Abstract
Cells of Chlamydomonas reinhardtii undergo gametogenesis to produce sexually competent gametes under nitrogen-starved conditions. By using a synchronized system for gametogenesis of early G1 cells, several previously identified marker genes and 18 novel nitrogen-starved gametogenesis (NSG) genes isolated by macroarray analysis were placed into at least three temporal classes of expression. Early genes are induced transiently in the first 2 h after transfer to nitrogen-free medium. Middle genes are strongly induced between 3 h and 4 h after nitrogen removal, a time corresponding to the acquisition of mating competency, suggesting their involvement in the gamete program. Late genes are induced between 5 h and 8 h after nitrogen removal, a time after the completion of gametic differentiation, suggesting that they are not directly involved in the formation of sexually competent gametes. All of the 18 NSG genes examined are induced in both mating-type plus and minus gametes and about two-thirds of the genes are also expressed in the mitotic cell cycle, especially at S/M phases.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Nitrogen starvation triggers the formation of specialized cell types in many organisms. In lower eukaryotes such as yeasts (Davey 1998; Kassi et al. 2003), Aspergillus (Skromne et al. 1995), and Neurospora (Nelson and Metzenberg 1992), nitrogen deprivation in the growth medium induces the morphogenesis of asexual or sexual spores. Higher plants respond to a reduced availability of nitrogen with changes in their developmental pattern, including the root architecture and early flowering (Zhang and Forde 2000).
In the unicellular green alga Chlamydomonas reinhardtii, removal of a utilizable nitrogen source, especially ammonium ions (Matsuda et al. 1992), from the growth medium induces gametic differentiation and the resulting mating-type plus (mt+) and minus (mt−) gametes undergo mating, when they are mixed, to form zygotes (Sager and Granick 1954). Some laboratory strains of C. reinhardtii also require light for this gametic differentiation (Beck and Haring 1996; Saito et al. 1998).
When nitrogen is depleted, the vegetative cells undergo two critical programs (Beck and Haring 1996; Goodenough 1991). First, they acclimate to nitrogen starvation through a variety of metabolic changes, including the synthesis of nitrogen-scavenging enzymes (Quesada and Fernández 1994; Vallon et al. 1993), the degradation and renewal of ribosomes (Martin et al. 1976; Siersma and Chiang 1971), and a decrease in photosynthetic activity (Bulté and Wollman 1992). Second, cells express a gamete program that produces cells competent for mating. Changes that occur during this gametogenesis program include the formation of mating-type specific agglutinins responsible for cell–cell recognition and adhesion (Adair et al. 1982; Saito and Matsuda 1984), increased synthesis of a matrix metalloprotease (gametolysin) and its activation enzyme responsible for removal of gametic cell walls (Kinoshita et al. 1992; Matsuda 1998; Snell et al. 1989), and formation of mating structures (which in mt+ gametes contain the gamete membrane protein FUS1) responsible for protoplasmic fusion (Ferris et al. 1996; Friedmann et al. 1968; Goodenough et al. 1982; Misamore et al. 2003).
The transcriptional activation of a number of genes by nitrogen starvation has been analyzed (Beck and Haring 1996). The NIA1, NRT2;1 (Quesada and Fernández 1994), NIT2 (Schnell and Lefebvre 1993), and NCG2 (Merchán et al. 2001) genes are involved in the adaptation program and they are induced quite rapidly after transfer to nitrogen-free (−N) medium. The FUS1 (Ferris et al. 1996), MID (Ferris and Goodenough 1997), MTA1, and MTA2 (Ferris et al. 2001, 2002) genes are involved in the gamete program, although their temporal pattern of gene expression during gametogenesis has not yet been analyzed. What types of regulatory networks of transcription exist among the genes for the adaptation program and the gamete program are still unknown.
Gene expression during gametogenesis in Chlamydomonas has been analyzed using a heterogeneous population of cells (Merchán et al. 2001; Rodriguez et al. 1999; von Gromoff and Beck 1993). However, in order to investigate the transcriptional networks of gametogenesis, a large and rather homogeneous population of cells is required, as in the case of the study on the transcriptional program of sporulation using a synchronized population of budding yeast (Chu et al. 1998). We found previously that only vegetative cells in the early G1 phase can directly undergo gametogenesis in −N medium (Matsuda et al. 1990). In the present work, we synchronized the vegetative cells by exposing them to alternating periods of light and darkness (Harris 1989) and the resulting early G1 cells were transferred to −N medium to analyze gene expression during synchronized gametogenesis. Besides using genes which have been identified previously (see above) as probes, we used 18 novel gametogenesis genes designated nitrogen-starved gametogenesis (NSG; Table 1), isolated by expressed sequence tag (EST)-based macroarray analysis.
Materials and methods
Cells and culture conditions
C. reinhardtii strains 11-32b (mt+) and C-9 (mt−) were used (Kubo et al. 2002). The 11-32b mt+ strain is known to be suitable for synchronous culture (Schlösser 1981, 1984) and for synchronous gametogenesis (Matsuda et al. 1990). Cells were synchronized in minimal (M) salt medium containing 3.7 mM ammonium nitrate (Sager and Granick 1953) under a cycle of 12 h light and 12 h dark (Matsuda et al. 1995). Synchronized vegetative cells at the early G1 stage (i.e., cells at the beginning of the light period, L-0 cells) were harvested and resuspended either in nitrogen-containing (+N) M medium to culture vegetatively at 25°C under continuous illumination or in nitrogen-free (−N) M medium to induce gametes (Matsuda et al. 1990). Gametic differentiation of cells was monitored by mixing them in equal numbers with tester gametes of the opposite mating type and incubating at 25°C for 30 min. The mating efficiency (the cell fusion) was calculated after counting the proportion of biflagellated and quadriflagellated cells (Matsuda et al. 1978).
Identification of genes for nitrogen-starved gametogenesis using cDNA macroarray analyses
A Chlamydomonas cDNA macroarray was constructed from Kazusa EST libraries representing cells grown under 25 different conditions, including gametogenesis and zygote formation (Asamizu et al. 1999, 2004; Miura et al. 2004). A total of 10,368 PCR-amplified EST clones were spotted in duplicate on Biodyne-A Nylon membranes. Then, [32P]-labeled targets were prepared from two mRNA pools derived from +N (target A) and −N (target B) cell mixtures, respectively. The L-0 cells of both mating-types were cultured separately in either +N or −N medium at 1.2×107 cells/ml at 25°C in the light. Aliquots (20 ml) were taken at 1 h intervals until 8 h incubation, the cells being harvested quickly by centrifugation and then frozen in liquid nitrogen. They were then mixed together and total RNA was prepared from the +N and −N cell mixtures by the methods described by Kubo et al. (2001). Polyadenylated [poly(A)+] RNA was isolated using a PolyATtract mRNA isolation system (Promega). The poly(A)+ mRNAs were labeled by incorporation of [32P]dCTP during first-strand cDNA synthesis and labeled cDNA products were purified with a CHROMA Spin-200 column (Clontech). Hybridization of macroarray filters with the labeled targets was carried out using an ExpressHyb hybridization solution (Clontech) at 68°C for 12–16 h (Sambrook and Russell 2001). After incubation and washing, the membranes were exposed to an imaging plate (Fuji Film) for detection. Quantification of signals was carried out using a FLA-2000 high-resolution scanner (Fuji Film) and ArrayVision software (Amersham). From two independent experiments, 102 ESTs were primarily isolated, whose average expression ratio of target B (−N) to target A (+N) exceeded 2.5-fold. To select genes that are expressed specifically upon a shift to −N medium, we generated contigs, assayed Northern blotting of the isolated EST clones, and identified 18 novel genes whose expression ratio of −N/+N exceeded 10-fold (cf. Fig. 3). These were designated as NSG1–NSG18 (Table 1).
Northern blot analyses
Total RNA (10 μg RNA per lane) was analyzed by Northern blotting (Sambrook and Russell 2001). The NSG gene probes for the hybridizations were prepared by a Gene Images random-prime labeling module (Amersham) using cDNA clones (Table 1) as the templates. The primer pairs used for PCR were T3-kobe (5′-CGCAATTAACCCTCACTAAAGGGAAC-3′) and M13-20-kobe (5′-GACGTTGTAAAACGACGGCCAGT-3′). The following fragments of previously identified genes were used to prepare probes for hybridization: (1) NIA1 (Fernández et al. 1989), the 0.35-kb 3′UTR region (positions 2,921–3,265, GenBank accession no. AF203033), (2) NRT2;1 (Quesada et al. 1994), the 0.69-kb 3′UTR region (positions 2,143–2,832, accession no. Z25438), (3) NCG2 (Pozuelo et al. 2000), the 2.05-kb ORF plus 3′UTR regions (positions 189–2,239, accession no. AF195795), (4) FUS1 (Ferris et al. 1996), the 0.53-kb 3′UTR region (positions 4,022–4,545, accession no. U49864), (5) GAS28 (Rodriguez et al. 1999), the 0.37-kb 3′UTR region (positions 1,695–2,065, accession no. AF015883), and (6) MTA1 and MTA2 (Ferris et al. 2002), the 0.55-kb 5′UTR plus ORF regions of MTA1 (positions 2,778–3,629, accession no. AF417571). The L27a gene, encoding a Chlamydomonas 60S ribosomal protein, was used as a standard because of its relatively constant level of expression during gametogenesis and the cell cycle. The probe was generated by PCR using L27a cDNA (accession no. AV640287). The expression profiles of each gene were shown in a tabular form using Cluster and TreeView (Eisen et al. 1998).
cDNA sequencing
The cDNA clones were subcloned into pBluescript II SK− (Strategene) and their sequence determined using a Thermo Sequence cycle sequencing kit (Amersham) with T3 and T7 primers (Nisshinbo) and a LiC-4200 DNA sequencer (Li-Cor).
Results
Expression profiles of previously identified marker genes during synchronized gametogenesis
Using the Chlamydomonas synchronized system for gametogenesis (Fig. 1), we first examined the expression profiles of several marker genes (NIA1, NRT2;1, NCG2, FUS1, MTA1, MTA2, GAS28) which had been identified previously.
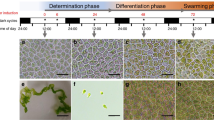
Time-course of synchronized gametogenesis of early G1 cells in −N medium. Synchronously grown mt+ cells (11-32b) at the beginning of the light period (L-0 cells) were transferred to −N or +N medium at 1.2×107 cells/ml and incubated for 8 h in the light. At the times indicated, these cells were mixed with tester mt− (C-9) gametes in equal numbers and the mating efficiency determined after 30 min incubation
The NIA1, NRT2;1, and NCG2 genes encode proteins for acclimation to nitrogen starvation and are induced rapidly and transiently after transfer to −N medium (Fernández et al. 1989; Merchán et al. 2001; Quesada and Fernández 1994). Our time-course study also showed (Fig. 2) that the transcripts of these genes were detectable 1 h after transfer to −N medium, then dropped sharply after 2 h and accumulated again, although slightly, after 7 h incubation. In +N medium containing ammonium nitrate, expression of these genes was undetectable during 6 h incubation (Fig. 2). It has been reported that ammonium and nitrate repress the expression of NIA1, NRT2;1, and NCG2 genes (Fernández et al. 1989; Pozuelo et al. 2000).
Temporal classification of gene expression during synchronized gametogenesis using previously identified marker genes NIA1, NRT2;1, NCG2, FUS1, MTA1, MTA2, and GAS28. Cell samples were taken from the synchronous culture transferred into −N or +N medium (see Fig. 1) at the indicated time-points. Total RNA (10 μg/lane) was used for Northern blot analysis. Probes are described in the Materials and methods. The size of the RNA is indicated on the right in kilobases. The L27a gene (encoding ribosomal protein L27a) was used as a loading control
The FUS1 gene is unique to the mt+ chromosome (Ferris and Goodenough 1994) and encodes a sex recognition protein called fringe (Ferris et al. 1996). In our gametogenetic system, the levels of FUS1 mRNA increased markedly at around 3 h incubation (Fig. 2), the time corresponding to when cells were just developing into gametes (Fig. 1). The transcripts for the two other gamete program genes, MTA1 and MTA2 (Ferris et al. 2001, 2002), were also accumulated transiently between 3 h and 4 h.
The GAS28 gene is expressed in the late phase of gametogenesis and is assumed to encode a hydroxyproline-rich glycoprotein of the zygotic cell wall (Rodriguez et al. 1999; von Gromoff and Beck 1993). We found that the steady-state levels of GAS28 mRNA increase between 6 h and 7 h (Fig. 2), the time corresponding to when cells have already become fully competent gametes (Fig. 1).
The above results indicate that gametogenesis in Chlamydomonas is characterized by the sequential expression of at least three sets of genes—early, middle, and late (Fig. 2). Early genes such as NIA1, NRT2;1, and NCG2 are induced in the first 2 h after transfer to −N medium. Middle genes such as FUS1, MTA1, and MTA2 are induced between 3 h and 4 h, paralleling the acquisition of mating competency. Late genes like GAS28 are induced between 5 h and 8 h, corresponding to the period after completion of gametic differentiation.
Expression profiles of novel NSG genes during synchronized gametogenesis
Figure 3 shows the steady-state levels of mRNAs for novel NSG genes, isolated by EST-based macroarray analysis, during synchronized gametogenesis. Similar to previously identified genes described above (Fig. 2), all of the 18 NSG genes showed a strong expression in cells placed under −N conditions, whereas no or very weak expression was observed in cells incubated under +N conditions for at least 6 h (Fig. 3). In −N cells, many NSG genes exhibited transient accumulation of mRNAs, but some genes such as NSG3, NSG13 and NSG18 showed sustained accumulation through the rest of the time-course.
Expression of newly isolated NSG genes during synchronized gametogenesis. Northern blot analysis was performed as in Fig. 2. Probes used are full-length cDNAs
To order genes on the basis of the time of the most significant accumulation during gametogenesis, the measured changes in mRNA levels were displayed in a graphical format (Fig. 4a). The NSG genes were then grouped according to similarities in their accumulation patterns to those of previously identified marker genes, which can be defined by three temporal classes. Class 1, the early genes: as with NIA1 and NRT2;1, NSG17 was induced transiently within 1 h after transfer to −N medium. Class 2, the middle genes: 14 NSG genes (NSG1, NSG2, NSG4–NSG12, NSG14–NSG16) followed the accumulation patterns of mRNAs characterized by FUS1, MTA1, and MTA2 genes. They were accumulated markedly and transiently between 3 h and 4 h after transfer to −N medium. In contrast, NSG3 exhibited a middle–late pattern of induction. The mRNA was accumulated significantly after 4 h incubation and the accumulation was sustained through the rest of the time-course. Class 3, the late genes: two NSG genes (NSG13, NSG18) followed a pattern of induction and accumulation characterized by GAS28, detectable strongly between 6 h and 7 h after transfer to −N medium.
The global pattern of gene expression during a gametogenesis and b mitotic cell cycle. Data shown in Figs. 2, 3, 6 are graphically displayed with color to represent the relative changes in the expression of each gene. Increases in mRNA levels are shown as shades of red. The genes that showed the most significant expression during gametogenesis are ordered so that genes with similar expression are grouped into three temporal stages: early, middle, and late (see Expression profiles of previously identified marker genes during synchronized gametogenesis, above)
Expression-specificity of NSG genes
We found that all of the NSG genes examined are also expressed during gametogenesis of mt− cells (data not shown), with similar temporal patterns to mt+ cells (Fig. 3). It was suggested, therefore, that NSG1–NSG18 are transcribed as a common process for gametic differentiation between the two mating-type cells and not as a sex-limited or sex-specific process for differentiation (Goodenough et al. 1995).
To further evaluate the expression-specificity of the NSG genes, their mRNA levels were examined during the mitotic cell cycle under +N conditions. As shown in Fig. 5, early G1 cells in synchronous culture grow during the first 12 h, undergo S and M phases between 12 h and 16 h, and liberate daughter cells after 18 h. Total RNA was isolated from cells collected every 2 h throughout the 18-h light period and used for Northern blot analysis, using previously identified genes and NSG genes as probes (Fig. 6). Figure 4b displays the global patterns of accumulation of transcripts during the mitotic cell cycle, where each gene is ordered according to the three temporal classes of mRNA accumulation during gametogenesis (Fig. 4a).
Time-course of synchronous growth and division of early G1 cells in +N medium. Synchronously grown mt+ cells at the beginning of the light period (L-0 cells) were transferred to M medium at a density of 1.6×106 cells/ml and incubated for 18 h in the light. Note that the cell density was about 8-fold less than that in Fig. 1, so that the cells could proceed through one round of the mitotic cell cycle with no apparent limitation of nutrients. Open circles Cell number, open squares dividing cells (%)
Expression of marker genes and NSG genes during the mitotic cell cycle. Cell samples were taken at the time-points indicated in Fig. 5 and used for Northern blot analysis
None of the early genes (NIA1, NRT2;1, NCG2, NSG17) showed significant expression during the mitotic cell cycle. Among the middle genes, no significant expression was observed in FUS1, MTA1, MTA2, NSG3, NSG6, and NSG7. In contrast, NSG4, NSG8, NSG9, NSG11, NSG12, and NSG14 mRNAs accumulated weakly at middle/late G1 phase (4–10 h) and significantly at S/M phase (12–14 h). NSG1, NSG2, NSG5, NSG10, and NSG16 appeared to be induced restrictively during S/M phase. NSG15 was accumulated weakly through G1 and S/M phases. Among the late genes, NSG13 mRNA was not detected at all throughout the cell cycle. In contrast, GAS28, which has been reported to be a gametogenesis-specific gene (Rodriguez et al. 1999; von Gromoff and Beck 1993), was transcribed during M phase (14–16 h). The NSG18 transcript was accumulated weakly through G1 and S/M phases.
Table 1 summarizes the transcriptional specificity of NSG genes during gametogenesis and the mitotic cell cycle.
Functional features of NSG genes
To annotate polypeptides encoded by NSG genes, we performed a BLAST analysis (Altschul et al. 1997) of each cDNA (largely 5′ end sequence) against the GenBank database and a genome analysis against the JGI Chlamydomonas genome database (Grossman et al. 2003; www.jgi.doe.gov/chlamy). All NSG genes localize to different genomic loci without linking to each other or to the mating-type locus (Table 1). Thirteen out of 18 NSG genes were annotated proteins with a BLAST e value less than 6.00×e−25, while one early gene (NSG17), four middle genes (NSG1, NSG6, NSG7, NSG11), and one late gene (NSG13) showed no matches with any entry in the databases (Table 1).
Several middle genes encode putative polypeptides related to nucleotide/protein metabolism: a small subunit (NSG2) and a large subunit (NSG5) of ribonucleotide reductase, the RPT4 (NSG8) and RPT2 (NSG9) subunits of the 26S proteasome triple-A ATPase, a dUTP pyrophosphatase (NSG10), a DEAD box RNA helicase (NSG12), a valosin-containing protein (NSG14), and a clathrin heavy chain (NSG15). Putative regulatory proteins are a reverse transcriptase homologue (NSG3) and a cyclin-dependent kinase regulatory subunit (NSG16). NSG4 encodes a Chlamydomonas actin, which functions in elongating a fertilization tubule from the mt+ gamete during mating (Sugase et al. 1996). A late gene, NSG18, encodes a Chlamydomonas glutamine synthetase1 (Chen and Silflow 1996), suggesting its involvement in the assimilation of alternative nitrogen sources.
Since we could not annotate proteins for six NSG genes by a database search using the partial nucleotide sequence (Table 1), their full-length cDNA clones were isolated and the entire nucleotide sequences determined. As diagrammed in Fig. 7, NSG1, NSG6, NSG7, NSG11, NSG13, and NSG17 were found to code for polypeptides of 382 (Mr 41,357), 363 (Mr 38,799), 770 (Mr 82,886), 312 (Mr 33,235), 587 (Mr 57,582), and 533 (Mr 53,834) amino acids, respectively. A search of the genome database revealed that the six NSG genes have 6, 7, 21, 6, 13, and 2 introns, respectively. The ORFs were then analyzed using Pfam (Bateman et al. 2002), SMART (Schultz et al. 1998), SOSUI (Hirokawa et al. 1998), PredictProtein (Rost and Liu 2003), and PREDATOR (Frishman and Argos 1997) programs.
Predicted structural features of the NSG1, NSG6, NSG7, NSG11, NSG13, and NSG17 proteins deduced from the full-length cDNA sequences. For NSG1, a predicted signal peptide (SP), a P-rich region, a T-rich region, and a potential N-linked glycosylation site (marked by a circle) are indicated. For NSG7, a P/Q/A/V-rich region and four ankyrin repeats (AR, marked I–IV) are indicated. For NSG11, a P/A-rich region and a putative actin-depolymerizing factor (ADF) domain are indicated. For NSG13, three helical transmembrane (TM) regions and four armadillo (Arm) repeats (marked I–IV) are indicated. For NSG17, a basic helix-loop-helix (bHLH) domain is indicated
NSG1 contains a predicted signal peptide (positions 1–26), as expected for a secreted protein (von Heijne 1985), a P-rich region (positions 77–90), and a T-rich region (positions 279–318; Fig. 7). No significant homology was found by a database search. NSG6 was characterized as a soluble protein with totally unknown function (Fig. 7). NSG7 contains a P/Q/A/V-rich region (positions 34–132) and four presumptive ankyrin repeats (positions 606–638, 641–673, 674–706, 708–740) at the C-terminus (Fig. 7). NSG11 has a P/A-rich region (positions 79–141) and a C-terminal domain (positions 172–306) which is highly homologous to an actin-depolymerizing factor (ADF; Fig. 7). Comparison with other members of the ADF/cofilin family demonstrated an amino acid identity of 36–45% and similarity of 60–70%, with the highest identity to Acanthamoeba actophorin (Quirk et al. 1993). Furthermore, the deduced secondary structure and a putative phosphorylation site (Ser-172) within the ADF domain of NSG11 are highly conserved among the ADF/cofilin family. NSG13 is predicted to contain three helical transmembrane regions (positions 115–136, 156–178, 182–203) and four armadillo/beta-catenin (Arm) repeats (positions 243–285, 286–326, 327–368, 452–494) toward the C-terminus (Fig. 7). Finally, NSG17 codes for a putative transcription factor with a basic helix-loop-helix (bHLH) DNA-binding domain (positions 137–192; Fig. 7). The bHLH domain showed highest identity (54%) to that of a putative DNA-binding protein of Arabidopsis thaliana (accession no. NP_177064).
Discussion
Utilizing a synchronized system, important information on the temporal program of gene expression during gametic differentiation was obtained. Using several previously identified marker genes and 18 newly identified NSG genes, we determined their temporal patterns of induction and classified all of them into three temporal classes (Table 1). Since genes with related functions appear to be induced in similar patterns, we speculate about possible roles for the NSG genes, based on their temporal association with the marker genes of known functions. All NSG genes that are described in this report may be “autosomal” (Goodenough et al. 1995), since they are expressed in gametes of both mating-types in response to nitrogen starvation.
Properties of an early NSG gene
NSG17 was classified as an early gene that followed the expression pattern of NIA1 and NRT2;1 during gametogenesis with no apparent expression during the vegetative cell cycle (Fig. 4, Table 1). This gene codes for a putative transcription factor with a bHLH domain similar to those that bind to DNA and exert a determinative influence in various developmental pathways of eukaryotes (Morgenstern and Atchley 1999). In the fission yeast, the esc1+ gene is induced early in response to nitrogen starvation and codes for a putative transcription factor with a bHLH domain that promotes sexual differentiation of Schizosaccharomyces pombe (Benton et al. 1993). It is of interest to examine whether NSG17 also promotes the early stage of sexual differentiation in C. reinhardtii.
Properties of middle NSG genes
Fifteen NSG genes were identified as middle genes (Fig. 4, Table 1) and considered to be of great importance in generating the gametic phenotypes. One of the genes, NSG4, encodes actin (Sugase et al. 1996). It is known in C. reinhardtii that actin polymerization accompanies elongation of the fertilization tubule from the mt+ gametes during mating with mt− gametes (Detmers et al. 1983, 1985). The Chlamydomonas genome has only a single actin-encoding gene (Sugase et al. 1996) and its mutation, originally marked by the ida5 mutation (Kato et al. 1993), causes slow swimming due to a lack of functional flagellar inner-arm dynein and also causes deficient growth of the fertilization tubule during mating (Kato-Minoura et al. 1997). The cytoplasmic actin displays dynamic reorganization during mitosis and cytokinesis in the cell cycle (Harper et al. 1992). Not surprisingly, the actin-encoding gene, NSG4, is expressed throughout the Chlamydomonas life cycle, as clearly seen by its expression both during the middle stage of gametogenesis and also during the S/M phase of the vegetative cell cycle (Fig. 4). It is of interest that a putative ADF gene, NSG11 (Fig. 7), follows the accumulation patterns of NSG4 in both gametic and vegetative stages (Fig. 4). ADF/cofilins are a family of actin-binding proteins in all eukaryotic cells and have the ability to remodel the cytoskeleton by severing and depolymerizing actin filaments (Maciver and Hussey 2002). Detmers et al. (1983) observed a rapid breakdown of the fertilization tubule cytoskeleton in the zygote after gametic cell fusion is completed, suggesting a possible role of NSG11 in actin remodeling after cell fusion. The ADF domain of NSG11 most closely resembles the Acantamoeba actopholin and higher plant ADF/cofilins, although NSG11 contains a unique, long N-terminal extension (170 amino acids) with unknown function (Fig. 7).
It is known that specific changes occur in the nucleotide/protein synthesis machinery during Chlamydomonas gametogenesis (Bulté and Bennoun 1990; Martin et al. 1976; Picard-Bennoun and Bennoun 1985; Siersma and Chiang 1971). Changes at the morphological level during gametogenesis have also been examined by Martin and Goodenough (1975), who observed the modification of ER and the appearance of new Golgi-derived vesicles (named gametic vesicles) prior to the appearance of mating structures in the mt+ and mt− gametes. The functions of many middle genes—NSG2 and NSG5 encoding two subunits of ribonucleotide reductase (Jordan and Reichard 1998), NSG8 and NSG9 encoding two subunits of 26S proteasome (Voges et al. 1999), NSG10 encoding a dUTP pyrophosphatase (Mol et al. 1996), NSG12 encoding a DEAD box RNA helicase (Aubourg et al. 1999), NSG14 encoding a valosin-containing protein/p97/Cdc48p (Latterich et al. 1995; Ye et al. 2001), and NSG15 encoding a clathrin heavy chain (Brodsky et al. 2001)—suggest they play a role in the specific changes that occur in the protein-synthesizing machinery during gametogenesis. Chlamydomonas cells acclimate to diverse environmental stresses, including nitrogen starvation, by the specific expression of a number of genes encoding polypeptides involved in proteolysis (Shrager et al. 2003).
NSG16 may encode a cell cycle regulatory protein, since the deduced amino acid sequence shows high homology with a subunit of cyclin-dependent kinases such as Cks1 and Suc1 in yeast (Harper 2001). We do not know at present the functional roles of NSG16 in gametes. However, since this gene and also other middle genes such as NSG 1, NSG2, NSG4, NSG5, NSG8, NSG9, NSG10, NSG11, NSG12, and NSG14 are transcribed mainly during the S/M phase in the cell cycle (Fig. 6), they all may play pivotal roles both in asexual and sexual stages of the Chlamydomonas life cycle.
In contrast, three middle genes, NSG 3, NSG6, and NSG7 appear to be specifically expressed under −N conditions, since no transcripts were detected in the vegetative cell cycle (Figs. 4, 6). The deduced amino acid sequence of NSG3 showed the highest homology to a reverse transcriptase from the copia/ty1-like retrotransposon named Lusen, from a related alga, V. carteri (Lindauer and Schmitt, unpublished data). The copia-like retrotransposons are present universally among higher and lower plants, insects, fungi, and protists (Lindauer et al. 1993; Voytas et al. 1992; Xiong and Eickbush 1990). Only a small number of plant retrotransposable elements have been shown to be active; and this activity seems to be restricted to unusual stress situations called “genomic shock” (Grandbastein 1992; McClintock 1984). It is therefore likely that NSG3, which is restrictively expressed under −N conditions, may be involved in facilitating genomic restructuring and, hence, adaptation to extreme environmental conditions (McClintock 1984). NSG6 and NSG7 encode novel proteins that are expressed only in gametes. Furthermore, the deduced amino acid sequence of a full-length cDNA clone of NSG7 contains four tandem ankyrin repeats proximal to the C-terminus (Fig. 7). Ankyrin repeats are 33-residue motifs involved in protein–protein interactions (Sedwick and Smerdon 1999). In this alga, the ZYS3 gene encodes a protein containing ankyrin repeats and is expressed only in the young zygotes (Kuriyama et al. 1999). Thus, these two proteins with ankyrin repeats may function as developmental regulators at different steps in the Chlamydomonas sexual cycle. A BLAST search of the Chlamydomonas genome database using the nucleotide sequences of NSG6 and NSG7 indicated that genes for each protein were present in the database and that ESTs for the sequences were present in libraries enriched for gamete and zygote genes and also in libraries from carbon-stressed cells. It is therefore possible that these genes could be related to stress rather than sexual development.
Properties of late NSG genes
Rodriguez et al. (1999) showed that GAS28 is expressed in the late phase of gametogenesis and hypothesized that this transcript is stored until activated for translation as a constituent of the zygote cell wall. Similarly, one of the late genes, NSG13, that encodes a putative transmembrane protein containing Arm repeats and is expressed only in gametes might be related to the zygotic stage of sexual cell cycle. Arm repeat domains are ubiquitous among eukaryotes and mediate specific protein–protein interactions for diverse functions such as cell–cell adhesion and intracellular signalling (Coates 2003; Peifer et al. 1994). In this alga, an Arm repeat protein, PF16, is required for cellular motility brought about by the flagellum (Smith and Lefebvre 1996, 2000).
NSG18 encodes a glutamine synthetase (Chen and Silflow 1996) and may be induced for acclimation of cells to a long-term starvation of nitrogen. Quesada and Fernández (1994) reported that nitrate assimilation genes such as NIA1 and NRT2;1 are transcribed transiently at the early stage and again at the late stage of gametogenesis (see also Fig. 2), while NRT2;2, a gene sharing sequence similarity with NRT2;1, is only accumulated after a long incubation period in −N medium.
The three sets of NSG genes reported here that are expressed at distinct points during synchronized gametogenesis of C. reinhardtii may be essential tools for defining their regulatory networks of gene expression, by analyzing their expression in mutants for gametic differentiation like dif (Saito and Matsuda 1991).
References
Adair WS, Monk BC, Cohen R, Hwang C, Goodenough UW (1982) Sexual agglutinins from the Chlamydomonas flagellar membrane: partial purification and characterization. J Biol Chem 257:4593–4602
Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Asamizu E, Nakamura Y, Sato S, Fukuzawa H, Tabata S (1999) A large scale structural analysis of cDNAs in a unicellular green alga, Chlamydomonas reinhardtii: generation of 3433 non-redundant expressed sequence tags. DNA Res 6:363–373
Asamizu E, Nakamura Y, Miura K, Fukuzawa H, Fujiwara S, Hirono M, Iwamoto K, Matsuda Y, Minagawa J, Shimogawara K, Takahashi Y, Tabata S (2004) Establishment of publicly available cDNA material and information resource of Chlamydomonas reinhardtii (Chlorophyta), to facilitate gene function analysis. Phycologia (in press)
Aubourg S, Kreis M, Lecharny A (1999) The DEAD box RNA helicase family in Arabidopsis thaliana. Nucleic Acids Res 27:628–636
Bateman A, Birney E, Cerruti L, Durbin R, Etwiller L, Eddy SR, Griffiths-Jones S, Howe KL, Marshall M, Sonnhammer EL (2002) The Pfam protein families database. Nucleic Acids Res 30:276–280
Beck CF, Haring MA (1996) Gametic differentiation of Chlamydomonas. Int Rev Cytol 168:259–302
Benton BK, Reid MS, Okayama H (1993) A Schizosaccharomyces pombe gene that promotes sexual differentiation encodes a helix-loop-helix protein with homology to MyoD. EMBO J 12:135–143
Brodsky FM, Chen C-Y, Knuchl C, Towler MC, Wakeham DE (2001) Biological basket weaving: formation and function of clathrin-coated vesicles. Annu Rev Cell Dev Biol 17:517–568
Bulté L, Bennoun P (1990) Translational accuracy and sexual differentiation in Chlamydomonas reinhardtii. Curr Genet 18:155–160
Bulté L, Wollman F-A (1992) Evidence for a selective destabilization of an integral membrane protein, the cytochrome b6/f complex, during gametogenesis in Chlamydomonas reinhardtii. Eur J Biochem 204:327–336
Chen Q, Silflow CD (1996) Isolation and characterization of glutamine synthetase genes in Chlamydomonas reinhardtii. Plant Physiol 112:987–996
Chu S, DeRisi J, Elsen M, Mulholland J, Botstein D, Brown PO, Herskowitz I (1998) The transcriptional program of sporulation in budding yeast. Science 282:699–705
Coates JC (2003) Armadillo repeat proteins: beyond the animal kingdom. Trends Cell Biol 13:463–471
Davey J (1998) Fusion of a fission yeast. Yeast 14:1529–1566
Detmers PA, Goodenough UW, Condeelis J (1983) Elongation of the fertilization tubule in Chlamydomonas: new observations on the core microfilaments and the effect of transient intracellular signals on their structural integrity. J Cell Biol 97:522–532
Detmers PA, Carboni JM, Condeelis J (1985) Localization of actin in Chlamydomonas using antiactin and NBD-phallacidin. Cell Motil 5:415–430
Eisen MB, Spellman PT, Brown PO, Botstein D (1998) Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA 95:14863–14868
Fernández E, Schnell R, Ranum LPW, Hussey SC, Silflow CD, Lefebvre PA (1989) Isolation and characterization of the nitrate reductase structural gene in Chlamydomonas reinhardtii. Proc Natl Acad Sci USA 86:6449–6453
Ferris PJ, Goodenough UW (1994) The mating type locus of Chlamydomonas reinhardtii contains highly rearranged DNA sequences. Cell 76:1135–1145
Ferris PJ, Goodenough UW (1997) Mating type in Chlamydomonas is specified by mid, the minus dominance gene. Genetics 146:859–869
Ferris PJ, Woessner JP, Goodenough UW (1996) A sex recognition glycoprotein is encoded by the plus mating-type gene fus1 of Chlamydomonas reinhardtii. Mol Biol Cell 7:1235–1248
Ferris PJ, Woessner JP, Waffenschmidt S, Kilz S, Drees J, Goodenough UW (2001) Glycosylated polyproline II rods with kinks as a structural motif in plant hydroxyproline-rich glycoproteins. Biochemistry 40:2978–2987
Ferris PJ, Armbrust EV, Goodenough UW (2002) Genetic structure of the mating-type locus of Chlamydomonas reinhardtii. Genetics 160:181–200
Friedmann I, Colwin AL, Colwin LH (1968) Fine structural aspects of fertilization in Chlamydomonas reinhardti. J Cell Sci 3:115–128
Frishman D, Argos P (1997) Seventy-five percent accuracy in protein secondary structure prediction. Proteins 27:329–335
Goodenough UW (1991) Chlamydomonas mating interactions. In: Dworkin M (ed) Microbial cell–cell interactions. American Society for Microbiology, Washington, D.C., pp 71–112
Goodenough UW, Detmers PA, Hwang C (1982) Activation for cell fusion in Chlamydomonas. Analysis of wild-type gametes and nonfusing mutants. J Cell Biol 92:378–386
Goodenough UW, Armbrust EV, Cambell AM, Ferris PJ (1995) Molecular genetics of sexuality in Chlamydomonas. Annu Rev Plant Physiol Plant Mol Biol 46:21–44
Grandbastein M-A (1992) Retroelements in higher plants. Trends Genet 8:103–108
Gromoff ED von, Beck CF (1993) Genes expressed during sexual differentiation of Chlamydomonas reinhardtii. Mol Gen Genet 241:415–421
Grossman AR, Harris EH, Hauser C, Lefebvre PA, Martinez D, Rokhsar D, Shrager J, Silflow CD, Stern D, Vallon O, Zhang Z (2003) Chlamydomonas reinhardtii at the crossroads of genomics. Eukaryot Cell 2:1137–1150
Harper JW (2001) Protein destruction: adapting roles for Cks proteins. Curr Biol 11:R431–R435
Harper JDI, McCurdy DW, Sanders MA, Salisbury JL, John PCL (1992) Actin dynamics during the cell cycle in Chlamydomonas reinhardtii. Cell Motil Cytoskeleton 22:117–126
Harris EH (1989) The Chlamydomonas sourcebook. Academic, San Diego
Heijne G von (1985) Signal sequences: the limits of variation. J Mol Biol 184:99–105
Hirokawa T, Boon-Chieng S, Mitaku S (1998) SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics 14:378–379
Jordan A, Reichard P (1998) Ribonucleotide reductases. Annu Rev Biochem 67:71–98
Kassi Y, Adir N, Boger-Nadjar E, Raviv NG, Rubin-Bejerano I, Sagee S, Shenhar G (2003) Transcriptional regulation of meiosis in budding yeast. Int Rev Cytol 224:111–171
Kato T, Kagami O, Yagi T, Kamiya R (1993) Isolation of two species of Chlamydomonas reinhardtii flagellar mutants, ida5 and ida6, that lack a newly identified heavy chain of the inner dynein arm. Cell Struct Funct 18:371–377
Kato-Minoura T, Hirono M, Kamiya R (1997) Chlamydomonas inner-arm dynein mutant, ida5, has a mutation in an actin-encoding gene. J Cell Biol 137:649–656
Kinoshita T, Fukuzawa H, Shimada T, Saito T, Matsuda Y (1992) Primary structure and expression of a gamete lytic enzyme in Chlamydomonas reinhardtii: similarity of functional domains to matrix metalloproteases. Proc Natl Acad Sci USA 89:4693–4697
Kubo T, Saito T, Fukuzawa H, Matsuda Y (2001) Two tandemly located matrix metalloprotease genes with different expression patterns in the Chlamydomonas sexual cell cycle. Curr Genet 40:136–143
Kubo T, Abe J, Saito T, Matsuda Y (2002) Genealogical relationships among laboratory strains of Chlamydomonas reinhardtii as inferred from matrix metalloprotease genes. Curr Genet 41:115–122
Kuriyama H, Takano H, Uchida H, Kawano S, Kuroiwa H, Kuroiwa T (1999) Characterization of Chlamydomonas reinhardtii zygote-specific cDNAs that encode novel proteins containing ankyrin repeats and WW domains. Plant Physiol 119:873–884
Latterich M, Fröhlich KU, Schekman R (1995) Membrane fusion and the cell cycle: Cdc48p participates in the fusion of ER membranes. Cell 82:885–893
Lindauer A, Fraser D, Brüderlein M, Schmitt R (1993) Reverse transcriptase families and a copia-like retrotransposon, Osser, in the green alga Volvox carteri. FEBS Lett 319:261–266
Maciver SK, Hussey PJ (2002) The ADF/cofilin family: actin-remodeling proteins. Genome Biol 5:1–12
Martin NC, Goodenough UW (1975) Gametic differentiation in Chlamydomonas reinhardti. I. Production of gametes and their fine structure. J Cell Biol 67:587–605
Martin NC, Chiang K-S, Goodenough UW (1976) Turnover of chloroplast and cytoplasmic ribosomes during gametogenesis in Chlamydomonas reinhardti. Dev Biol 51:190–201
Matsuda Y (1998) Gametolysin. In: Barrett A, Woessner F, Rawlings N (eds) Handbook of proteolytic enzymes. Academic, New York, pp 1140–1143
Matsuda Y, Tamaki S, Tsubo Y (1978) Mating type specific induction of cell wall lytic factor by agglutination of gametes in Chlamydomonas reinhardtii. Plant Cell Physiol 19:1253–1261
Matsuda Y, Saito T, Koseki M, Shimada T (1990) The Chlamydomonas non-synchronous and synchronous gametogenesis as analyzed by the activities of cell body-agglutinin and cell wall lytic enzyme (Life Sci Adv). Plant Physiol 9:1–6
Matsuda Y, Shimada T, Sakamoto Y (1992) Ammonium ions control gametic differentiation and dedifferentiation in Chlamydomonas reinhardtii. Plant Cell Physiol 33:909–914
Matsuda Y, Koseki M, Shimada T, Saito T (1995) Purification and characterization of a vegetative lytic enzyme responsible for liberation of daughter cells during the proliferation of Chlamydomonas reinhardtii. Plant Cell Physiol 36:681–689
McClintock B (1984) The significance of responses of the genome to challenge. Science 236:792–801
Merchán F, Ende H van den, Fernández E, Beck CF (2001) Low-expression genes induced by nitrogen starvation and subsequent sexual differentiation in Chlamydomonas reinhardtii, isolated by the differential display technique. Planta 213:309–317
Misamore MJ, Gupta S, Snell WJ (2003) The Chlamydomonas Fus1 protein is present on the mating type plus fusion organelle and required for critical membrane adhesion event during fusion with minus gametes. Mol Biol Cell 14:2530–2542
Miura K, Yamano T, Yoshioka S, Kohinata T, Inoue Y, Taniguchi F, Asamizu E, Nakamura Y, Tabata S, Yamato KT, Ohyama K, Fukuzawa H (2004) Expression profiling-based identification of CO2-responsive genes regulated by CCM1 controlling a carbon-concentrating mechanism in Chlamydomonas reinhardtii. Plant Physiol 135:1595–1607
Mol CD, Harris JM, McIntosh EM, Tainer JA (1996) Human dUTP pyrophosphatase: uracil recognition by a beta hairpin and active sites formed by three separate subunits. Structure 4:1077–1092
Morgenstern B, Atchley WR (1999) Evolution of bHLH transcription factors: modular evolution by domain shuffling? Mol Biol Evol 16:1654–1663
Nelson MA, Metzenberg RL (1992) Sexual development genes of Neurospora crassa. Genetics 132:149–162
Peifer M, Berg S, Reynolds AB (1994) A repeating amino acid motif shared by proteins with diverse cellular roles. Cell 76:789–791
Picard-Bennoun M, Bennoun P (1985) Change in cytoplasmic ribosome properties during gametogenesis in the alga Chlamydomonas reinhardtii. Curr Genet 9:239–243
Pozuelo M, Merchán F, Macías MI, Beck CF, Galván A, Fernández E (2000) The negative effect of nitrate on gametogenesis is independent of nitrate assimilation in Chlamydomonas reinhardtii. Planta 211:287–292
Quesada A, Fernández E (1994) Expression of nitrate assimilation related genes in Chlamydomonas reinhardtii. Plant Mol Biol 24:185–194
Quesada A, Galván A, Fernández E (1994) Identification of nitrate transporter genes in Chlamydomonas reinhardtii. Plant J 5:407–419
Quirk S, Maciver SK, Ampe C, Doberstein SK, Kaiser DA, Van Damme J, Vandekerckhove JS, Pollard TD (1993) Primary structure of and studies on Acanthamoeba actophorin. Biochemistry 32:8525–8533
Rodriguez H, Haring MA, Beck CF (1999) Molecular characterization of two light-induced, gamete-specific genes from Chlamydomonas reinhardtii that encode hydroxyproline-rich proteins. Mol Gen Genet 261:267–274
Rost B, Liu J (2003) The PredictProtein server. Nucleic Acids Res 31:3300–3304
Sager R, Granick S (1953) Nutritional studies with Chlamydomonas reinhardti. Ann N Y Acad Sci 56:831–838
Sager R, Granick S (1954) Nutritional control of sexuality in Chlamydomonas reinhardti. J Gen Physiol 37:729–742
Saito T, Matsuda Y (1984) Sexual agglutinin of mating-type minus gametes in Chlamydomonas reinhardtii: purification and characterization of minus agglutinin and comparison with plus agglutinin. Arch Microbiol 139:95–99
Saito T, Matsuda Y (1991) Isolation and characterization of Chlamydomonas temperature-sensitive mutants affecting gametic differentiation under nitrogen-starved conditions. Curr Genet 19:65–71
Saito T, Inoue M, Yamada M, Matsuda Y (1998) Control of gametic differentiation and activity by light in Chlamydomonas reinhardtii. Plant Cell Physiol 39:8–15
Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual, 3rd edn. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
Schlösser UG (1981) Algal wall-degrading enzymes—autolysines. In: Tanner W, Loewus FA (eds) Encyclopedia of plant physiology new series, vol 13B. Springer, Berlin Heidelberg New York, pp 333–351
Schlösser UG (1984) Species-specific sporangium autolysins (cell wall-dissolving enzymes) in the genus Chlamydomonas. In: Irvine DEG, John DM (eds) Systematics of the green algae. Academic, London, pp 409–418
Schnell RA, Lefebvre PA (1993) Isolation of the Chlamydomonas regulatory gene NIT2 by transposon tagging. Genetics 134:737–747
Schultz J, Milpetz F, Bork P, Ponting CP (1998) SMART, a simple modular architecture research tool: identification of signaling domains. Proc Natl Acad Sci USA 95:5857–5864
Sedwick SG, Smerdon SJ (1999) The ankyrin repeat: a diversity of interactions on a common structural framework. Trends Biochem Sci 24:311–316
Shrager J, Hauser C, Chang C-W, Harris EH, Davies J, McDermott J, Tamse R, Zhang Z, Grossman AR (2003) Chlamydomonas reinhardtii genome project: a guide to the generation and use of the cDNA information. Plant Physiol 131:401–408
Siersma PW, Chiang K-S (1971) Conservation and degradation of cytoplasmic and chloroplast ribosomes in Chlamydomonas reinhardtii. J Mol Biol 58:167–185
Skromne I, Sánchez O, Aguirre J (1995) Starvation stress modulates the expression of the Aspergillus nidulans brlA regulatory gene. Microbiology 141:21–28
Smith EF, Lefebvre PA (1996) PF16 encodes a protein with armadillo repeats and localizes to a single microtubule of the central apparatus in Chlamydomonas flagella. J Cell Biol 132:359–370
Smith EF, Lefebvre PA (2000) Defining functional domains within PF16: a central apparatus component required for flagellar motility. Cell Motil Cytoskeleton 46:157–165
Snell WJ, Eskue WA, Buchanan MJ (1989) Regulated secretion of a serine protease that activates an extracellular matrix-degrading metalloprotease during fertilization in Chlamydomonas. J Cell Biol 109:1689–1694
Sugase Y, Hirono M, Kindle KL, Kamiya R (1996) Cloning and characterization of the actin-encoding gene of Chlamydomonas reinhardtii. Gene 168:117–121
Vallon O, Bulté L, Kuras R, Olive J, Wollman FA (1993) Extensive accumulation of an extracellular l-amino-acid oxidase during gametogenesis of Chlamydomonas reinhardtii. Eur J Biochem 215:351–360
Voges D, Zwickl P, Baumeister W (1999) The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu Rev Biochem 68:1015–1068
Voytas DF, Cummings MP, Konieczny A, Ausubel FM, Rodermel SR (1992) copia-like retrotransposons are ubiquitous among plants. Proc Natl Acad Sci USA 89:7124–7128
Xiong Y, Eickbush TH (1990) Origin and evolution of retroelements based upon their reverse transcriptase sequences. EMBO J 9:3353–3362
Ye Y, Meyer HH, Rapoport TA (2001) The AAA ATPase Cdc48/p97 and its partners transport proteins from the ER into the cytosol. Nature 414:652–656
Zhang H, Forde BG (2000) Regulation of Arabidopsis root development by nitrate availability. J Exp Bot 51:51–59
Acknowledgements
We thank Drs. Erika Asamizu and Satoshi Tabata (Kazusa DNA Institute) for providing EST clones and Drs. Patrick Ferris and Ursula Goodenough (Washington University) for MTA1. We also thank Patrick Ferris for his critical reading of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by F.-A. Wollman
Rights and permissions
About this article
Cite this article
Abe, J., Kubo, T., Takagi, Y. et al. The transcriptional program of synchronous gametogenesis in Chlamydomonas reinhardtii. Curr Genet 46, 304–315 (2004). https://doi.org/10.1007/s00294-004-0526-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00294-004-0526-4