Abstract
The mitotic kinetochore of the budding yeast contains a number of proteins which are required for chromosome transmission but are non-essential for vegetative growth. We show that one such protein, Iml3, is essential for meiosis, in that the absence of this protein results in reduced spore viability, precocious sister chromatid segregation of artificial and natural chromosomes in meiosis I and chromosome non-disjunction in meiosis II.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The life cycles of most eukaryotic organisms include a meiotic phase, in which diploid parental cells produce haploid gametes. During meiosis, a single round of DNA replication is followed by two rounds of chromosome segregation. In the first, or reductional division (meiosis I), homologous chromosomes segregate from one another, whereas in the second, or equational division (meiosis II), sister chromatids segregate away. Therefore, during meiosis, sister chromatids separate in a two-step process. At anaphase of meiosis I, cohesion between sister chromatids, provided by a protein complex called cohesin, is lost along the arms but is maintained at the centromeres of sister chromatids. At anaphase of meiosis II, cohesion at the centromeres is broken and sister chromatids separate away from each other. In contrast, prior to mitotic anaphase, cohesion is lost along the entire chromosome, including the centromeres, releasing sister chromatids from one another and allowing them to segregate to opposite poles. Pairing of homologues, sister chromatid cohesion and recombination play a central role in the fidelity of chromosome segregation in meiosis I. (for reviews, see Nasmyth 2001; Uhlmann 2001, 2003; Petronczki et al. 2003; Page and Hawley 2003; McKee 2004).
In the budding yeast Saccharomyces cerevisiae, the cohesin complex that holds sister chromatids together consists of four proteins, Scc1/Mcd1, Scc3, Smc1 and Smc3. Another protein, Pds5p, is also found to be loosely associated with the complex. In metaphase, the spindle checkpoint protein, Pds1p (also called securin), inhibits the destruction of cohesion between sister chromatids by binding to a cysteine protease, Esp1p (also called separase) that cleaves Scc1p. At the onset of anaphase, Pds1p is itself proteolyzed due to ubiquitination by the anaphase-promoting complex. This releases the separase Esp1p, which destroys the cohesion to trigger sister chromatid separation (for reviews, see Biggins and Murray 1999; Nasmyth 2001; Uhlmann 2001; Petronczki et al. 2003). In meiosis, the cohesin complex is the same as that in mitosis, except that Scc1p is replaced by the meiosis-specific variant Rec8p (Klein et al. 1999). Rec8p is found localized along sister chromatid arms and centromeres. At the onset of anaphase I, Rec8p is cleaved by separase and cohesion is destroyed, but only along the arms. However, Rec8p stays protected against proteolysis at and in the vicinity of centromeres. Thus, mono-oriented sister chromatids, still bound to each other at the centromeres, move together to one pole. They finally separate from each other at the onset of anaphase II when they are bi-oriented and separase destroys the cohesion at the centromeres (for recent reviews, see Petronczki et al. 2003; Page and Hawley 2003). Several proteins have been identified which help in the mono-orientation of sister kinetochores and bi-orientation of homologues in meiosis I, including the monopolins Mam1, Csm1 and Lrs4 (Tóth et al. 2000; Rabitsch et al. 2003) and the Aurora B kinase Ipl1p (Tanaka et al. 2002). More recently, another protein, Sgo1p, has been implicated in the bi-orientation of homologues in meiosis I (Katis et al. 2004). Spo13p, whose absence causes a single meiotic division with mixed segregation of chromosomes (Klapholz and Esposito 1980), has been shown to be important for cohesion at sister centromeres in meiosis I (Klein et al. 1999; Lee et al. 2002; Shonn et al. 2002). Recent studies (Marston et al. 2004; Katis et al. 2004) identified three more proteins, Sgo1, Iml3 (subject of this work) and Chl4, which also regulate the retention of centromeric cohesion after metaphase I and until the start of anaphase II. In the sgo1 mutant, the loss of cohesion leads to precocious separation of sister chromatids in meiosis I and their random segregation in meiosis II. Increased non-disjunction of sister chromatids in meiosis II has also been observed in iml3 and chl4 mutants. Spo12 and Slk19 proteins, whose absence leads to a single meiotic division (Klapholz and Esposito 1980; Zeng and Saunders 2000; Kamieniecki et al. 2000) along with separase, are all implicated in exit from meiosis I by causing the release of Cdc14p from the nucleolus in early anaphase I. In their absence, the cells continue to progress in meiosis II, leading to a single meiotic division on anaphase I spindles with equational segregation of sister chromatids (Buonomo et al. 2003).
The difference in the dynamics of chromosome segregation between meiosis and mitosis is suggestive of a meiosis-specific kinetochore that may differ from its mitotic counterpart in structure and regulation. We argue that, due to this difference, several trans-acting factors at the centromere may play a different role during meiosis, as compared with that in mitosis. A number of proteins that are required for centromere functions during mitosis have also been shown to play a role in meiosis. Cbf1p, the CDE I-binding protein of S. cerevisiae and Abp1p, the centromere-binding protein of Schizosaccharomyces pombe, do not play essential roles during mitosis (Cai and Davis 1990). However, both cbf1 and abp1 mutant strains have pronounced meiotic defects (Masison and Baker 1992). In addition, deletions and rearrangements spanning cis-acting CDE I and CDE II elements can have a considerably greater effect on meiotic than on mitotic functions of the centromere (Carbon and Clarke 1984; Cumberledge and Carbon 1987; Gaudet and Fitzgerald-Hayes 1989).
It has been shown that the mitotic kinetochore protein Iml3/Mcm19, although not essential for cell growth, is involved in the precise segregation of minichromosomes and natural chromosomes (Ghosh et al. 2001). This protein is localized at the mitotic kinetochore (Pot et al. 2003). Does this protein have any role in meiosis? We address this question in the following work. Using genetics and cell biology we show that mutations in the IML3 gene cause premature separation of sister chromatids in meiosis I and their increased non-disjunction in meiosis II. While this work was in progress, a report by Marston et al. (2004) established that Iml3p is required for the retention of Rec8p at the centromeres from anaphase I to anaphase II and for the proper segregation of sister chromatids in meiosis II. Our work provides genetic and cytological evidence for the role of Iml3p in maintaining sister chromatid cohesion from meiosis I to anaphase II. In addition, we show that Iml3p is essential for meiosis, in that its absence leads to poor spore viability. Furthermore, Iml3p does not affect meiotic recombination frequency.
Materials and methods
Materials
All media, chemicals and enzymes etc., were described by Poddar et al. (1999) and Ghosh et al. (2001).
Yeast strains and plasmid
The homozygous wild-type diploid (Table 1, diploid number 1) was constructed by crossing 301-2B (MATα leu2 ura3 his4 trp1) with AB1380 (MATa ade2 trp1 ura3 his5 can1 lys2). The isogenic homozygous mutant diploid (Table 1, diploid number 2) was constructed by crossing IML3-deleted strains 301-2BΔ19 and AB1380Δ19 (Ghosh et al. 2001). M31-L/6A was M31/6A (MATα leu2 ura3 trp1 his4 mcm19-1) carrying YCp121-L and was obtained as described by Roy et al. (1997). The heterozygous wild-type diploid (Table 1, diploid number 3) was constructed by crossing PS31-5A (MATa leu2 his3 mcm19-1; Ghosh et al. 2001) with M31I-L/6A, where M31I-L/6A was obtained by integrating the plasmid pM31-2, carrying IML3/MCM19 and URA3 (Ghosh et al. 2001) in M31-L/6A. The isogenic mutant diploid (Table 1, diploid number 4) was constructed by crossing M31-L/6A with PS31-5A. The heterozygous wild-type diploid (Table 1, diploid number 5) was constructed by crossing SL1 (MATα ade1 leu2 trp1 ura3 his4 MCM19; this study) with SG1 (MATa ADE1 LEU2 TRP1 his3 mcm19-1; this study). The isogenic mutant diploid (Table 1, diploid number 6) was constructed by crossing SL1Δ19 (SL1 deleted for IML3) with SG1. US3329 [MATa leu2∷LEU2∷tetR-GFP tetOX224∷HIS3 (inserted 1.5 kb left of CEN5) ura3 trp1 leu2 his3 ade2] in a W303 background, was obtained from U. Surana. This strain had an array of Tet operators integrated about 1.5 kb to the left of CEN5. It also carried a fusion of Tet repressor with green fluorescent protein (GFP), driven by the URA3 promoter, integrated at leu2. In this way, chromosome V was marked with GFP near its centromere and was visible as a dot during fluorescence microscopy (Straight et al. 1996; Michaelis et al. 1997). This strain was used for scoring the segregation pattern of the GFP-marked chromosome V. US3329Δ19 was derived from US3329 by the deletion of its IML3 gene. SL7 (MATα ura3 his4) was a segregant of the cross between #2 (MATa ho∷LYS2 lys2 leu2∷hisG his4 ura3) and 757 (MATα ho∷lys2 tetR-GFP∷LEU2 tetOX224∷URA3; both in a SK1 background, obtained from F. Klein). #2Δ19 was constructed by deleting #2 for IML3. SL7-2Δ19 (MATα leu2 ura3 his4 mcm19∷URA3) was obtained as a segregant of the diploid SL7 crossed with #2Δ19. The spo13 mutant strain (spo13-1 sir4 leu2 ura3 trp1 his6) was originally from P. Briza and was obtained from K. Muniyappa. Deletions of IML3/MCM19 using URA3 were constructed as described by Ghosh et al. (2001).
Sporulation and disome analysis
For routine studies, sporulation of the diploids and haploids was carried out at 28°C in liquid medium containing 2% potassium acetate and 0.05% yeast extract. The spores obtained from the tetrads or dyads were allowed to germinate at 28°C for 4 days. Disomy for chromosome I (ADE1/ade1) was analyzed in the mutant spores by scoring the number of red colonies that appeared due to the loss of ADE1-containing chromosome. Since the mutant (mcm19) shows a high rate of mitotic chromosome loss (ca. 2×10−3 for chromosome III; Ghosh et al. 2001), disomy for chromosome I was analyzed in the mutant spores by growing them in yeast extract/peptone/dextrose medium (YEPD) supplemented with 0.05 mg/ml adenine until saturated and plating for single colonies on YEPD plates. The disomic segregants gave about 20–50 red (ade1) colonies from about 2,000 colonies of each segregant spread on two to three YEPD plates. For the wild type, the putative disomic spores (explained in the text) were mated with a haploid (ADE1), the resulting diploid was sporulated and the spores were analyzed for the appearance of ade1 segregants (expected to appear with a frequency of about 25%). Precocious sister chromatid segregation of chromosome III should produce disomic MATa/MATα spores if there is no recombination between the MAT locus and its centromere and, if there is recombination, 50% of the disomic spores should still be of the non-mater MATa/MATα type. The frequency of recombination between the MAT locus and its centromere is about 35% in these crosses. Therefore, at least 80% of the spores (0.65+0.5 of 0.35) will be non-maters in the event of precocious sister chromatid segregation of chromosome III. Tetratype (T-type) tetrads or random spores were analyzed for disomy of chromosome III by scoring for non-mating phenotype.
Synchronization of cells for sporulation was carried out in liquid cultures as described in Cha et al. (2000), except that yeast extract/peptone/acetate medium contained 2% potassium acetate, instead of 1%. Synchronization was carried out for experiments on the estimation of sporulation efficiencies and for scoring GFP dots in diploids by fluorescence microscopy.
Fluorescence microscopy
Diploids were induced to sporulate synchronously as described above. DNA was stained with 4′,6-diamidino-2-phenylindole (DAPI; Sigma) by taking 500-μl aliquots of sporulating cultures, spinning the cells down and resuspending the cell pellet for 30 min at 23°C in 20 μl of DAPI solution (0.5 μg/ml DAPI in 90% glycerol, containing 1 mg/ml of the anti-fade dye p-phenylenediamine; Sigma). GFP dots and nuclear morphology were scored using a Zeiss Axiovert 200M fluorescence microscope with Axiovision software.
Results
iml3 mutations affect spore viability
To study the role of Iml3 protein in meiosis, we reasoned that segregational defects during meiosis I or II would produce disomes and nullisomes, causing inviability of spores. Therefore, if the homozygous mutant diploid mcm19/mcm19 were to give poor spore viability, it would indicate a role of Iml3p in meiosis. Tetrads were dissected and the spores were allowed to germinate at 28°C. Table 1 shows that the spore viability was significantly reduced in tetrads obtained from the homozygous mutant diploids, as compared with parental wild-type diploids. Less than 3% of the tetrads obtained from the null mutant diploid gave four-spore germination. Thus, almost every meiotic division occurring in the absence of the Iml3p appeared to cause spore death, suggesting its role in meiosis. Spores which failed to grow into visible colonies were observed under the microscope. Most of these (ca. 70%) failed to germinate, while the remainder grew only up to a single-budded cell stage.
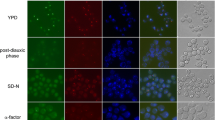
It was found that both the mutant and wild-type diploids described above did not sporulate efficiently. Less than 20% tetrads were produced from asynchronous cells after 4 days of incubation in sporulation medium. In contrast, the wild-type diploid US3329 × SL7 (used for scoring the segregation pattern of the GFP-marked chromosome V in a later experiment) gave a high frequency (>60%) of sporulation, even without meiotic synchronization. Upon synchronization, US3329 × SL7 gave more than 90% sporulation within 12 h of transfer to sporulation medium. The sporulation efficiency of this diploid was compared with its mutant derivative US3329Δ19 × SL7-2Δ19. Both diploids were synchronized for sporulation. Figure 1 shows that the wild-type cells proceeded through meiosis at a faster rate than the mutant cells. Binucleate and tetranucleate cells accumulated later in the mutant culture, which is consistent with a delayed progress of cells lacking Iml3p through metaphase I (Marston et al. 2004). After 10 h, about 45% of mutant cells reached the tetranucleate stage. In contrast, about 86% of the wild-type cells were tetranucleate at this point of time. After 24 h, both mutant and wild-type cells showed a plateau of sporulation. Nearly 70% of the mutant cells had formed tetranucleate cells, 12% were still binucleate and 5% had formed binucleate dyads. By this time, the wild-type cells were 87% tetranucleate and 10% binucleates, of which 7% were dyads. Therefore, mutant cells proceeded through sporulation more slowly than wild-type cells and, when left for longer times in the sporulation medium, formed tetrads with an efficiency that was about 20% lesser than the wild type.
The iml3 mutation causes a slower progression through meiosis. Wild-type [wt, US3329 × SL7 (MCM19 × MCM19)] and mutant [mt, US3329Δ19 × SL7-2Δ19 (mcm19-Δ1 × mcm19-Δ1)] diploids were induced to sporulate synchronously. Aliquots of cultures were taken at various time-points after the transfer of cells to sporulation medium. DNA was stained with DAPI for visualization of nuclei. Dyads were included in the binucleate population of cells
spo13 rescues poor spore viability caused by the iml3 mutation
The spo13 mutation causes a single division in meiosis where chromosomes show mixed segregation: one chromosome of a bivalent may segregate equationally while the other may segregate reductionally (Klapholz and Esposito 1980; Hugerat and Simchen 1993; Shonn et al. 2002). This mutation slows down meiosis I, due to some defect (perhaps in kinetochore-microtubule connections) that is sensed by the spindle checkpoint proteins (Shonn et al. 2002). It is suggested that the time allotted for the completion of meiosis is fixed and a slowing down of this process by spo13 allows for only one meiotic division. spo13 has been used to identify mutations which cause spore death due to aberrations in the early events of meiosis, including those in the reductional segregation of chromosomes in meiosis I (Malone and Esposito 1981; Rockmill and Roeder 1988). A haploid cell that allows silent copies of MATa and MATα to express (such as sir4/ste9) can also attempt meiosis, leading to the formation of immature and inviable spores due to random segregation of chromosomes in meiosis I. The introduction of the spo13 mutation in this haploid allows viable spores to be formed (Wagstaff et al. 1982). Both spores of a dyad are viable only if all the chromosomes in the haploid cell undergo equational, meiosis II-like segregation. It was reasoned that, if the spore inviability caused by the iml3 mutation was due to defects in the equational segregation of chromosomes, which normally occurs in meiosis II, then the spo13 mutation would not suppress spore death. That is, if equational segregation is affected, the spo13 sir4 haploid would give a higher spore viability than the spo13 sir4 mcm19 haploid. This conclusion is based on the assumption that the events preceding the equational division induced by the spo13 mutation are mechanistically the same as those which precede a normal meiosis II in the iml3 mutant (addressed in the Discussion). Dyads obtained from haploids spo13 sir4 and spo13 sir4 mcm19-Δ1 were dissected. Table 2 shows that both spo13 sir4 and spo13 sir4 mcm19-Δ1 give similar spore viabilities. In particular, the frequency of occurrence of two-spore viable dyads is about the same in the two haploids. This suggests that spore death in the iml3/mcm19 mutant diploid is not due to mistakes occurring in the equational segregation of chromosomes, with the stipulation that equational segregation caused by the absence of Spo13p completely mimics meiosis II.
The iml3 mutation does not affect meiotic recombination frequency
Genetic recombination plays a key role in maintaining the fidelity of chromosome segregation in meiosis I. Several mutations which cause reduced genetic recombination display poor spore viability, due to aberrations in chromosome segregation in meiosis I (Malone and Esposito 1981; Klapholz et al. 1985). To determine whether iml3 had an effect on genetic recombination, recombination frequencies were measured in the interval between HIS4 and LEU2 and between LEU2 and MATa in the mutant (301-2BΔ19 × AB1380Δ19) and wild-type (301-2B × AB1380) diploids. Random spore analysis was done, since the spore viability in tetrads was very poor. HIS4 leu2 and his4 LEU2 recombinant spores were obtained with a frequency of 18% (36 out of 200 randomly-picked spores) from the wild-type diploid and 17% (31 out of 184) from the mutant diploid, while LEU2 MATα and leu2 MATa recombinant spores were obtained with a frequency of 38% from the wild type and 41% from the mutant diploid. Thus, there is no significant change in the meiotic recombination frequencies between mutant and wild-type diploids; and iml3/mcm19 does not appear to inhibit normal pairing and exchanges between non-sister chromatids prior to chromosome segregation in meiosis I.
The iml3 mutation causes precocious segregation of sister chromatids of an artificial unpaired circular chromosome
Precocious separation of sister chromatids in meiosis I is one of the reasons for missegregation of chromosomes in meiosis. This has been demonstrated before for the cbf1 mutant, using artificial plasmids (Cumberledge and Carbon 1987; Masison and Baker 1992). We showed earlier that a LEU2-carrying 60-kb artificial midichromosome, YCp121-L, is significantly more stable in mcm19 mutant cells, perhaps due to its larger size (Roy et al. 1997). Unlike its smaller counterpart, YCp121, which is present in elevated copies in mutant cells (Ghosh et al. 2001), YCp121-L is present in single copies in both mcm19 mutant and wild-type cells (Poddar and Sinha, unpublished data). We used YCp121-L to determine whether it undergoes precocious sister chromatid segregation in the mutant diploid. The trp1 marker, tightly linked to the centromere of chromosome IV, was used to study the segregation of YCp121-L in four-spore viable tetrads. Table 3 shows that, in both wild-type (M31I-L/6A × PS31-5A) and mutant (M31-L/6A × PS31-5A) diploids, tetrads carrying YCp121-L showed predominantly 2+:2− segregation of the LEU2 marker, again suggesting that YCp121-L was present mostly in one copy per diploid cell in both the wild type and the iml3 mutant. Figure 2 explains that, in 2+:2− tetrads, correct segregation of the LEU2 marker on YCp121-L gives either parental ditype (PD) or nonparental ditype (NPD) tetrads with trp1. When there is precocious sister segregation in meiosis I, T-type tetrads are obtained for these two markers. T-type tetrads arose with a high frequency from the mutant diploid, whereas the wild-type diploid gave only PD or NPD tetrads (Fig. 2c). No T-type tetrads were obtained in this case. These results are indicative of a high rate of precocious sister segregation of this midichromosome in the iml3 mutant. Thus, Iml3p is required to prevent sister chromatid separation of artificial circular chromosomes in meiosis I.
Precocious separation of sister chromatids in mutant cells generates T-type tetrads for leu2 and trp1. Segregation of chromosome IV is depicted, where one copy carries the centromere-linked TRP1 (T) gene and the other its mutant allele trp1 (t). a Wild-type cells: after replication and recombination, the sister chromatids are held together by a cohesin complex (shown as ellipses) at the arms and the centromeres. In anaphase I, cohesion of the arms is destroyed but sister chromatids are still held together at their centromeres and move to the same pole. At anaphase II, cohesion at the centromeres is destroyed and the sisters disjoin to opposite poles. The unpaired ring midichromosome is YCp121-L, carrying the LEU2 (L) gene. During meiosis I, YCp121-L can segregate either with the TRP1 (as shown) or with trp1 (not shown) to generate NPD or PD tetrads, respectively, for TRP1 and LEU2 markers. b Mutant cells: Sister kinetochores of YCp121-L could be mono-oriented at metaphase, but separate at the onset of anaphase I due to the destruction of cohesin at the sister centromeres. One of the separated chromatids could lose attachment to the spindle pole and drift to the other nucleus in anaphase I. Alternatively, the iml3 mutation could result in sister kinetochores getting attached to opposite poles. At anaphase I, the sister chromatids would move away from each other. c The observed frequency of occurrence of PD, NPD and T-type tetrads in mutant and wild-type diploids is given
The iml3 mutation causes precocious segregation of sister chromatids of paired native chromosomes
We also studied the effect of the iml3 mutation on the segregation of native chromosomes. The rates of precocious sister chromatid segregation of chromosomes I and III were determined experimentally by tetrad analysis of diploids 5 and 6 (Table 1). One copy of chromosome I was marked with ade1, while one copy of chromosome III was marked with leu2. We focused on the precocious segregation of ADE1-containing chromosome I and LEU2-containing chromosome III homologues. Precocious segregation of sister chromatids would give rise to spore death; and so it was not analyzed in four-spore viable tetrads. Among three-spore viable tetrads, precocious segregation of ADE1-containing sister chromatids would give rise to tetrads which are T-types with respect to ADE1 and TRP1, containing two Ade+ spores where one of the spores would be a disome of the type ADE1/ade1 (Fig. 3; the genotype of the dead spore was inferred from the genotypes of three viable spores). T-type tetrads could also arise due to genetic recombination and due to precocious segregation of chromosome IV sister chromatids. Ade+ spores from only those two-spore viable tetrads were analyzed which were destined to be T-types with respect to ADE1 and TRP1 [containing one parental and one recombinant spore of these types: (1) ADE1 TRP1 and ADE1 trp1, (2) ADE1 TRP1 and ade1 TRP1, (3) ade1 trp1 and ADE1 trp1]. A subset of only such tetrads would contain Ade+ spores disomic for chromosome I, due to precocious segregation of ADE1-containing sister chromatids. One-spore viable tetrads were not analyzed, since sister spores could not be identified in these. Table 4 shows that, in the mutant diploid precocious sister chromatid segregation of chromosome I occurred with a frequency of at least 10% of all meiotic events (8 of 73 tetrads; 33 one-spore viable tetrads were not analyzed). In contrast, none of the 71 tetrads from the wild-type diploid gave a disomic spore. Our results are very similar to those obtained by Masison and Baker (1992), who showed that, in the absence of the CDE I-binding protein Cbf1p, an unpaired chromosome I undergoes a high rate of precocious sister chromatid separation (7.6% in the mutant, as compared with <1% in the heterozygous wild-type diploid). Likewise, the occurrence of precocious segregation of the LEU2-containing copy of chromosome III was analyzed. In this case, only one disomic spore was recovered from the mutant diploid, with a frequency of 1.4% (Table 4).
Precocious sister chromatid separation of chromosome I in meiosis I generates spore death, T-type tetrads and disomy of chromosome I. One homologue of chromosome I was marked with ade1 (a) and the other with ADE1 (A), while one homologue of chromosome IV was marked with its tightly-linked centromere marker trp1 (t) and the other with TRP1 (T). Precocious separation of ADE1-carrying sister chromatids of chromosome I in meiosis I gives T-type tetrads, as opposed to normal segregation, which gives PD or NPD tetrads (the latter not shown in the figure). The broken arrow suggests the drifting of ADE1-containing precociously separated sister chromatid of chromosome I towards the non-sister spindle pole
GFP-marked chromosome V in mutant cells shows precocious sister chromatid segregation in meiosis I and increased sister chromatid non-disjunction in meiosis II
A failure by the mutant to disjoin sister chromatids in meiosis II leads to the lowering of spore viability. Our work with the spo13 mutant suggested that the iml3 deletion does not cause defects in the equational segregation of chromosomes. Using fluorescence microscopy to follow the segregation of a GFP-tagged chromosome, Marston et al. (2004) showed that Iml3p is required to prevent random disjunction of sister chromatids in meiosis II. We, too, used fluorescence microscopy to determine whether there are any meiosis II defects and, in addition, to see whether sister chromatids show precocious separation in meiosis I. This was expected to give better estimates of chromosome missegregation frequency, since dead spores would also be available for analysis.
Each of the diploids US3329 × SL7 (MCM19 × MCM19) and US3329Δ19 × SL7-2Δ19 (mcm19-Δ1 × mcm19-Δ1) carried one homologue of chromosome V marked with GFP, 1.5 kb from its centromere (see Materials and methods). Correct segregation of the GFP chromosome in meiosis I should produce binucleate cells with one green dot in one nucleus, representing a pair of sister chromatids in close association with each other (class I). At the onset of anaphase II, the sister chromatids should disjoin to produce tetranucleate cells with two nuclei having one dot each (class III). Precocious separation of sister chromatids at the onset of, or during anaphase I, should generate binucleate cells having two split GFP dots, each dot representing a sister chromatid. If both the sisters stay together, there will be two dots in one nucleus of the binucleated cell. If one sister moves away to the opposite pole, one dot will be present per nucleus. In addition, we studied meiosis II defects in mutant and wild-type cells. For this, the frequency of occurrence of tetranucleate cells having only one dot in one nucleus (meiosis II non-disjunction) was compared with the occurrence of those tetrads which displayed the normal segregation of two dots segregated from each other in two nuclei. Figure 4 shows that both meiosis I and meiosis II defects were present in the mutant diploid. The mutant diploid gave a significantly higher percentage of binucleate cells where two dots separated precociously before anaphase II, moving either together to the same pole or away from each other to opposite poles (class II segregation). This observation is consistent with the decreased retention of Rec8p at sister centromeres before the onset of anaphase II in the iml3 mutant diploid (Marston et al. 2004). This is also consistent with our genetic results, described above, on the precocious segregation of sister chromatids. In addition to the defect caused in meiosis I, the iml3 mutant diploid also exhibited a defective segregation of sister chromatids in meiosis II. The mutant diploid gave a higher percentage of tetranucleate cells where only one GFP dot was visible in one spore (class IV), suggesting meiosis II non-disjunction of sister chromatids. In the mutant diploid, about one-third of this class of cells had two GFP dots of lower intensities in one spore (data not shown). This could be due to precocious sister chromatid separation before anaphase II, followed by random segregation to the same spore (Klein et al. 1999). These observations are similar to those reported by Amon and colleagues (Marston et al. 2004). The experiment described in this section was repeated twice more. In one of these repeats, the wild-type diploid (US3329 × SL7-2Δ19; MCM19 × mcm19-Δ1) was heterozygous for the iml3 mutation, while the mutant diploid (US3329Δ19 X SL7-2Δ19; mcm19-Δ1 × mcm19-Δ1) was the same as above. Similar results were obtained in all the experiments, in that the mutant diploid gave higher frequencies of both class II (premature separation of sister chromatids before anaphase II) and class IV (meiosis II non-disjunction) segregation of the GFP-marked chromosome V homologue (data not shown).
Segregation of a GFP-marked homologue of chromosome V during meiosis in the iml3 mutant and wild-type diploids. Each of the diploids US3329 × SL7 and US3329Δ19 × SL7-2Δ19 carried a homologue of chromosome V marked with GFP. The segregation of chromosome V was followed by observing the segregation of GFP dots in the nuclei of cells stained with DAPI. To determine the percentage of cells displaying a particular segregation type, 100–140 cells were counted. About 1–2% of the tetrads from both diploids gave aberrant segregation of GFP dots. Three dots were visible in these tetrads in two sacs, where one was in one sac and two, visible as split dots, were in another sac; and this class is not shown
Discussion
The work presented here shows that the absence of Iml3p causes missegregation of chromosomes in meiosis, which results in poor spore viability of an iml3 mutant diploid. Using genetics, we show that an unpaired, non-essential 60-kb midichromosome segregates sister chromatids with a high frequency: as high as 60% of tetrads were T-types with respect to TRP1, a tightly linked centromere marker. Genetics was used again to show that chromosomes I and III separate sister chromatids precociously in meiosis I. It should, however, be mentioned that the 10% and 1% missegregation rates observed for chromosomes I and III are very likely underestimates, as some of the disomes could be in dead spores. This work suggests that all chromosomes do not separate with same frequency. Whether the length of the chromosome plays an important role remains to be determined. The mutant cells are recombination competent; and therefore the defect appears to be kinetochore-related. Iml3 protein may be required in meiosis I either to prevent bipolar attachment of sister chromatids or to hold sister kinetochores together. In principle, non-disjunction of homologous chromosomes in meiosis I also gives rise to spore death. We do not think this defect is of significance in the iml3 mutant, for two reasons. First, a higher fraction of T-type tetrads (10/15) was obtained in the case of YCp121-L segregation, which suggests that precocious segregation of sister chromatids is a major defect. Second, non-disjunction of chromosome I homologue in meiosis I should generate an excess of two-spore and zero-spore viable tetrads, which was not observed. Furthermore, the two-spore viable tetrads should be PD or NPD with respect to ADE1 and TRP1, having both the spores as Ade+. Only three such tetrads were obtained from 29 two-spore viable tetrads. However, none of the Ade+ spores from these tetrads was disomic (ADE1/ade1) for chromosome I, suggesting that non-disjunction in meiosis I was not a major cause of spore death. Work done in parallel by Marston et al. (2004) showed that Iml3p is required for the maintenance of centromere cohesion at sister centromeres in meiosis I and until the onset of anaphase II. Consequentially, the mutant cells should exhibit precocious separation of sister chromatids in meiosis I. Our work provides genetic evidence for this prediction. Furthermore, using fluorescence microscopy to follow a homologue of chromosome V tagged with GFP, we show that sister chromatids indeed separate precociously in binucleate cells. Either they are segregated from each other at opposite poles or they appear as separated dots in a single nucleus of the binucleate cell. It is unlikely that class II cells in the mutant diploid arise from carryover aneuploidy (2n+1, where the extra chromosome is another copy of the GFP-tagged chromosome V) in mitosis, for two reasons:
-
1
The level of chromosome non-disjunction that iml3 mutants show in mitosis is about 1.5×10−3 for chromosome III (Ghosh et al. 2001). The presence of two separate dots in 26% of binucleate cells cannot be entirely due to this low level of aneuploidy.
-
2
Assuming that 12% of binucleate cells having a single dot in each nucleus represent two copies of the GFP chromosome, rather than precociously separated sister chromatids of a single chromosome, there should be more tetrads with a GFP dot in three to four spores.
The expected frequency would be at least 8–9% (70% of 12%, where 70% is the frequency of sister chromatid disjunction in meiosis II; Fig. 4). No tetranucleate cell having a dot in three or four of its spores was observed from either of the diploids (98 tetranucleate cells were analyzed from the mutant and 111 cells from the wild type). A small number of tetranucleate cells (three from the wild type and one from the mutant) did, however, give three GFP dots, but only in two spores. One dot was in one nucleus and two separated dots in another (data not shown). The origin of these tetrads is not known, but if they represent aneuploidy, the number is too small to affect the validity of our conclusions.
The work of Marston et al. (2004) suggests that Iml3p is also involved in chromosome segregation in meiosis II; and a GFP-marked homologue of chromosome V shows non-disjunction in meiosis II, as deduced by the occurrence of a single GFP dot in only one of four spores. Our results on chromosome V segregation using fluorescence microscopy support these observations. However, the spo13 sir4 mcm19 mutant gave the same spore viability in haploid meiosis as the spo13 sir4 mutant, suggesting that there may be no defect in the equational segregation of sister chromatids. How do we resolve this discrepancy? One possible explanation is that the events that precede a normal meiosis II (in the wild type or the iml3 mutant) are not exactly the same as those that precede the single division in a spo13 mutant. Iml3p, being a kinetochore protein (Marston et al. 2004), could be involved in the execution of these or some as yet unknown processes, required for the disjunction of chromatids in meiosis II but not crucial for the spo13 division. For example, Iml3p regulates centromeric cohesion from anaphase I until the start of anaphase II (Marston et al. 2004). It is quite conceivable that the equational division caused by the spo13 mutation in the iml3 mutant occurs prior to a substantial loss of sister centromere cohesion, thus preventing spore death. Future work can resolve this issue.
At least four other non-essential proteins that reside at the mitotic kinetochores, namely Slk19, Cbf1, Sgo1 and Chl4, have requirements in meiosis I. Slk19p appears to have no role to play in kinetochore–microtubule attachment in mitosis, but is required to exit from meiosis I by causing the release of Cdc14p from the nucleolus (Zeng et al. 1999; Kamieniecki et al. 2000; Buonomo et al. 2003). Cbf1p, which binds to centromere box CDE I, is required for mitotic kinetochore function and prevents precocious sister chromatid separation in meiosis I (Cai and Davis 1990; Masison and Baker 1992). Both Sgo1 (a conserved protein) and Chl4 are involved in the maintenance of sister centromere cohesion between the two meiotic divisions (Marston et al. 2004; Katis et al. 2004). Sgo1p is additionally required for the bi-orientation of homologues in meiosis I (Katis et al. 2004).
Iml3p binds to a network of proteins which link the CDE III box of the centromere to the outer kinetochore (for a review, see McAinsh et al. 2003). Several proteins of this network (such as Ctf3, Mcm16, Chl4, Ctf19, Mcm21, Mcm22 etc.) behave like Iml3p, in that they are required for high-fidelity chromosome transmission but are non-essential for vegetative growth (Kouprina et al. 1993; Roy et al. 1997; Sanyal et al. 1998; Poddar et al. 1999; Hyland et al. 1999; Pot et al. 2003). It has been shown in the fission yeast that a highly specialized heterochromatic centromeric domain is required for the loading of the cohesin complex at the centromeres and that this loading depends upon the heterochromatin Swi6 protein (Bernard et al. 2001; Nonaka et al. 2002). It is possible that, in the budding yeast also, higher order chromatin structure at centromeres has a role to play in sister chromatid cohesion. This structure may be required to protect centromeric cohesin in meiosis I until anaphase II. Iml3p, a component of the kinetochore complex, may be required for the formation and maintenance of this higher order chromatin structure. This work should contribute towards understanding the structure and function of the meiotic kinetochore.
References
Bernard P, Maure JF, Partridge JF, Genier S, Javerzat JP, Allshire RC (2001) Requirement of heterochromatin for cohesion at centromeres. Science 294:2539–2542
Biggins S, Murray AW (1999) Sister chromatid cohesion in mitosis. Curr Opin Genet Dev 9:230–236
Buonomo SB, Rabitsch KP, Fuchs J, Gruber S, Sullivan M, Uhlmann F, Petronczki M, Toth A, Nasmyth K (2003) Division of the nucleolus and its release of CDC14 during anaphase of meiosis I depends on separase, SPO12, and SLK19. Dev Cell 4:727–739
Cai M, Davis RW (1990) Yeast centromere binding protein CBF1, of the helix-loop-helix protein family, is required for chromosome stability and methionine prototrophy. Cell 61:437–446
Carbon J, Clarke L (1984) Structural and functional analysis of a yeast centromere (CEN3). J Cell Sci [Suppl] 1:43–58
Cha RS, Weiner BM, Keeney S, Dekker J, Kleckner N (2000) Progression of meiotic DNA replication is modulated by interchromosomal interaction proteins, negatively by Spo11p and positively by Rec8p. Genes Dev 14:493–503
Cumberledge S, Carbon J (1987) Mutational analysis of meiotic and mitotic centromere function in Saccharomyces cerevisiae. Genetics 117:203–212
Gaudet A, Fitzgerald-Hayes M (1989) Mutations in CEN3 cause aberrant chromosome segregation during meiosis in Saccharomyces cerevisiae. Genetics 121:477–489
Ghosh SK, Poddar A, Hajra S, Sanyal K, Sinha P (2001) The IML3/MCM19 gene of Saccharomyces cerevisiae is required for a kinetochore-related process during chromosome segregation. Mol Genet Genomics 265:249–257
Hugerat Y, Simchen G (1993) Mixed segregation and recombination of chromosomes and YACs during single division meiosis in spo13 strains of Saccharomyces cerevisiae. Genetics 135:297–308
Hyland KM, Kingsbury J, Koshland D, Hieter P (1999) Ctf19p: a novel kinetochore protein in Saccharomyces cerevisiae and a potential link between the kinetochore and mitotic spindle. J Cell Biol 145:15–28
Kamieniecki RJ, Shanks RM, Dawson DS (2000) Slk19p is necessary to prevent separation of sister chromatids in meiosis I. Curr Biol 10:1182–1190
Katis VL, Galova M, Rabitsch KP, Gregan J, Nasmyth K (2004) Maintenance of cohesin at centromeres after meiosis I in budding yeast requires a kinetochore-associated protein related to MEI-S332. Curr Biol 14:560–572
Klapholz S, Esposito RE (1980) Recombination and chromosome segregation during the single division meiosis in spo12-1 and spo13-1 diploids. Genetics 96:589–611
Klapholz S, Waddell CS, Esposito RE (1985) The role of the SPO11 gene in meiotic recombination in yeast. Genetics 110:187–216
Klein F, Mahr P, Galova M, Buonomo SB, Michaelis C, Nairz K, Nasmyth K (1999) A central role for cohesins in sister chromatid cohesion, formation of axial elements, and recombination during yeast meiosis. Cell 98:91–103
Kouprina N, Kirillov A, Kroll E, Koryabin M, Shestopalov B, Bannikov V, Zakharyev V, Larionov V (1993) Identification and cloning of CHL4 gene controlling chromosome in yeast. Genetics 135:327–341
Lee BH, Amon A, Prinz S (2002) Spo13 regulates cohesin cleavage. Genes Dev 16:1672–1681
Malone RE, Esposito RE (1981) Recombinationless meiosis in Saccharomyces cerevisiae. Mol Cell Biol 1:891–901
Marston AL, Tham WH, Shah H, Amon A (2004) A genome-wide screen identifies genes required for centromeric cohesion. Science 303:1367–1370
Masison DC, Baker RE (1992) Meiosis in Saccharomyces cerevisiae mutants lacking the centromere-binding protein CP1. Genetics 131:43–53
McAinsh AD, Tytell JD, Sorger PK (2003) Structure, function, and regulation of budding yeast kinetochores. Annu Rev Cell Dev Biol 19:519–539
McKee BD (2004) Homologous pairing and chromosome dynamics in meiosis and mitosis. Biochim Biophys Acta 1677:165–180
Michaelis C, Ciosa R, Nasmyth K (1997) Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell 91:35–45
Nasmyth K (2001) Disseminating the genome: joining, resolving, and separating sister chromatids during mitosis and meiosis. Annu Rev Genet 35:673–745
Nonaka N, Kitajima T, Yokobayashi S, Xiao G, Yamamoto M, Grewal SI, Watanabe Y (2002) Recruitment of cohesin to heterochromatic regions by Swi6/HP1 in fission yeast. Nat Cell Biol 4:89–93
Page SL, Hawley RS (2003) Chromosome choreography; the meiotic ballet. Science 301:785–789
Petronczki M, Siomos MF, Nasmyth K (2003) Un ménage à quatre: the molecular biology of chromosome segregation in meiosis. Cell 112:423–440
Poddar A, Roy N, Sinha P (1999) MCM21 and MCM22, two novel genes of the yeast Saccharomyces cerevisiae are required for chromosome transmission. Mol Microbiol 31:349–360
Pot I, Measday V, Snydsman B, Cagney G, Fields S, Davis TN, Muller EG, Hieter P (2003) Chl4p and Iml3p are two new members of the budding yeast outer kinetochore. Mol Biol Cell 14:460–476
Rabitsch KP, Petronczki M, Javerzat JP, Genier S, Chwalla B, Schleiffer A, Tanaka TU, Nasmyth K (2003) Kinetochore recruitment of two nucleolar proteins is required for homolog segregation in meiosis I. Dev Cell 4:535–548
Rockmill B, Roeder GS (1988) RED1: a yeast gene required for the segregation of chromosomes during the reductional division of meiosis. Proc Natl Acad Sci USA 85:6057–6061
Roy N, Poddar A, Lohia A, Sinha P (1997) The mcm17 mutation of yeast shows a size-dependent segregational defect of a minichromosome. Curr Genet 32:182–189
Sanyal K, Ghosh SK, Sinha P (1998) The MCM16 gene of the yeast Saccharomyces cerevisiae is required for chromosome segregation. Mol Gen Genet 260:242–250
Shonn MA, McCarroll R, Murray AW (2002) Spo13 protects meiotic cohesin at centromeres in meiosis I. Genes Dev 16:1659–1671
Straight AF, Belmont AS, Robinett CC, Murray AW (1996) GFP tagging of budding yeast chromosomes reveals that protein–protein interactions can mediate sister chromatid cohesion. Curr Biol 6:1599–1608
Tanaka TU, Rachidi N, Janke C, Pereira G, Galova M, Schiebel E, Stark MJ, Nasmyth K (2002) Evidence that the Ip11-Sli15 (Aurora kinase-INCENP) complex promotes chromosome bi-orientation by alternating kinetochore–spindle pole connections. Cell 108:317–329
Tóth A, Rabitsch KP, Gálová M, Schleiffer A, Buonomo SB, Nasmyth K (2000) Functional genomics identifies monopolin: a kinetochore protein required for segregation of homologs during meiosis I. Cell 103:1155–1168
Uhlmann F (2001) Chromosome cohesion and segregation in mitosis and meiosis. Curr Opin Cell Biol 13:754–761
Uhlmann F (2003) Chromosome cohesion and separation: from men and molecules. Curr Biol 13:104–114
Wagstaff JE, Klapholz S, Esposito RE (1982) Meiosis in haploid yeast. Proc Natl Acad Sci USA 79:2986–2990
Zeng X, Saunders WS (2000) The Saccharomyces cerevisiae centromere protein Slk19p is required for two successive divisions during meiosis. Genetics 155:577–587
Zeng X, Kahana JA, Silver PA, Morphew MK, McIntosh JR (1999) Slk19p is a centromere protein that functions to stabilize mitotic spindles. J Cell Biol 146:415–425
Acknowledgements
This work was supported by a Council of Scientific and Industrial Research (CSIR) grant [37(1094)/02-EMR-II] to P.S. and a grant (63/127/2001-BMS) by the Indian Council of Medical Research (ICMR) to A.L. and P.S. S.K.G. and S.L. were supported by CSIR fellowships 9/15(175)/96 EMR-I and 9/15(226)/2001 EMR-I, respectively. S.S. was supported by a University Grants Commission (UGC) fellowship [F.2-8/2002(SA-I)]. We are grateful to F. Klein, U. Surana and K. Muniyappa for strains. We are thankful to Sujata Hajra for a critical reading of the manuscript and to Gursharan Kaur for help with the fluorescence microscopy of GFP dots. The laboratory assistance of Md. Asraf Ali Molla is gratefully acknowledged. The experiments comply with the current laws of India.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by S. Hohmann
Rights and permissions
About this article
Cite this article
Ghosh, S.K., Sau, S., Lahiri, S. et al. The Iml3 protein of the budding yeast is required for the prevention of precocious sister chromatid separation in meiosis I and for sister chromatid disjunction in meiosis II. Curr Genet 46, 82–91 (2004). https://doi.org/10.1007/s00294-004-0516-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00294-004-0516-6