Abstract
The intense use of pesticides in agricultural activities for the last several decades has caused contamination of the ecosystems connected with crop fields. Despite the well-documented occurrence of pesticide biodegradation by microbes, natural attenuation of atrazine (ATZ), and its effects on ecological processes in subtropical forested areas, such as Iguaçu National Park located in Brazil, has been poorly investigated. Subtropical environments sustain a great degree of fungal biodiversity, and the patterns and roles of these organisms should be better understood. This work aimed to evaluate nine ligninolytic-producer fungi isolated from the INP edge to degrade and detoxify ATZ solutions. ATZ degradation and the main metabolites produced, including deisopropylatrazine and deethylatrazine (DEA), were analyzed using dispersive liquid–liquid microextraction followed by gas chromatography-mass spectrometer. Four fungi were able to degrade ATZ to DEA, and the other five showed potential to grow and facilitate ATZ biodegradation. Furthermore, two strains of Fusarium spp. showed an enhanced potential for detoxification according to the Allium cepa (onion) test. Although the isolates produced ligninolytic enzymes, no ligninolytic activity was observed in the biodegradation of ATZ, a feature with ecological significance. In conclusion, Ascomycota fungi from the INP edge can degrade and detoxify ATZ in solution. Increasing the knowledge of biodiversity in subtropical protected areas, such as ecosystem services provided by microbes, enhances ecosystem conservation.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Current indiscriminate use of pesticides in agriculture has caused many applied products to remain and disseminate within soil, surface water, and groundwater of associated ecosystems, endangering the health of microorganisms, plants, animals, and human beings [1]. Although the environmental and human impact of atrazine (ATZ) has been reported, and alternatives for ATZ use have been documented, the relevance of ATZ biodegradation in environmental protection areas in the natural process is largely unknown.
ATZ is an herbicide widely used in cultures like corn (Zea mays), sugar cane (Saccharum officinarum), sorgo (Sorghum bicolor), among other less economically relevant crops, to eliminate pre- or post-scrub germination [2]. Although its application has been restricted (or banned) by the European Union, USA, and Australia, it is widely used in most Latin American and Caribbean countries (maximum contaminant level (MCL) of 2 µg L−1 to potable water) [3]. ATZ has been recognized, in researches from 2012 to 2017, as the second-most concentrated pollutant in surface water and groundwater worldwide [4, 5].
This herbicide is considered moderately toxic for humans and very dangerous for the environment [6] since significant levels persist in the environment and are highly toxic even in low concentrations [7]. Fungi, algae, and several animal groups are affected when exposed to ATZ. For example, hermaphroditism and demasculinization occur in frogs and sexual reproduction difficulties in rats. ATZ exposure mainly causes endocrine system effects in animals, including humans, but the chemical also affects the central nervous system, reproductive system, immune system, and cardiovascular function [8,9,10].
ATZ fate in soil depends on many factors, including its composition (sorption to organic carbon compounds, especially humic acids and clay minerals), physicochemical characteristics (e.g. pH, temperature), chemical, and biological degradation, plant absorption, and water flows (e.g. leaching, run-off) [11, 12]. The half-life of the herbicide mainly depends on microbes capable of degrade the compound influenced by oxygen and water availability, depth, temperature, ATZ concentration, history of herbicide application, and agricultural practices [13]. Smith and Walker [14] showed that the half-life of ATZ was 338 days in superficial dark brown chernozem soil (70% clay, 25% silt, and 5% sand) where 2 µg g−1 of ATZ were applied, considering 8% of humidity and a temperature of 20 °C.
Current studies have shown the widespread use of bacteria for the biodegradation of ATZ since they can mineralize the compound. However, fungi are responsible for the N-dealkylation of ATZ side-chains, producing principally (deisopropylatrazine) DIA and (deethylatrazine) DEA. Fungi activity allows bacteria to subsequently open the heterocyclic ring [7, 15, 16]. Thus, despite the significant focus on the genetics of ATZ biodegradation by bacteria [17,18,19], microbial consortia and associations can directly affect degradation in natural environment. Therefore, the fungal role in these processes requires further study. Fungi catalyze the biodegradation of pesticides using intracellular and extracellular enzymatic compounds [20]. The major biodegradation potential of basidiomycetes and ascomycetes is associated with the extracellular ligninolytic complex, which is composed of biotechnologically and industrially important molecules [21].
The primary goal associated with the creation of Brazilian National Parks is the preservation of ecosystems of great ecological importance and scenic beauty [22]. These conservation areas are recurrently immersed in large agricultural and livestock land proprieties, and the protection of valuable natural resources coexists very closely with intense anthropic impacts. Particularly, Iguaçu National Park (INP), the biggest forestall remaining of the semideciduous seasonal forest of south Brazil, is a green island submerged in an agricultural area where pesticide use is intense. Soil is made up of sandy ground with high iron content and is classified as basaltic volcanic lithotype [23, 24]. The impacts of pesticide in edge areas are largely unknown. Consequently, knowledge regarding the biodegradation of pesticides by microorganisms and their effects on ecological processes has heretofore been scarce [25].
Although the macroecological richness pattern of saprotrophic fungi, parasites, and pathogens, as well as the Ascomycota phylum, is in accordance with latitudinal gradients (higher levels of richness of fungal groups occur in tropical areas and decrease toward the poles) and there is a high fungal endemicity in the tropics according to Rapoport’s rule, fungal biodiversity remains poorly studied [26]. There is an abysmal difference between the 100,000 described fungal species and the estimated existing number (0.8 million to 5.1 million globally) [27]. The taxonomic and functional diversity of microbes and their role in ecosystemic processes require a depth investigation, especially with respect to the biodegradation potential of pesticides.
The Allium cepa (onion) toxicity test is a low-cost, easy to handle, and rapid test used to assess acute and chronic toxicity. In this work, it is used to evaluate the toxic effects of ATZ using fungi treated with ATZ solutions. Cytotoxic, genotoxic, and mutagenic assays in onion correlate adequately with other test systems, e.g. mammals, and can serve as a bioindicator used to assess toxicity to the environment and human beings [28, 29]. According to Datta et al. [30], the onion test is a standard technique used in toxicological studies. Cytotoxic assays, e.g. root growth and mitotic index (MI), exhibit sensitive and reliable effects even when small concentrations of pollutants are used.
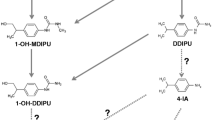
The capacity of fungal strains with ligninolytic degradation potential to degrade ATZ is still unknown in terms of biodiversity of microbes (especially with respect to functional diversity). Thus, this work aimed to evaluate the ability of potential ligninolytic-producer fungi from the INP to degrade and detoxify ATZ solutions. Furthermore, polyphasic characterization approaches (internal transcribed spacer (ITS) region of rDNA and morphological features) were carried out in the isolates with the most significant potential to degrade herbicides (Fig. 1).
Materials and Methods
Microorganisms
In order to evaluate the ATZ biodegradation, nine fungi isolated from an artificial edge of the INP in contact with agricultural areas used to grow corn and soybean crops were selected (Codes CCLM_GW, CCLM_GU, CCLM_IB, CCLM_NH, CCLM_IC, CCLM_DF, CCLM_BP, CCLM_BO, and CCLM_AD). The points used to collect fungal samples were 8 m from the park edge and were separated from agricultural areas by a road that was 7 m wide (Figure S7). The fungi collected were potential lignin degraders in a qualitative study that used guaiacol (2-methoxyphenol) as an enzymatic inductor and colorimetric indicator (supplementary material). Isolates were preserved at Culture Collection of Microbiology Laboratory (CCLM) from the Federal University of Latin American Integration (UNILA).
Fungal Preparation and Growth
Fungi were reactivated in the malt extract agar (MA2) for 7 days at 28 ºC. Three discs placed 6 cm from the perimeter of mycelium were transferred to a 125 mL Erlenmeyer flask containing 25 mL MA2, and 10 mg L−1 ATZ PESTANAL® (Sigma-Aldrich) prepared in ultrapure water [31, 32]. MA2 did not contain peptone and dextrose. Flasks were randomly distributed in the shaker at set at 28 ºC and 150 rpm for 7 days. When the incubation period ended, the culture medium was filtered, and the supernatant solution was stored at – 4 °C for chromatographic and toxicological assays.
Analysis of ATZ Degradation and the Production of Principal Metabolites Using GC–MS
Sample preparation and chromatographic analysis of both samples containing treated fungi and controls (without fungi) were performed according to procedures detailed in Della-Flora et al. [33]. To extract ATZ and its metabolites from the culture medium using the dispersive liquid–liquid microextraction approach (DLLME), 0.10 g mL−1 NaCl was dissolved and combined with ATZ and ethion solution at 2.0 μg L−1 in 50 mL of ultrapure water (working solution). Ethion was used as the internal standard. ATZ solution at 10 mg L−1 was diluted by transferring 10 μL of the supernatant obtained from the culture medium into the working solution, also 0.50 mg L−1 ethion solution was diluted by transferring 200 μL. The solution was adjusted to pH 7.0 and 5-mL aliquots were placed in 15 mL glass centrifuge tubes with a conical base. 150 μL HPLC grade dichloromethane (DCM) (extractive solvent) and 550 μL HPLC grade acetone (dispersing solvent) were quickly added. In the formed emulsion, DCM and acetone were precipitated by carefully bumping the external tube surface, and the tube was centrifuged at 2000 rpm for 5 min. Approximately 150 μL of the bottom phase was removed, avoiding contamination between the bottom and water phases, and deposited into 300 μL inserts and stored at – 20 °C prior to gas chromatography-mass spectrometer (GC–MS) analysis.
The GC–MS system was equipped with a TRACE 1300 GC–MS and an ISQ Single Quadrupole MS mass analyzer. Analytes were separated using a TR-5MS capillary column (30 m × 0.25 mm × 0.25 μm) (ThermoScientific). The temperature of the injector was set at 250 °C, and the samples were manually injected (2 μL) in splitless mode using Helium as running gas with a constant flow rate of 1 mL min−1. The initial temperature was set at 50 °C (for 2 min) and increased to 140 °C at 10 °C min−1. The temperature was then raised to 158 °C at a rate of 1 °C min−1; 250 °C at a rate of 10 °C min−1; and 290 °C at a rate of 7 °C min−1. The temperature of the ion source and line transfer was set at 300 °C and 250 °C, respectively. Selective ion monitoring mode using an intense ion and two qualifying ions were selected (Table S1). Thermo Xcalibur software, version 2.2 (Thermo Scientific) was used for processing data.
DIA and DEA formation were expressed in terms of concentration (mg L−1), while ATZ degradation was calculated according to the following formula:
where Ccontrol is the concentration of an analyte in the control treatment and Cfungus is the concentration of an analyte in the culture media containing fungi.
Toxicity Evaluation of ATZ and Fungi-Treated ATZ by Allium cepa Test
Supernatants obtained from cultures (CCLM_GW, CCLM_GU and CCLM_DF) according to topic 2.2, were assessed using the onion test, as described by MARINHO et al. [34], which was based on Fiskesjo [28]. Laves, roots, and external layers of onions at the same stage of growth were carefully removed from the ones of similar shape and form. They were placed in plastic cups with deionized water and stored in a location protected from light and low humidity up to 72 h at approximately 20 °C. Then, onions were removed from the water, and root growth was measured with a ruler. All the roots from each bulb were extracted and measured, and average root lengths were determined. Onions that had roots that grew similarly were selected for use in subsequent toxicity tests.
As experimental controls, MA2 culture medium plus ATZ at 10 mg L−1 (ATZbt), and deionized water plus ATZ at 10 mg L−1 (ATZc), both without biological treatment, were incubated at 28 °C for 7 days. Deionized water (without ATZ) was used as positive control for root growth. All treatments were tested in triplicate. Incubation and measurement of onions were performed as described above. Percentage growth inhibition was calculated by comparing the obtained root growth values for each treatment with the blank.
Statistical Analyses
Regarding ATZ degradation and the production of principal metabolites by INP-derived fungi, the independent variables were the fungal isolates. The dependent variables corresponded to levels of ATZ degradation, and DEA and DIA production. When testing the toxicity of ATZ solutions, the independent variables were CCLM_GW, CCLM_GU, CCLM_DF, ATZbt, and ATZc solutions, and dependent variables corresponded to growth inhibition (%).
Bio-stat 5.3 was used to carry out the Shapiro Wilk normality test, outliers test, and ANOVA one-way (parametric) or Kruskal Wallis (non-parametric) tests to determine if there was a significant difference between treatments. All analyses were assessed using a p < 0.05 significance level. Figures were prepared by plotting data using RStudio 1.1.456 software. The logarithm (log (x + 1)) was applied to standardize heteroscedastic data, but homoscedasticity was not always achieved. Thus, parametric analysis of variance was used for ATZ, and non-parametric analysis was used to assess DEA and DIA levels and growth inhibition (%) of onion roots.
Morphological Characterization
Morphological characterization based on reproductive structures was analyzed using the microculture technique described by WEBER and PITT [35], with minor modifications. Inside a petri dish, a small square of MA2 culture media (1 × 1 cm) was placed on a slide. The square was inoculated on all four sides using mycelium transfer of reactivated isolates. The isolates used were the same ones that were used to assess ATZ degradation, and DIA and DEA formation. The plates were incubated at 28 °C for 2 to 4 days until reproductive structures were visible. Before observation using an optical microscope, the coverslip was removed and placed on a clean slide and stained with 2% methylene blue.
Molecular Characterization
Strains were grown in MA2 medium for 7 days at 28 °C, and total DNA was extracted using Wizard Genomic DNA Purification Kit (Promega, USA) according to the manufacturer’s instructions. The ITS1-5.8S-ITS2 region of rDNA was amplified using ITS-1 (5ʹTCCGTAGGTGAACCTGCGG-3ʹ) and ITS-4 (5ʹTCCTCCGCTTATTGATATGC-3ʹ) primers [36]. PCRs were performed using reaction volumes of 50 µl reaction containing 1 × PCR buffer (Invitrogen, Brazil), 20 ng template DNA, 0.2 µM of each primer, 200 µM of each dNTP, and 2.5 units of Easy Taq DNA polymerase (Invitrogen, Brazil). Amplifications were performed an Applied Biosystems™ Veriti 96 Well Thermal Cycler (Foster City, CA, USA). The amplification parameters consisted of initial denaturation at 94 °C for 5 min followed by 30 cycles at 94 °C for 30 s, 55 °C for 30 s and 72 °C for 1 min, and a final extension step of 72 °C for 10 min. After electrophoresis, the fragments were purified and sequenced at DSMA Biotecnologia (Mogi das Cruzes, SP, Brazil).
Phylogenetic analysis was performed using high-quality sequences (80% of bases with quality > 20) using Phred software. Trimmed sequences were used for comparison with sequences within the GenBank database (http://www.ncbi.nlm.nih.gov/BLAST) via BLASTn (nr/nt). The best hits corresponding well-characterized strains were retrieved from databases and subsequently aligned and edited using the Clustal Omega website from the European Bioinformatics Institute (EMBL-EBI) and the BioEdit program, version 7.0.5.3, respectively. The phylogenetic tree was built with MEGA 7.0 version software [37], and the evolutionary history was inferred using the Neighbor-Joining method [38]. Evolutionary distances were computed using the Kimura 2 evolutionary distances method [39]. The partial DNA sequences of the ITS1-5.8S-ITS2 region were deposited in the GenBank database under the accession numbers MT062480 (Fusarium sp. CCLM_DF), MT062481 (Fusarium sp. CCLM_GU), MT062482 (Fusarium sp. CCLM_GW), and MT062483 (Fusarium sp. CCLM_IB).
Results and Discussion
ATZ Degradation
Nine potential ligninolytic producer fungi (Codes CCLM_GW, CCLM_GU, CCLM_IB, CCLM_NH, CCLM_IC, CCLM_DF, CCLM_BP, CCLM_BO, and CCLM_AD) isolated from the INP were evaluated regarding their ability to biodegrade ATZ. Figure 2 shows levels of fungal ATZ degradation (%) in terms of average and standard deviation, except for CCLM_DF and CCLM_AD isolates that do not show considerable degradation activity (many negative replications).
Based on one-way ANOVA (p > 0.05), there was no significant difference between isolates in the ATZ biodegradation level. Therefore, the fungal isolates had a similar capacity to degrade ATZ (Fig. 2). Four fungi produced DEA: CCLM_GW, CCLM_GU, CCLM_IB, and CCLM_DF (6.98, 7.23, 3.97, and 0.21 mg L−1, respectively), but there was no significant difference between the four DEA-producing fungi (Kruskal Wallis; p > 0.05). These data result in an average concentration of 4.6 mg L−1. DIA formation was not observed.
These results suggested that some fungal had the capacity to form DEA (CCLM_GW, CCLM_DF, CCLM_IB, and CCLM_GU), while other did not (CCLM_NH, CCLM_IC, CCLM_BP, CCLM_BO, and CCLM_AD). The absence of relationship between ATZ biodegradation and metabolite production could be experimentally associated with ATZ adsorption/absorption. Nevertheless, our results suggested that of the nine isolates studied, four (CCLM_GU, CCLM_DF, CCLM_GW, CCLM_IB) were capable of degrading ATZ, and the other five showed potential to grow and possibly be involved in ATZ biodegradation. Fungal isolates capable of producing DEA were considered to be the ones with the greatest potential to biodegrade ATZ, since DEA formation indicates ATZ transformation, at least in cases where DEA is not absorbed.
Results revealed that DIA formation (mg L−1) did not occur in comparison to DEA formation (mg L−1). The main mechanisms of the biodegradation of s-triazine herbicides by soil fungi occur through N-alkyl side chain removal, which causes N-dealkylation, deamination, or both. This results in DEA and DIA formation, and both metabolites are independently produced through the hydrolytic, oxidative pathway [7, 16]. DEA is the most commonly identified metabolite in ATZ-contaminated groundwater in Australia. The relative degradability of DIA may be the reason it is detected at lower levels than either ATZ or DEA [40]. In addition, there has been no evidence showing that the s-triazine ring breakage occurs, according to records from 1965 to the present, as our results suggest. However, other analytes have been found, such as hydroxyatrazine (HA), deisopropylhydroxytrazine (DIHA), desethyldesisopropylatrazine (DEDIA), and desethylhydroxyatrazine (DEHA) [32, 41]. Analyzing all formed compounds could help researchers establish comparisons with other studies and uncover the metabolic pathway of fungal ATZ degradation, the details of which are unknown [16].
Several studies have evaluated the capacity of fungal isolates to biodegrade ATZ, both in a solid and liquid medium, e. g. using mycelium diameter as a response variable, and evaluating ATZ decreases and/or the presence of metabolic products [21, 32, 34]. Initial concentrations of herbicides, duration of incubation, which was frequently more than 7 days, and percentage of ATZ degradation are examples of variables used in these studies [31, 42, 43]. Considering an initial concentration of 10 mg L−1, 71% degradation of ATZ was obtained after incubation for 15 days [32], and 86.2% degradation was achieved after 42 days [31] using the fungal strains Pleurotus ostreatus UFLA and Coreolus versicolor L., respectively. Notably, these were among the isolates that had an increased capacity to degrade ATZ compared to the 7-day ATZ degradation assay of this study examining leaf litter fungi.
Toxicity Evaluation
Regarding the monitoring of toxicity, onion root growth assays were performed to compare positive controls (deionized water), with roots exposed to supernatants from fungal cultures (CCLM_GW, CCLM_GU, and CCLM_DF) and negative controls (ATZ before treatment ATZbt and the control assay ATZc). According Table S2, 10 mg L−1 ATZ inhibited the growth of onion roots. Values obtained for ATZbt (95.4%) and ATZc (100%) indicate a high toxicity level. Although the capacity of strains CCLM_DF and CCLM_GU to detoxify ATZ was not statistically different (p > 0.05) than non-treated ATZ (ATZbt and ATZc), as shown in Fig. 3, a different result was obtained using strain CCLM_GW (p < 0.05). The solution from isolate CCLM_GW was 30.3% less toxic after 72 h of treatment according to onion root growth assays. Also, the detoxification capacity of fungal CCLM_GU (27.5%) was significantly different from control (ATZc), which had the highest growth inhibition potential (100%). These results are in accordance with those of Marinho et al. [34] that revealed that biological treatment with Aspergillus niger AN 400 for 72 h resulted in a 54% decrease in the toxicity of a solution of 30 mg L−1 ATZ. In the presence of non-toxic water, cell proliferation in the root apical meristem causes root growth; however, the presence of inhibitors, namely ATZ, causes a delay (or the total inhibition) of cell division, affecting the root growth [44]. ATZ produced both cytotoxic and genotoxic effects, according to Silveira et al. [44]. They showed that root growth inhibition of 18.7 mm was observed in onion exposed to the herbicide after exposure to 30 mg L−1 ATZ for 120 h. In the study, ATZ treatment resulted in higher chromosomal aberration (CA) frequencies and increased DNA damage (99%) in onion root tips exposed to ATZ solution for 48 h. In addition, Datta et al. [30] reported that root length and concentration of an ATZ-treated soil (PTS) were negatively correlated after roots were exposed to the chemical for 24 and 48 h, while chromosomic aberrations were positively correlated after both periods. The mitotic index was reduced from 26.1 ± 1.6 to 10.3 ± 0.9 after 24 h, and from 26.1 ± 1.3 to 9.7 ± 0.6 after 48 h in 100% PTS, revealing a decrease in the proliferation status of meristematic onion cells.
Onion root growth inhibition (%) of onion roots either treated with 10 mg L−1 ATZ (ATZbt) before and after treatment with fungal isolates CCLM_GW, CCLM_GU, and CCLM_DF, and roots subjected to deionized water-containing control (ATZc) treatments for a 7-day period. Data are represented with boxplots that show the three quartiles and the upper and lower limits. The arithmetic mean of the treatments that do not share the same letters are significantly different (p < 0.05)
As described above, the capacity of fungi to biodegrade ATZ has been established, but the potential of native fungal strains from environmental protection areas to naturally promote pesticide detoxification, and promote sustainability has not been well investigated. Zablotowicz, Weaver, and Locke [45] reported a quickly degradation of ATZ (50% to 70%) in soils of the Mississippi Delta that had been exposed to ATZ for a short time. According to Krutz, Zablotowicz and Reddy [46] when the application of ATZ had terminated, ATZ biodegradation activity gradually declined over the years to a low level, which was comparable to natural attenuation in ‘non adapted’ soils that had not received ATZ in reference to the raised biodegradation of ATZ in areas with a history of ATZ exposure (‘adapted’) soils. The authors suggested that the number of soil-borne ATZ-degraders declines in the absence of ATZ to a steady, uncultivable level until ATZ is available again. Therefore, according to Douglass, Radosevich and Tuovinen [47], although the effect of the history of ATZ application has been well documented, its biological basis remains cryptic. An evaluation of potential microorganisms to biodegrade herbicides in uncontaminated environments may provide insights into the discussion. Further, agricultural frontiers appear to be unique in terms of the conflicts between production and conservation, and necessary risk management and risk reduction strategies regarding pesticide use can only be achieved through the responsible sharing of information between diverse stakeholders. This will increase knowledge regarding local biodiversity, and its functionality, and, in this case, ecosystem services provided by fungal strains.
Taxonomic Characterization
CCLM_DF, CCLM_GU, CCLM_IB, and CCLM_GW fungi were chosen for taxonomic characterization based on their capacity to produce DEA, which is an indication of ATZ biodegradation (Fig. 4).
Phylogenetic analysis was performed using high-quality sequences. Sequences were trimmed using 0.07 error probability limits using Geneious version 7.0.6 by Biomatters (http://www.geneious.com/). The ITS1-5.8S-ITS2 trimmed sequences were aligned with the sequences of selected reference taxa of the genus Fusarium within the GenBank database (https://blast.ncbi.nlm.nih.gov) using the ClustalW algorithm of MEGA 7.0 software. The alignment was inspected and adjusted manually where necessary. Evolutionary history was inferred using the Maximum Likelihood (ML) method and the GAMMA distribution model (1,000 bootstraps) using MEGA 7.0 software. The robustness of the inference was assessed by bootstrap support (values greater than 70 were considered significant, SBMV > 70) [37]
All isolates belonged to the genus Fusarium, containing a 99% probability of compatibility with the genus, class Sordariomycetes, which contained a clade with the greatest richness in tropics. Strains CCLM_DF and CCLM_GU display a close level of phylogenetic proximity with F. incamatum and F. camptoceras. CCLM_GW and CCLM_IB fungal strains were similar to many Fusarium species (F. equiseti, F. incamatum, F. camptoceras, F. oxysporum. F. verticillioides, F. proliferatum, F. chlamydosporum, F. acuminatum, and F. aveneceum), table S3. Several DNA databases are available for DNA-based identification. There was a difference in the percentage of similarity and in the level of species when comparing the databases of American government-funded National Center for Biotechnology Information (NCBI) and Centraalbureau voor Schimmelcultures (CBS), hosted by the Westerdijk Institute in the Netherlands. However, the similarity percentage is considerable enough (higher of 94%) to relate our isolates with Fusarium species. The molecular analysis was based on ITS region, a part of a ribosomal DNA gene that corresponds to one of the most frequently used regions for the fungal identification to the genus, and often species, level(s). Since it underwent a rapid evolution rate within the eukaryotes, CCLM_DF, CCLM_GU, CCLM_IB, and CCLM_GW strains could not be identified at the species level [48]. Waalwijk et al. [49] emphasized that ITS region may not be sufficient for Fusarium genus identification due to the presence of two non-homologous copies within the region, causing divergence. Instead, a widely used region for this genus is elongation factor 1-alpha or TEF-1 alpha. According to morphological characterization, all isolates presented asexual reproduction structures, a slightly curved to a hooked apical cell or fusiform of conidia (Fig. 5), which are characteristics associated principally with the Fusarium genus.
The Fusarium genus contains a large proportion of the filamentous fungi, which are widely distributed in soil and associated with plants worldwide [50]. In these symbiotic relationships, fungi are pathogens of ornamental and economically important crops, with high survival rates in the environment. Taxonomic characterization of the Fusarium genus is difficult since there exist a great variability and instability of characteristics used in its classification, such as conidial size and conidial septation (Fig. 5). In a general, Fusarium is a monilybaceous, hyphomycete genus characterized by hyaline mycelium and conidiophores. These structures can be branched and finish in tuftsor may consist of sporodoquios that give rise to macroconidia with one or several transverse septa, which are hyaline, fusoides, acrogens, and frequently pedicelled. Hyaline microconidia are produced in isolation and form wet head or chains, usually without septa. They have terminal, intercalary spores and are unicellular or have setae and thick walls [51].
Although ATZ-degrading fungi were selected based on their capacity to degrade lignin in a culture medium and their potential to produce ligninolytic enzymes according to the guaiacol colorimetric test, the results suggest that ATZ biodegradation was not achieved by laccase enzyme activity, a result also observed by Pereira et al. [32] and Bending, Friloux and Walker [31]. As a result of the observation that the production of ligninolytic enzymes in C. versicolor (a wood- or litter-inhabiting fungus) was low, mechanisms of degradation are not clearly related to ligninolytic potential. Further, P. ostreus did not produce significant levels of laccase, which demonstrated that ATZ did not act as an inductor of laccase production [31, 32]. Also, considering the absence of peptone and dextrose in the culture medium, ATZ biodegradation in MA2 is likely co-metabolic. Fungi might use ATZ as the primary food substrate.
A laccase-type intracellular enzyme has been isolated from Fusarium sp. that is capable of oxidizing guaiacol [52], which is a result that was observed in this study. In the present study, enzymatic assays used for the quantification of laccase using a synthetic lignin substrate, ABTS (2,2′-azino-bis (3-ethylbenzothiazoline-6-sulphonic acid)), revealed that no enzymatic activity was present in any biologically treated ATZ solutions (supplementary material). This result indicates that although the literature suggests that ligninolytic enzymes may have roles in pesticide degradation, the mechanism is different for our isolates that were not climatized to ATZ. Fusarium fungal isolates have the potential to degrade many herbicides, such as lindane, monocrotophos, chlorpyrifos, DDT, malathion, metolachlor, and paraquat. However, metabolic pathways involved in degradation are practically unknown [1, 7, 15]. DDT degradation by Fusarium occurs via lignin peroxidases [53]. In Fusarium, degradation of monocrotophos occurs by the release of extracellular alkaline phosphatases, inorganic phosphates, and ammonia [53, 54]. Also, the capacity of Fusarium to secrete a group of enzymes, homologous to the arylamine N-acetyltransferase (NAT) of humans, allows the organisms to tolerate aromatic amines present in environmental pollutants, like ATZ [55]. NAT is associated with pathogen fitness since they are responsible for detoxifying the defense compound 2-benzoxazolinone produced by host species [56]. N-acetyltransferase and N-malonyltransferase activities are of particular importance to acetyl coenzyme A- and malonyl coenzyme A-dependent detoxification of aromatic amines, respectively [55, 57]. Additionally, modification and detoxification of ATZ by cytochrome P450 monooxygenase genes in plants and animals may also function in fungal species [58, 59]. According to Cocaign et al. [57], ascomycetes may detoxify aromatic amines using another, unidentified metabolic pathway (aside from acetylation). Further research will be needed to fully understand the natural attenuation of ATZ and its effects on ecological processes.
Conclusion
In the present study, leaf litter fungi isolated from an environmental protection area and previously selected for guaiacol screening (potential to produce ligninolytic enzymes) were assessed to determine whether they could biodegrade ATZ. Four fungal isolates (CCLM_GU, CCLM_DF, CCLM_GW, CCLM_IB) were able to degrade ATZ in terms ofDEA formation, and the other five showed potential to grow and possibly be involved with the biodegradation of ATZ. CCLM_GW and CCLM_GU strains most effectively reduced the toxicity of ATZ according to onion assays.
A taxonomic analysis of DEA-producing fungi using morphological (reproduction structures) and molecular (ITS1 and ITS4 ribosomal DNA sequence) characteristics revealed that CCLM_DF, CCLM_GU, CCLM_IB, and CCLM_GW strains belong to the Fusarium genus. Although the fungal strains were isolated using a culture medium designed to select lignin-degrading fungi, ATZ degradation was not achieved via laccase activity. This finding underscored the importance of non-ligninolytic enzymes in the native biodegradation of ATZ, which have been neglected. All isolates studied may potentially function in the degradation of pesticides, and future research will be needed to understand their impacts fully. Though the importance of the soil history of ATZ application has been well documented, its biological basis is not well understood. Finally, increasing our understanding of biodiversity in subtropical protected areas, especially those involving ecosystem services provided by microbes, will help enhance conservation efforts.
References
Maqbool Z et al (2016) Perspectives of using fungi as bioresource for bioremediation of pesticides in the environment: a critical review. Environ Sci Pollut Res 23:16904–16925
Moore DR et al (2017) A weight-of-evidence approach for deriving a level of concern for atrazine that is protective of aquatic plant communities. Integr Environ Assess Manag 13(4):686–701
Hansen AM et al (2013) Atrazina: Un herbicida polémico. Rev Int Contam Ambiental 29:65–84
Sousa JCG et al (2018) A review on environmental monitoring of water organic pollutants identified by EU guidelines. J Hazard Mater 344:146–162
WHO (World Health Organization) (2018) A global overview of national regulations and standards for drinking-water quality. WHO, Geneva
NORTOX 500 SC. Prescripción de agrotóxico. Atrazina (2017). http://www.nortox.com.br/wp-content/uploads/2017/05/Atrazina-Nortox-500-SC-BulaVER-04-17.08.2017.pdf. Accessed 20 May 2018
Singh B, Singh K (2016) Microbial degradation of herbicides. Crit Rev Microbiol 42(2):245–261
Hayes TB et al (2002) Hermaphroditic, demasculinized frogs after exposure to the herbicide atrazine at low ecologically relevant doses. Proc Natl Acad Sci USA 99(8):5476–5480
Hirano LQL et al (2019) Effects of egg exposure to atrazine and/or glyphosate on bone development in Podocnemis unifilis (Testudines, Podocnemididae). Ecotoxicol Environ Saf 182:109400
García-Espiñeira M, Tejeda-Benitez L, Olivero-Verbel J (2018) Toxicity of atrazine- and glyphosate-based formulations on Caenorhabditis elegans. Ecotoxicol Environ Saf 156:216–222
Andleeb S et al (2016) Influence of soil pH and temperature on atrazine bioremediation. J Northe Agric Univ 23(2):12–19
Salazar-Ledesma M et al (2018) Mobility of atrazine in soils of a wastewater irrigated maize field. Agr Ecosyst Environ 255:73–83
Koskinen WC, Clay SA (1997) Factors affecting atrazine fate in North Central U.S. soils. Rev Environ Contam Toxicol 151:117–165
Smith AE, Walker A (1989) Prediction of the persistence of the triazine herbicides atrazine, cyanazine, and metribuzin in regina heavy clay. Can J Soil Sci 69:587–595
Kaufman DD, Blake J (1970) Degradation of atrazine by soil fungi. Soil Biol Biochem 2(2):73–80
Singh S et al (2018) Toxicity, degradation and analysis of the herbicide atrazine. Environ Chem Lett 16:211–237
Martinez B et al (2001) Complete nucleotide sequence and organization of the atrazine catabolic plasmid pADP-1 from Pseudomonas sp. strain ADP. J Bacteriol 183(19):5684–5697
Ma L et al (2017) Rapid biodegradation of atrazine by Ensifer sp. strain and its degradation genes. Int Biodeterior Biodegrad 116:133–140
Yang X et al (2018) Biodegradation of atrazine by the novel Citricoccus sp. strain TT3. Ecotoxicol Environ Saf 147:144–150
Deshmukh R, Khardenavis AA, Purohit HJ (2016) Diverse metabolic capacities of fungi for bioremediation. Indian J Microbiol 56(3):247–264
Fan X, Song F (2014) Bioremediation of atrazine: recent advances and promises. J Soils Sediments 14:1727–1737
Brasil Ministério do Meio Ambiente. Snuc – Sistema Nacional de Unidades de Conservação da Natureza: Lei no. 9.985, de 18 de julho de 2000; Decreto no. 4.340, de 22 de agosto de 2002; Decreto no. 5.746, de 5 de abril de 2006. Plano Estratégico Nacional de Áreas Protegidas: Decreto no. 5.758, de 13 de abril de 2006. Brasília: MMA, 2011. 76 p.
ICMBio – Instituto Chico Mendes de Conservação da Biodiversidade. Parque Nacional do Iguaçu. 2019. Accessed 06 Jun 2019. http://www.icmbio.gov.br/parnaiguacu/.
Salamuni R et al (2002) Parque Nacional do Iguaçu, PR - Cataratas de fama mundial. In: Schobbenhaus C et al (edn) C. Sítios Geológicos e Paleontológicos do Brasil. 1st edn. Brasilia, DNPM/CPRM - Comissão Brasileira de Sítios Geológicos e Paleobiológicos (SIGEP), pp 313–321. http://sigep.cprm.gov.br/sitio011/sitio011.htm. Accessed 2 Jul 2018
Schiesari L et al (2013) Pesticide use and biodiversity conservation in the Amazonian agricultural frontier. Philosoph Trans R Soc B Biol Sci 368:1619
Tedersoo L et al (2014) Global diversity and geography of soil fungi. Science 346:6213
Blackwell M (2011) The fungi: 1, 2, 3 ... 5.1 million species? Am J Bot 98:426–438
Fiskesjo G (1985) The Allium test as a standard in environmental monitoring. Hereditas 102:99–112
Leme DM, Marin-Morales MA (2009) Allium cepa test in environmental monitoring: a review on its application. Mutat Res 682:71–81
Datta S et al (2018) (2018) Assessment of genotoxic effects of pesticide and vermicompost treated soil with Allium cepa test. Sustain Environ Res 28:171–178
Bending GD, Friloux M, Walker A (2002) Degradation of contrasting pesticides by white rot fungi and its relationship with ligninolytic potential. FEMS Microbiol Lett 212(1):59–63
Pereira PM et al (2013) Optimized atrazine degradation by Pleurotus ostreatus INCQS 40310: an alternative for impact reduction of herbicides used in sugarcane crops. J Microb Biochem Technol S12:006
Della-Flora A, et al (2018) Fast, cheap and easy routine quantification method for atrazine and its transformation products in water matrixes using a DLLME-GC/MS method. Anal Methods
Marinho G et al (2017) Potential of the filamentous fungus Aspergillus niger AN 400 to degrade Atrazine in wastewaters. Biocatal Agric Biotechnol 9:162–167
Weber RWS, Pitt D (2000) Teaching techniques for mycology: 11. Riddell’s slide cultures. Mycologist 14(3):118–120
Gardes M, Bruns TD (1993) ITS primers with enhanced specificity for basidiomycetes-application to the identification of mycorrhizae and rusts. Mol Ecol 2(2):113–118
Kumar S, Stecher G, Tamura K (2016) Mega7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33(7):1870–1874
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4(4):406–425
Kimura M (1980) A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 16(1):11–120
Australian Government, Review of Atrazine - Technical report: environmental assessment. https://apvma.gov.au/node/14356. Accessed Jan 2018.
Kaufman DD, Kearney PC, Sheets TJ (1965) microbial degradation of simazine microbial. J Agric Food Chem 13(3):238–242
Jeffery S, Burgess LW (1990) Growth of Fusarium graminearum Schwabe group 1 on media amended with atrazine, chlorsulfuron or glyphosate in relation to temperature and osmotic potential. Soil Biol Biochem 22(5):665–670
Oliveira BR et al (2015) Biodegradation of pesticides using fungi species found in the aquatic environment. Environ Sci Pollut Res 22(15):11781–11791
Silveira GL et al (2017) (2017) Chemosphere toxic effects of environmental pollutants: comparative investigation using Allium cepa L. and Lactuca sativa L. Chemosphere 178:359–367
Zablotowicz RM, Weaver MA, Locke MA (2006) Microbial adaptation for accelerated atrazine mineralization/degradation in Mississippi Delta soils. Weed Sci 54:538–547
Krutz LJ, Zablotowicz RM, Reddy KN (2012) Selection pressure, cropping system, and rhizosphere proximity affect atrazine degrader populations and activity in s-triazine–adapted soil. Weed Sci 60(3):516–524
Douglass JF, Radosevich M, Tuovinen OH (2017) Microbial attenuation of atrazine in agricultural soils: biometer assays, bacterial taxonomic diversity, and catabolic genes. Chemosphere 176:352–360
Brien HEO et al (2005) Fungal community analysis by large-scale sequencing of environmental samples. Appl Environ Microbiol 71(9):5544–5550
Waalwijk C et al (1996) Discordant groupings of Fusarium spp. from sections Elegans Liseola and Dlaminia based on ribosomal ITS1 and ITS2sequences. Mycology 88(3):361–368
Srivastava S, Kadooka C, Uchida JY (2018) Fusarium species as pathogen on orchids. Microbiol Res 207:188–195
Tortora GJ, Funke BR, Case CL (2005) Microbiologia, 8a edn. Porto Alegre, Artemed
Janshekar H, Fiechter A (1983) Lignin: biosynthesis, application, and biodegradation. Adv Biochem Eng Biotechnol Berl 27:119–178
Gaur N, Narasimhulu K, Pydisetty Y (2018) Recent advances in the bio-remediation of persistent organic pollutants and its effect on environment. J Clean Prod 198:1602–1631
Jain R, Garg V, Yadav D (2014) In vitro comparative analysis of monocrotophos degrading potential of Aspergillus flavus,Fusarium pallidoroseum and Macrophomina sp. Biodegradation 25:437–446
Karagianni EP et al (2015) Homologues of xenobiotic metabolizing N-acetyltransferases in plant-associated fungi: novel functions for an old enzyme family. Sci Rep 5:1–14
De Lima DP et al (2018) Fungal bioremediation of pollutant aromatic amines. Curr Opin Green Sustain Chem 11:34–44
Cocaign A et al (2013) Biotransformation of Trichoderma spp. and their tolerance to aromatic amines, a major class of pollutants. Appl Environ Microbiol 79(47):19–26
Tan LR et al (2015) A collection of cytochrome P450 monooxygenase genes involved in modification and detoxification of herbicide atrazine in rice (Oryza sativa) plants. Ecotoxicol Environ Safety 119:25–34
Xing H et al (2014) Effects of atrazine and chlorpyrifos on cytochrome P450 in common carp liver. Chemosphere 104:244–250
Acknowledgements
We want to thank to UNILA for the facilities to develop our projects and all support of DELABEN team members while the execution of this research.
Funding
The research was supported by UNILA University and we received no specific Grant from any funding agency to carry out this work.
Author information
Authors and Affiliations
Contributions
SBE, CKP, MB and RCB planned this research. SBE and MB worked analysis of ATZ degradation and the production of principal metabolites using GC–MS. SBE and RCB worked on morphological characterization, wrote the article and designed figures. GFS made the toxicity evaluation of ATZ and fungi-treated ATZ by Allium cepa test, DCD, WLA and RCB developed isolates’ molecular characterization.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Esparza-Naranjo, S.B., da Silva, G.F., Duque-Castaño, D.C. et al. Potential for the Biodegradation of Atrazine Using Leaf Litter Fungi from a Subtropical Protection Area. Curr Microbiol 78, 358–368 (2021). https://doi.org/10.1007/s00284-020-02288-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-020-02288-6