinpoint functionally relevant residues for such highly conserved, generic domains, sequence comparison across the entire eukaryotic kingdom is potentially more informative. It should be stressed that, in all cases, the information extracted from comparative studies either supports interpretation of previous functional or structural analyses or generates hypotheses for future experimental studies.
Protein modification motifs
The analysis of protein modification motifs exemplifies the utility and predictive power of comparative genomics. Although both the Hot1 and Sko1 transcription factors are phosphorylated by Hog1 concomitantly with transcriptional activation, these phosphorylations seem relevant for Sko1 only. Consistently, these sites are well conserved in Sko1 but not in Hot1.
While Hot1 is phosphorylated at five positions by the Hog1 kinase, mutation of all these five sites did not affect function of Hot1 nor transcriptional activation of target genes (Alepuz et al. 2003). In fact, none of these five potential sites (sequence Ser-Pro or Thr-Pro) is positionally conserved, and Hot1 from S. kluyveri does not have a single potential Hog1 phosphorylation site. Hence, sequence comparison alone would have strongly suggested that phosphorylation of Hot1 by Hog1 is not important for function. This observation also indicates that many protein phosphorylations in the proteome may not have functional significance but rather are a result of “vicinity” of a protein with a kinase; this aspect should be considered in global protein phosphorylation analyses.
In the case of Sko1, which is targeted by both the HOG and PKA pathways, the Hog1 phosphorylation sites are well conserved while the PKA sites are not. Active Hog1 is known to convert Sko1 from a repressor to an activator by phosphorylation at all three sites (Proft and Struhl 2002), while the role of PKA is somewhat unclear. PKA seems to affect subcellular localisation of Sko1 but has little effect on Sko1-dependent transcriptional regulation (Pascual-Ahuir et al. 2001; Proft et al. 2001). Together with the poor conservation of phosphorylation sites this suggests that PKA-dependent regulation of Sko1 may play only a minor role.
Similarly, the lack of function of the putative glycosylation sites in the extracellular domain of Sln1 (Reiser et al. 2003) could have been predicted from the fact that these sites are not conserved.
Interaction domains
This study identified several sequence elements that do not have any known function but which are, on the basis of their conservation within otherwise poorly conserved domains, of potential functional importance. Examples are elements described in Results section for the N-terminal domain of Ssk1, the linker domain of Sho1 as well as two elements in the Sko1 N-terminal part. All these elements might play roles in protein–protein interaction but this remains to be studied experimentally. In any case, these observations highlight the value of protein sequence comparison for the discovery of potentially relevant domains.
Sites for protein interactions that have been studied previously and re-investigated here include docking sites on Pbs2 for Ssk2 and Nbp2. In both instances the sites characterised in S. cerevisiae are only conserved in closely related species. There are two possible interpretations for this observation. Either Ssk2 and Nbp2 orthologues employ other sites in Pbs2 of other organisms or the sequence requirements for protein interaction have not been properly interpreted. In any case, sequence comparison calls for re-investigation of these interaction domains.
In well-characterised proteins such as Ypd1, conservation and functionality can be directly compared for individual residues. The crystal structure of this phosphotransfer protein has been reported, individually and in complex with the Sln1 response regulator, and detailed mutational analysis has been performed to characterise the interaction interface with the three yeast-response regulators, Sln1, Ssk1 and Skn7 (Xu and West 1999; Porter et al. 2003; Xu et al. 2003; Porter and West 2005). Although the correlation between conservation and functional importance is high, Asp60 stands out as a notable exception. It has been suggested that Asp60 and Arg90 are involved in the stabilisation of the structure via electrostatic interactions, but as neither of the two is conserved or replaced by residues that otherwise could form electrostatic or hydrogen–bond interactions, their role should probably be re-interpreted. The same is true for other charged residues such as Glu58, which is not conserved either.
Clues to the cues
In spite of over a decade of intense studies, the input of the HOG pathway has so far eluded characterisation. Although input signals for both branches most probably are generated at the plasma membrane, the mechanism of perception is unknown. While sequence comparisons alone are inconclusive as to the fundamental question of osmosensing, they highlight features that may play important roles in signal transmission.
In the case of Sln1, we noticed a conserved stretch of hydrophobic residues in the extracellular domain. Presumably, this domain provides a leucine zipper for dimerisation, as previously reported for its bacterial counterpart, EnvZ (Yaku and Mizuno 1997). This is consistent with the observation that a leucine zipper introduced in place of TMD1 and the extracellular domain renders Sln1 active (Ostrander and Gorman 1999), which in turn led to the suggestion that the extracellular domain is indeed needed for dimerisation. The fact that the zipper domain found in Sln1 stretches out over 41 residues and contains ten hydrophobic amino acids in conserved positions indicates that it may have a more specific function beyond dimerisation alone. Perhaps, the zipper domains of two Sln1 molecules can interact at different registers upon osmotic changes, which in turn may result in tilts of the TMDs and a conformational change at the inside of the cell, directly influencing the activation state of the dimer. A detailed analysis of the putative zipper might reveal more functional details and alternative explanations. Hence, sequence comparison emphasises the possible importance of a domain previously recognised and assists the design of a mutational analysis.
Along a similar argumentation the domain of Sho1 containing four TMDs may have a more specific function than previously thought. The arrangement with four TMDs is highly conserved across fungi that possess Sho1 (and those extend into the basidiomycetes), although this conservation is not apparent at the level of overall sequence identity (only four conserved residues over 160 amino acids). It is not obvious why a function for membrane anchoring should be such well conserved, and it stands in stark contrast to the findings that the function of Sho1 can be taken over by an engineered protein consisting of any membrane anchor that directs Pbs2 to the cell surface (Raitt et al. 2000).
Recent data have demonstrated that in Aspergillus nidulans Sho1 does not interact with Pbs2 (Furukawa et al. 2005). This is due to the fact that Pbs2 of this organism is lacking the proline-rich (PXXP) motif that is required for the interaction between Sho1 and Pbs2. However, Sho1 of A. nidulans is conserved and can interact with Pbs2 from S. cerevisiae. Interestingly, the sequence KPLPPLP in Pbs2 is well conserved only in yeasts, while filamentous fungi as well as C. albicans and Y. lipolytica, which are quite distinct from S. cerevisiae, do not have a domain in this position that could interact with Sho1. Experimental studies are needed to confirm if indeed Pbs2 is only activated via the Sln1 branch in such organisms. Such a scenario, strongly suggested by the studies in A. nidulans, would have two consequences. First, it suggests that the link of the Sho1 branch, which also plays a role in pseudohyphal development, to the osmosensing HOG pathway is not a generic feature of the architecture of this signalling system in fungi. This was already implicated by the observation that Sz. pombe lacks a Sho1 orthologue. The observation that fungi possessing Sho1 do not use it for osmostress signalling through the HOG pathway further suggests that the function of Sho1 is not primarily in osmosensing. It has already been suggested that the Sho1 branch in S. cerevisiae does not respond to turgor changes (Reiser et al. 2003). A potentially appealing speculation is that the Sho1 branch monitors mechanical stimuli dependent on surface growth of fungi and hence is involved in directing growth of fungal filaments. Such mechanical stimuli may be physically similar to those caused by osmotic changes and therefore yeasts might have developed a link of this pathway to the osmosensing system. It should be noted that the role of Sho1 and its different domains in yeast pseudohyphal growth are not well studied, while it is known that Sho1 is required for pseudohyphal development (O’Rourke and Herskowitz 1998).
Conclusion
Taken together, sequence comparison of orthologues from fungal species is a useful tool for functional analyses and will become even more interesting with further genome sequences becoming available. We suggest that functional protein analysis of fungal proteins should now always be aided by comparative studies both for the design of mutational studies as well as for the interpretation of the effect of mutations or of structural information.
References
Alepuz PM, Jovanovic A, Reiser V, Ammerer G (2001) Stress-induced MAP kinase Hog1 is part of transcription activation complexes. Mol Cell 7:767–777
Alepuz PM, de Nadal E, Zapater M, Ammerer G, Posas F (2003) Osmostress-induced transcription by Hot1 depends on a Hog1-mediated recruitment of the RNA Pol II. EMBO J 22:2433–2442
Alonso-Monge R, Navarro-Garcia F, Roman E, Negredo AI, Eisman B, Nombela C, Pla J (2003) The Hog1 mitogen-activated protein kinase is essential in the oxidative stress response and chlamydospore formation in Candida albicans. Eukaryot Cell 2:351–361
Aravind L, Ponting CP (1999) The cytoplasmic helical linker domain of receptor histidine kinase and methyl-accepting proteins is common to many prokaryotic signalling proteins. FEMS Microbiol Lett 176:111–116
Attwood TK, Croning MD, Flower DR, Lewis AP, Mabey JE, Scordis P, Selley JN, Wright W (2000) PRINTS-S: the database formerly known as PRINTS. Nucleic Acids Res 28:225–227
Bateman A, Birney E, Cerruti L, Durbin R, Etwiller L, Eddy SR, Griffiths-Jones S, Howe KL, Marshall M, Sonnhammer EL (2002) The Pfam protein families database. Nucleic Acids Res 30:276–280
Bell M, Capone R, Pashtan I, Levitzki A, Engelberg D (2001) Isolation of hyperactive mutants of the MAPK p38/Hog1 that are independent of MAPK kinase activation. J Biol Chem 276:25351–25358
Borkovich KA, Alex LA, Yarden O, Freitag M, Turner GE, Read ND, Seiler S, Bell-Pedersen D, Paietta J, Plesofsky N, Plamann M, Goodrich-Tanrikulu M, Schulte U, Mannhaupt G, Nargang FE, Radford A, Selitrennikoff C, Galagan JE, Dunlap JC, Loros JJ, Catcheside D, Inoue H, Aramayo R, Polymenis M, Selker EU, Sachs MS, Marzluf GA, Paulsen I, Davis R, Ebbole DJ, Zelter A, Kalkman ER, O’Rourke R, Bowring F, Yeadon J, Ishii C, Suzuki K, Sakai W, Pratt R (2004) Lessons from the genome sequence of Neurospora crassa: tracing the path from genomic blueprint to multicellular organism. Microbiol Mol Biol Rev 68:1–108
Brachat S, Dietrich FS, Voegeli S, Zhang Z, Stuart L, Lerch A, Gates K, Gaffney T, Philippsen P (2003) Reinvestigation of the Saccharomyces cerevisiae genome annotation by comparison to the genome of a related fungus: Ashbya gossypii. Genome Biol 4:R45
Catlett NL, Yoder OC, Turgeon BG (2003) Whole-genome analysis of two-component signal transduction genes in fungal pathogens. Eukaryot Cell 2:1151–1161
Cliften P, Sudarsanam P, Desikan A, Fulton L, Fulton B, Majors J, Waterston R, Cohen BA, Johnston M (2003) Finding functional features in Saccharomyces genomes by phylogenetic footprinting. Science 301:71–76
de Nadal E, Alepuz PM, Posas F (2002) Dealing with osmostress through MAP kinase activation. EMBO Rep 3:735–740
de Nadal E, Casadome L, Posas F (2003) Targeting the MEF2-like transcription factor Smp1 by the stress-activated Hog1 mitogen-activated protein kinase. Mol Cell Biol 23:229–237
Dietrich FS, Voegeli S, Brachat S, Lerch A, Gates K, Steiner S, Mohr C, Pohlmann R, Luedi P, Choi S, Wing RA, Flavier A, Gaffney TD, Philippsen P (2004) The Ashbya gossypii genome as a tool for mapping the ancient Saccharomyces cerevisiae genome. Science 304:304–307
Dujon B, Sherman D, Fischer G, Durrens P, Casaregola S, Lafontaine I, De Montigny J, Marck C, Neuveglise C, Talla E, Goffard N, Frangeul L, Aigle M, Anthouard V, Babour A, Barbe V, Barnay S, Blanchin S, Beckerich JM, Beyne E, Bleykasten C, Boisrame A, Boyer J, Cattolico L, Confanioleri F, De Daruvar A, Despons L, Fabre E, Fairhead C, Ferry-Dumazet H, Groppi A, Hantraye F, Hennequin C, Jauniaux N, Joyet P, Kachouri R, Kerrest A, Koszul R, Lemaire M, Lesur I, Ma L, Muller H, Nicaud JM, Nikolski M, Oztas S, Ozier-Kalogeropoulos O, Pellenz S, Potier S, Richard GF, Straub ML, Suleau A, Swennen D, Tekaia F, Wesolowski-Louvel M, Westhof E, Wirth B, Zeniou-Meyer M, Zivanovic I, Bolotin-Fukuhara M, Thierry A, Bouchier C, Caudron B, Scarpelli C, Gaillardin C, Weissenbach J, Wincker P, Souciet JL (2004) Genome evolution in yeasts. Nature 430:35–44
Dutta R, Inouye M (2000) GHKL, an emergent ATPase/kinase superfamily. Trends Biochem Sci 25:24–28
Dutta R, Qin L, Inouye M (1999) Histidine kinases: diversity of domain organization. Mol Microbiol 34:633–640
Elion EA (2000) Pheromone response, mating and cell biology. Curr Opin Microbiol 3:573–581
Fabre E, Muller H, Therizols P, Lafontaine I, Dujon B, Fairhead C (2004) Comparative genomics in hemiascomycete yeasts: evolution of sex, silencing and subtelomeres. Mol Biol Evol 22:856–873
Furukawa K, Hoshi Y, Maeda T, Nakajima T, Abe K (2005) Aspergillus nidulans HOG pathway is activated only by two-component signalling pathway in response to osmotic stress. Mol Microbiol 56 (in press)
Hiramoto F, Nomura N, Furumai T, Igarashi Y, Oki T (2005) Pradimicin resistance of yeast is caused by a mutation of the putative N-glycosylation sites of osmosensor protein sln1. Biosci Biotechnol Biochem 69:238–241
Hohmann S (2002) Osmotic stress signaling and osmoadaptation in yeasts. Microbiol Mol Biol Rev 66:300–372
Hull CM, Heitman J (2002) Genetics of Cryptococcus neoformans. Annu Rev Genet 36:557–615
Jones T, Federspiel NA, Chibana H, Dungan J, Kalman S, Magee BB, Newport G, Thorstenson YR, Agabian N, Magee PT, Davis RW, Scherer S (2004) The diploid genome sequence of Candida albicans. Proc Natl Acad Sci USA 101:7329–7334
Kellis M, Patterson N, Endrizzi M, Birren B, Lander ES (2003) Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature 423:241–254
Kellis M, Birren BW, Lander ES (2004) Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature 428:617–624
Koszul R, Caburet S, Dujon B, Fischer G (2004) Eucaryotic genome evolution through the spontaneous duplication of large chromosomal segments. EMBO J 23:234–243
Krantz M, Becit E, Hohmann S (2006) Comparative genomics of the HOG-signalling system in fungi. Curr Genet DOI 10.1007/s00294-005-0038-x
Lafontaine I, Fischer G, Talla E, Dujon B (2004) Gene relics in the genome of the yeast Saccharomyces cerevisiae. Gene 335:1–17
Langkjaer RB, Cliften PF, Johnston M, Piskur J (2003) Yeast genome duplication was followed by asynchronous differentiation of duplicated genes. Nature 421:848–852
Maeda T, Wurgler-Murphy SM, Saito H (1994) A two-component system that regulates an osmosensing MAP kinase cascade in yeast. Nature 369:242–245
Mapes J, Ota IM (2004) Nbp2 targets the Ptc1-type 2C Ser/Thr phosphatase to the HOG MAPK pathway. Embo J 23:302–311
Marles JA, Dahesh S, Haynes J, Andrews BJ, Davidson AR (2004) Protein-protein interaction affinity plays a crucial role in controlling the Sho1p-mediated signal transduction pathway in yeast. Mol Cell 14:813–823
Moller S, Croning MD, Apweiler R (2001) Evaluation of methods for the prediction of membrane spanning regions. Bioinformatics 17:646–653
Nelson B, Parsons AB, Evangelista M, Schaefer K, Kennedy K, Ritchie S, Petryshen TL, Boone C (2004) Fus1p interacts with components of the HOG1p mitogen-activated protein kinase and Cdc42p morphogenesis signaling pathways to control cell fusion during yeast mating. Genetics 166:67–77
O’Rourke SM, Herskowitz I (1998) The Hog1 MAPK prevents cross talk between the HOG and pheromone response MAPK pathways in Saccharomyces cerevisiae. Genes Dev 12:2874–2886
O’Rourke SM, Herskowitz I, O’Shea EK (2002) Yeast go the whole HOG for the hyperosmotic response. Trends Genet 18:405–412
Ostrander DB, Gorman JA (1999) The extracellular domain of the Saccharomyces cerevisiae Sln1p membrane osmolarity sensor is necessary for kinase activity. J Bacteriol 181:2527–2534
Pascual-Ahuir A, Posas F, Serrano R, Proft M (2001) Multiple levels of control regulate the yeast cAMP-response element-binding protein repressor Sko1p in response to stress. J Biol Chem 276:37373–37378
Pearson WR (1990) Rapid and sensitive sequence comparison with FASTP and FASTA. Methods Enzymol 183:63–98
Porter SW, West AH (2005) A common docking site for response regulators on the yeast phosphorelay protein YPD1. Biochim Biophys Acta 1748:138–145
Porter SW, Xu Q, West AH (2003) Ssk1p response regulator binding surface on histidine-containing phosphotransfer protein Ypd1p. Eukaryot Cell 2:27–33
Posas F, Saito H (1997) Osmotic activation of the HOG MAPK pathway via Ste11p MAPKKK: scaffold role of Pbs2p MAPKK. Science 276:1702–1705
Posas F, Saito H (1998) Activation of the yeast SSK2 MAP kinase kinase kinase by the SSK1 two-component response regulator. EMBO J 17:1385–1394
Posas F, Wurgler-Murphy SM, Maeda T, Witten EA, Thai TC, Saito H (1996) Yeast HOG1 MAP kinase cascade is regulated by a multistep phosphorelay mechanism in the SLN1-YPD1-SSK1 “two-component” osmosensor. Cell 86:865–875
Proft M, Struhl K (2002) Hog1 kinase converts the Sko1-Cyc8-Tup1 repressor complex into an activator that recruits SAGA and SWI/SNF in response to osmotic stress. Mol Cell 9:1307–1317
Proft M, Pascual-Ahuir A, de Nadal E, Arino J, Serrano R, Posas F (2001) Regulation of the Sko1 transcriptional repressor by the Hog1 MAP kinase in response to osmotic stress. EMBO J 20:1123–1133
Raitt DC, Posas F, Saito H (2000) Yeast Cdc42 GTPase and Ste20 PAK-like kinase regulate Sho1-dependent activation of the Hog1 MAPK pathway. EMBO J 19:4623–4631
Ramezani-Rad M (2003) The role of adaptor protein Ste50-dependent regulation of the MAPKKK Ste11 in multiple signalling pathways of yeast. Curr Genet 43:161–170
Reiser V, Salah SM, Ammerer G (2000) Polarized localization of yeast Pbs2 depends on osmostress, the membrane protein Sho1 and Cdc42. Nat Cell Biol 2:620–627
Reiser V, Raitt DC, Saito H (2003) Yeast osmosensor Sln1 and plant cytokinin receptor Cre1 respond to changes in turgor pressure. J Cell Biol 161:1035–1040
Rep M, Reiser V, Gartner U, Thevelein JM, Hohmann S, Ammerer G, Ruis H (1999a) Osmotic stress-induced gene expression in Saccharomyces cerevisiae requires Msn1p and the novel nuclear factor Hot1p. Mol Cell Biol 19:5474–5485
Rep M, Reiser V, Holzmüller U, Thevelein JM, Hohmann S, Ammerer G, Ruis H (1999b) Osmotic stress-induced gene expression in Saccharomyces cerevisiae requires Msn1p and the novel nuclear factor Hot1p. Mol Cell Biol 19:5474–5485
Rep M, Krantz M, Thevelein JM, Hohmann S (2000) The transcriptional response of Saccharomyces cerevisiae to osmotic shock. Hot1p and Msn2p/Msn4p are required for the induction of subsets of high osmolarity glycerol pathway-dependent genes. J Biol Chem 275:8290–8300
Rep M, Proft M, Remize F, Tamas M, Serrano R, Thevelein JM, Hohmann S (2001) The Saccharomyces cerevisiae Sko1p transcription factor mediates HOG pathway-dependent osmotic regulation of a set of genes encoding enzymes implicated in protection from oxidative damage. Mol Microbiol 40:1067–1083
Saito H, Tatebayashi K (2004) Regulation of the osmoregulatory HOG MAPK cascade in yeast. J Biochem (Tokyo) 136:267–272
Santos JL, Shiozaki K (2001) Fungal histidine kinases. Sci STKE 2001:RE1
Seet BT, Pawson T (2004) MAPK signaling: Sho business. Curr Biol 14:R708–710
Sharma P, Mondal AK (2005) Evidence that C-terminal non-kinase domain of Pbs2p has a role in high osmolarity-induced nuclear localization of Hog1p. Biochem Biophys Res Commun 328:906–913
Sheikh-Hamad D, Gustin MC (2004) MAP kinases and the adaptive response to hypertonicity: functional preservation from yeast to mammals. Am J Physiol Renal Physiol 287:F1102–1110
Smith DA, Nicholls S, Morgan BA, Brown AJ, Quinn J (2004) A conserved stress-activated protein kinase regulates a core stress response in the human pathogen Candida albicans. Mol Biol Cell 15:4179–4190
Suzuki M, Brenner SE (1995) Classification of multi-helical DNA-binding domains and application to predict the DBD structures of sigma factor, LysR, OmpR/PhoB, CENP-B, Rapl, and Xy1S/Ada/AraC. FEBS Lett 372:215–221
Tao W, Malone CL, Ault AD, Deschenes RJ, Fassler JS (2002) A cytoplasmic coiled-coil domain is required for histidine kinase activity of the yeast osmosensor, SLN1. Mol Microbiol 43:459–473
Tatebayashi K, Takekawa M, Saito H (2003) A docking site determining specificity of Pbs2 MAPKK for Ssk2/Ssk22 MAPKKKs in the yeast HOG pathway. Embo J 22:3624–3634
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Westfall PJ, Ballon DR, Thorner J (2004) When the stress of your environment makes you go HOG wild. Science 306:1511–1512
White SH, von Heijne G (2004) The machinery of membrane protein assembly. Curr Opin Struct Biol 14:397–404
Wolanin PM, Thomason PA, Stock JB (2002) Histidine protein kinases: key signal transducers outside the animal kingdom. Genome Biol 3:REVIEWS3013
von Heijne G (1989) Control of topology and mode of assembly of a polytopic membrane protein by positively charged residues. Nature 341:456–458
Wood V, Gwilliam R, Rajandream MA, Lyne M, Lyne R, Stewart A, Sgouros J, Peat N, Hayles J, Baker S, Basham D, Bowman S, Brooks K, Brown D, Brown S, Chillingworth T, Churcher C, Collins M, Connor R, Cronin A, Davis P, Feltwell T, Fraser A, Gentles S, Goble A, Hamlin N, Harris D, Hidalgo J, Hodgson G, Holroyd S, Hornsby T, Howarth S, Huckle EJ, Hunt S, Jagels K, James K, Jones L, Jones M, Leather S, McDonald S, McLean J, Mooney P, Moule S, Mungall K, Murphy L, Niblett D, Odell C, Oliver K, O’Neil S, Pearson D, Quail MA, Rabbinowitsch E, Rutherford K, Rutter S, Saunders D, Seeger K, Sharp S, Skelton J, Simmonds M, Squares R, Squares S, Stevens K, Taylor K, Taylor RG, Tivey A, Walsh S, Warren T, Whitehead S, Woodward J, Volckaert G, Aert R, Robben J, Grymonprez B, Weltjens I, Vanstreels E, Rieger M, Schafer M, Muller-Auer S, Gabel C, Fuchs M, Dusterhoft A, Fritzc C, Holzer E, Moestl D, Hilbert H, Borzym K, Langer I, Beck A, Lehrach H, Reinhardt R, Pohl TM, Eger P, Zimmermann W, Wedler H, Wambutt R, Purnelle B, Goffeau A, Cadieu E, Dreano S, Gloux S, Lelaure V, Mottier S, Galibert F, Aves SJ, Xiang Z, Hunt C, Moore K, Hurst SM, Lucas M, Rochet M, Gaillardin C, Tallada VA, Garzon A, Thode G, Daga RR, Cruzado L, Jimenez J, Sanchez M, del Rey F, Benito J, Dominguez A, Revuelta JL, Moreno S, Armstrong J, Forsburg SL, Cerutti L, Lowe T, McCombie WR, Paulsen I, Potashkin J, Shpakovski GV, Ussery D, Barrell BG, Nurse P, Cerrutti L (2002) The genome sequence of Schizosaccharomyces pombe. Nature 415:871–880
Xu Q, West AH (1999) Conservation of structure and function among histidine-containing phosphotransfer (HPt) domains as revealed by the crystal structure of YPD1. J Mol Biol 292:1039–1050
Xu Q, Porter SW, West AH (2003) The Yeast YPD1/SLN1 Complex. Insights into molecular recognition in two-component signaling systems. Struct (Camb) 11:1569–1581
Yaakov G, Bell M, Hohmann S, Engelberg D (2003) Combination of two activating mutations in one HOG1 gene forms hyperactive enzymes that induce growth arrest. Mol Cell Biol 23:4826–4840
Yaku H, Mizuno T (1997) The membrane-located osmosensory kinase, EnvZ, that contains a leucine zipper-like motif functions as a dimer in Escherichia coli. FEBS Lett 417:409–413
Zarrinpar A, Bhattacharyya RP, Nittler MP, Lim WA (2004) Sho1 and Pbs2 act as coscaffolds linking components in the yeast high osmolarity MAP kinase pathway. Mol Cell 14:825–832
Acknowledgements
This work was supported by the QUASI EC-funded project (contract LSHG-CT2003–530203). SH is a recipient of a research position from the Swedish Research Council.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by J. Hasek
Rights and permissions
About this article
Cite this article
Krantz, M., Becit, E. & Hohmann, S. Comparative analysis of HOG pathway proteins to generate hypotheses for functional analysis. Curr Genet 49, 152–165 (2006). https://doi.org/10.1007/s00294-005-0039-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00294-005-0039-9