Abstract
The diversity and community composition of archaea in soil samples from three wetlands (SP1, SP2, and SP3) of Ebinur Lake were studied by constructing 16S rDNA cloning library. The correlation between the diversity of archaea and soil environmental factors was analyzed by CANOCO software. The aim of this study was to reveal the differences of community structures of archaea in different sample sites, to provide a theoretical basis for further study on degradation and restoration of Ebinur Lake wetland. The results showed that Euryarchaeota accounted for 57.1% was the most dominant phylum observed, followed by Thaumarchaeota and Crenarchaeota for the three wetland soil analyzed. Compared with SP3 site, the proportions of Euryarchaeota were decreased by 16.70% and 31.78%, while Thaumarchaeota increased by 7.26% and 17.64% in the SP1 and SP2, respectively. Crenarchaeota was found only in SP3. Shannon–wiener diversity indices in SP1, SP2, and SP3 sites were 3.44, 3.87, and 3.94, respectively, indicating that the diversity of archaea in three plots was: SP3 > SP2 > SP1. Redundancy analysis (RDA) showed that electrical conductivity (EC), soil moisture (SM), hydrogen potential (pH), and soil organic matter content (SOM) may affect archaeal communities. Compared to EC and pH, SM and SOM may have a greater impact on the community composition of archaea.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Archaea is one of the hotspots of biogeochemistry research, and can exist under extreme conditions such as high temperature, strong acid, alkaline condition, high salinity, and hypoxia. They are diverse and play an important role in the biogeochemical process [1]. Research in this field is of great significance to clarify the basic laws of life movement, revealing the origin of life and evolution of species, and the interaction between the biosphere and the geosphere environment [2].
The Ebinur Lake Wetland, located in northern part of Xinjiang, is a climate regulation hub in the Ebinur Lake basin [3]. However, due to the large-scale agricultural reclamation and water diversion activities since the 1950s, a large number of reservoirs have been built in the upper reaches of the river, resulting in the continuous degradation of Ebinur Lake and the increasingly serious environmental problems [4]. These human factors have reduced the number of rivers flowing into Ebinur Lake, and now the main rivers flowing into Ebinur Lake are the Boltala River and the Jinghe River. Changes in these two rivers may directly affect the soil situation of the Ebinur Lake wetland. Changes in environmental factors in ecosystems can lead to changes in the composition of microbial communities [5]. Therefore, microbial diversity can be viewed as an important indicator for assessing soil changes caused by natural or man-made disturbances.
In this paper, soil samples were collected from three sampling sites in the Bortala and Jinghe River basins of Ebinur Lake Wetland in Xinjiang and the 16S rDNA clone library of archaea was constructed by gene clone library technology. Redundancy analysis (RDA) between diversity of archaea and soil environmental factors was carried out using CANACO software. The purpose of this study was to reveal the diversity of archaea in three different wetland, in order to explore the potential power of archaeal communities in the process of ecological function restoration of the continuously salinized Ebinur Lake Wetland, and to provide theoretical and data basis for the degradation and restoration of Ebinur Lake Wetland.
Materials and Methods
Site Description and Sample Collection
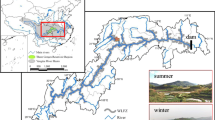
Ebinur Lake Wetland National Reserve is located in the northwest of Jinghe County, Xinjiang province (82°36′–83°50′E, 44°30′–45°09′N). It is the center of the lowest depression and saltwater in this area [6]. Sampling was carried out during July 2014 from three sampling sites: the main inlet lake of the Jinghe River in the Lake District of the Bird Island Protection Station (SP1), the meadows of the Bird Appreciation Station near the main inlet lake in Bole (SP2) and the salt works in the overflow area of the main inlet lake in Jinghe (SP3) (Table 1). Sampling methods of each sample sites were as follows: the horizontal direction, three sampling points were selected at intervals of 300 m, and three sites were randomly selected for each sampling point. After removing the plant residues and other impurities in the surface layer, equal amount of soil samples was collected in the 10–35 cm soil layer in the vertical direction. Three soil samples from the same sampling point were mixed evenly and divided into two portions. One portion was placed in a portable refrigerator and brought back to the laboratory for storage in an − 80 °C freezers for the extraction of soil microbial total DNA, and the other was naturally air dried in the laboratory for the determination of soil physicochemical indices.
Description of Measurements of Environmental Factors
Soil moisture content (SM), potential of hydrogen (pH), and soil organic content (SOM) were determined by dry specific gravity method, electric potential method, and the potassium dichromate method (external heating method), respectively. But soil sample for measuring SM was dried under natural conditions until stable weight was achieved. Electrical conductivity method was used to determine conductivity (EC) using Dj-320 conductivity meter. All soil physicochemical indicators were assayed as described by Lu [7]. The soil samples were provided with three biological replicates.
Total DNA Extraction, PCR, Clone Library Construction
Genomic DNA was extracted from soil using Power Soil® DNA Isolation Kit (MOBIO, USA) according to the manufacturer’s instructions. 16S rRNA genes of archaea were amplified using the archaea universal primers Arch 21F (5′-TTCCGGTTGATCCYGCCG GA-3′) and Arch 958R (5′-YCCGGCGTTGAMTCCAATT-3′). PCR was performed for each sample with the following thermal cycling: an initial denaturation at 94 °C for 5 min, followed by 35 cycles of 30 s at 94 °C, 30 s at 59 °C and 90 s at 72 °C, final extension carried out at 72 °C for 10 min. The presence and sizes of the PCR amplification products was determined by an agarose gel electrophoresis. PCR products were purified using Easy PureTM PCR purification kit (Axygen, USA) and connected into PMD18-T vector from Tangen company. The positive clones of PCR amplified products, screened for correct size and grouped by restriction enzymes Afa I and Msp I, were sent to a commercial sequencing company for cloning and sequencing (Sangon, Shanghai, China).
Analysis of Archaea Community Structure
The quality filtering, chimera removal, and operational taxonomic unit (OTU)-based clustering of the sequences were performed using the Dotur software [8]. The sequences of all microorganisms were aligned using the software ClustalX-1.83 [9]. Firstly, Operational Taxonomic Units (OTU) based on 97% DNA sequences identity were calculated using software Phylip and Dotur. Then nucleotide sequences were aligned using BLASTn. Search blast will be used to determine the sequence of genes in the NCBI database for online comparison, and to obtain the most similar gene reference sequences. Finally, the neighbor-joining method [10] was used to construct gene phylogenetic tree based on bootstrap method by Mega 7 with 1000 bootstrap replicates. Analysis of archaea community structure was based on a phylogenetic tree.
Statistical Analysis
The Shannon–Wiener diversity index was used to measure microbial diversity [11]. The estimated coverage of each clone library was calculated using the formula C = (1 − n/N) × 100%, where C is the homologous coverage. n is the number of OTUs with only one sequence, and N is the total number of clones analyzed [12]. Rarefaction curves calculated by the aRarefactWin program were used to determine that if libraries were saturated [13]. The diversity indices for each sample, including Shannon, were generated by Dotur software. Correlation between archaeal diversity and soil environmental factors was analyzed by the redundancy analysis (RDA) using the software CANOCO for Windows.
GenBank Accession Number
All sequences obtained from the database were deposited in GenBank under accession number: KT165096-KT165228.
Results
Characteristics of Environmental Variables
There are some differences in soil physiochemical indexes among the three sampling sites (Table 2). There was no significant difference in SOM content among the three sites. According to the second national soil census nutrient grading standard [14], the soil organic matter (SOM) content of SP1, SP2, and SP3 was 6, 5, and 6 grade standards, respectively, which showed that SOM content in the Ebinur Lake wetland was scarce. There was no significant difference of EC and pH levels between the sites. The pH values of the three plots were above 8, indicating that the wetland soil of Ebinur Lake was alkaline. SM content of the three sites was relatively low. But compared with SP2 and SP3, SM content in SP1 was the lowest.
Diversity Analysis of Archaea
In sum, 399 OTUs were divided in three sampling sites (Table 2). Compared with SP1 and SP2 sampling sites, the number of OTUs of SP3 was relatively large. The Shannon–wiener diversity index showed SP3 were higher than SP2 and SP1. And in combination with the data of the Schao 1 index, it should be that SP3 had higher diversity and richness. Coverage percentage ranged from 75.57% to 89.84%. The whole curve has tended to be flat, which showed that the sampling was reasonable and can reflect the archaeal community in soil samples more truthfully (Fig. 1). These results indicated that about 300 clones were basically sufficient to estimate archaeal diversity within one library in this study.
Rarefaction curves of the 16S rDNA clone libraries. For sample abbreviations, see Table 1
Community Structure Analysis of Archaea
Representative sequences obtained and known archaeal strains in soil environment samples were used to construct a phylogenetic tree (Fig S1, Fig S2, and Fig S3). The homology between the sequences and known sequences in Genbank ranged from 78 to 100%. All the tested clones were divided into 22 phylogenetic genera (6, 10, and 15 genera in SP1, SP2, and SP3, respectively) including one unclassified bacteria group (Fig. 2b). Accounting for 77.07% of the total sequencing clones, Euryarchaeota represented the dominant phylum in each soil. And Halobacteriaceae was the absolute dominant family in the three sites. With 2.02–19.66% of the clones in each soil, Thaumarchaeota was the second most abundant phylum in the SP1 and SP2 sites. The presence of Crenarchaeota was found only in SP3, accounting for 4.04% of the total clones, making it the second abundant phylum, while Thaumarchaeota was the third abundant phylum. Approximately 19.08% of the total clones remained unclassified (Fig. 2a).
Euryarchaeota and Thaumarchaeota were the most abundant phyla, but there were differences in the level of genera in three sample sites. Natronomonas (5.05%), Haloterrigena (5.05%), and Halovivax (5.05%) were the dominant genera for SP3. The dominant genera in SP1 and SP2 were Halalkalicoccus (11.34%) and Nitrososphaera (15.38%), respectively. In addition, more genera were found in SP3 when compared to three sampling sites. Haladaptatus (1.71%), Methanosarcina (2.56%), Methanosaeta (0.85%), Halolamina (0.85%), and Nitrososphaera (15.38%) were found only in SP2. Likewise, Halosimplex (5.13% and 1.01%, respectively), Haloterrigena (0.85% and 5.05%, respectively), and Halovivax (3.42% and 5.05%, respectively) were found only in SP2 and SP3, not found in SP1. Natrinema were not detected in SP2 and SP3, and Halorussus were not detected in SP2. In total, distribution of archaea was unbalanced in the three sampling sites of the Ebinur Lake Wetland.
Correlation Analysis Between the Diversity of Archaea and Environmental Factors
In order to further study the different responses of microbial communities in the soil environment, RDA analysis and CANOCO software were employed to group the microbial community, based on similarities in their 16S rDNA profiles (Fig. 3). Compared with other soil environmental factors, SM and SOM were the main environmental factors affecting the diversity and distribution of archaea. And the diversity of archaea in the SP2 and SP3 was relatively high when compared with SP1. Thus, in the increasingly desertified Ebinur Lake Wetland, SM and SOM were important environmental factors determining archaea diversity.
Discussion
Community Composition and Diversity of Archaea
In this study, soil samples from three plots of Ebinur Lake Wetland were collected, and 16S rDNA clone library of archaea was constructed to analyze the diversity of archaea and their community composition. Diversity results showed that the diversity index of SP3 was larger than the other two sites, and diversity index of archaea in three plots was: SP3 > SP2 > SP1. This may be due to the different vegetation types in the three sites, resulting in different soil environments. Su et al. [15] showed that plant community types were closely related to soil microbial diversity. Wang et al. [16] showed that Reed, a halophyte, could adapt to saline environment by increasing the root system's access to deep soil moisture and nutrients. In this study, roots of Salicornia and Reed are more developed than those of Halocnemum Strobilaceum. They may have a highly active root network system, which creates favorable conditions for rhizosphere microbial activities. Through the analysis of the diversity of the archaeal community structure in the Ebinur Lake Wetland, it was found that Halobacteriaceae belonging to Euryarchaeota was the absolute dominant family in three sites. This was similar to the results of archaeal diversity studied by Sandaa et al. [17], Dave et al. [18], and Zafrilla et al. [19] in different salt environments, which showed that Halobacteriaceae was present as a dominant family in most terrestrial high-salt environments. Halobacteriaceae maintained a dynamic balance with a high concentration of Na+ in the environment by accumulating a large amount of K+ in the cell, so that it can live in a high-salt environment and exist as a dominant microbiota [20, 21]. From the soil pH results, it can be seen that the soil solutions of the three sampling sites were alkaline. It may be that this adaptation mechanism makes Halobacteriaceae a dominant microbiota in three sites.
At the genus level, Halalkalicoccus, Nitrososphaera, and Natronomonas were dominant genera in SP1, SP2, and SP3 sites, respectively. Xu et al. [22] and Cui et al. [23] studied the diversity of archaea in the Ebinur Lake wetland by cultivable methods and found that Haloterrigena was the dominant genus, which was different from the results obtained in this experiment. The research method of this experiment was not culturable, so the difference may be caused by different culture methods. Torsten et al. [24] studied archaea diversity in three high-salt environments by non-culture and culturable methods. It was found that Halomicrobium was the dominant group, but the results obtained by non-culture method were quite different from those obtained by culturable method. Pan Hong et al. [25] studied the archaeal community composition in volcanic sediments by non-culture method, the results showed that Nitrosotalea was the dominant population, followed by Nitrososphaera. This is similar to the results of the study. At present, only 0.3% of the total soil microorganisms can be cultured [26]. Compared with the culturable method with some limitations, the results obtained by non-cultured method are more comprehensive and representative, which may be the reason for the similarities and differences between the results of this experiment and other research results.
Effects of Soil Physical and Chemical Factors on the Community Structure of Archaea
Preliminary analysis of soil physical and chemical properties showed that there was no significant difference between EC and pH values in the three sites, but the overall trend was decreasing. Various salts in the soil leachate generally exist in the form of ions, the content of total salt in the soil is generally expressed by soil conductivity [27]. The results of diversity index in three sites showed that the diversity of archaea was the highest in SP3 site. Li [28] and Sun et al. [29] showed that the microbial diversity will show a downward trend as the soil salinity increases. Cao et al. [30] also identified that the salt content in the soil has a more significant effect on the microbial community structure than the carbon and nitrogen content in soil. Sardinha et al. [31] and Li et al. [32] showed that the pH and salinity of the soil can not only greatly affect the microbial community structure, but also restrict the microbial diversity in the soil. This is different from the results of this experiment.
In this experiment, EC and pH values showed a decreasing trend in the three sample plots, but the difference between them was not obvious. As described by previous research results, the diversity of archaea in the three sample plots should have been almost the same, but in this experiment, diversity of archaea was SP3 > SP2 > SP1. This may be due to the dominant rhizosphere effect of plants. The vegetation roots of SP2 and SP3 plots are well developed, and the plant roots can secrete many secretions, such as sugar, organic acid, amino acid, hormone, extracellular enzyme, and so on. These substances can provide enough energy for microorganisms; thus, forming a rhizosphere environment suitable for the growth of microorganisms [33]. This may be the reason why the results of this experiment are different from those of previous studies.
RDA results showed a positive correlation between archaeal diversity and SM. The water in the soil exists in the form of a solution and has a certain osmotic pressure. When the water content in the soil is kept within a certain range, it will not have a great influence on the microorganisms; if the soil water content exceeds the soil field water holding capacity, the excessive osmotic pressure of soil solution will lead to water loss of microbial cells, which will make microorganisms face drought stress [34]. The Ebinur Lake Wetland National Nature Reserve is one of the most representative wetland desert ecosystems in temperate arid regions in China. SM content was generally low in wetland soil. However, compared with SP1, the SM content in SP2 and SP3 was relatively high, which may be one of the reasons for the high diversity of archaea in SP2 and SP3.
SOM provides nutrients and energy for the growth of microorganisms in the soil [35]. The higher the SOM content in the soil, the stronger the buffering effect of microorganisms on the environment [36]. RDA analysis showed that SOM content had greater influence on the diversity of archaea than pH and EC. This was consistent with the results of this study. Although SOM content in the three sites was relatively low, the SOM content greatly affects the diversity of archaea in the relatively barren wetland soil of Ebinur Lake.
In conclusion, this study provided the existence of archaeal diversity in the Ebinur wetland. The results of community composition of archaea indicated that all sequences obtained in the three sites were divided into three categories: Euryarchaeota, Thaumarchaeota, and Crenarchaeota, and most sequences in three sites belonged to Halobacteriaceae, which showed soil environment of SP1, SP2, and SP3 were suitable for growth of Halobacteriaceae. Crenarchaeota was relatively rare and found only in SP3, which indicated that the archaeal diversity in SP3 was relatively high and its soil environment was suitable for the growth of the Crenarchaeota phylum. In addition, among the four soil environmental factors, only SM and SOM were the two main environmental factors affecting microbial diversity. This showed that the content of SM and SOM in Ebinur Lake wetland can play a great role in the restoration of Ebinur Lake Wetland.
References
Deng F (2018) Advances in archaea research. J Anhui Agr Sci 46(28), 11–14+47.
Li SG, Pi YD, Zhang CL (2007) Archaea research and its prospects. J Univ Sci Technol China 37:830–838
Aller JY, Kemp PF (2008) Are archaea inherently less diverse than bacteria in the same environments? Fems Microbiol Ecol 65:74–87
Su XM, Liu ZH, Wei TF, Wang YJ, Liu Y (2016) Response of the area change of Ebinur Lake and its changes in runoff characteristics. Soil water conserv res 23:252–256
Clark JS, Campbell JH, Grizzle H, Acosta-Martìnez Veronica, Zak JC (2009) Soil microbial community response to drought and precipitation variability in the Chihuahuan Desert. Microb Ecol 57(2):248–260
Zhang Y, Zhang F, Wang J, Ren Y, Abduwasit G, Hsiang-te KUNG (2017) Temporal and spatial dynamics and landscape pattern changes of ecological interference in the Ebinur Lake Wetland Nature Reserve in Recent 40 Years. J Ecol 37:7082–7097
Lu RK (1999) Chemical analysis methods of soil agriculture. Agriculture Science and Technology Press, Beijing
Schloss PD, Handelsman J (2005) Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl Environ Microbiol 71(3):1501–1506
Li L, Lin Y, Liu Y, Zeng LL, Li CT (2003) Comparison between mtDNA sequencing results and Anderson standard sequences–Application of shared software such as Clustal X in forensic evidence. Chin Judic Ident 27–31.
Liu P, Zhong M, Xu JS, Guo QY, Liao CQ (2018) Life history and phylogenetic analysis of argopistes tsekooni. North Horticult 05:59–63
Huang HF (2015) Forest biodiversity assessment in Sanming City based on Shannon-Wiener index. J Fujian Sci Technol 42:121–124
Singleton DR, Furlong MA, Rathbun SL, Whitman WB (2001) Quantitative comparisons of 16S rRNA gene sequence libraries from environmental samples. Appl Environ Microbiol 67:4374–4376
Webster NS, Negri AP, Munro MM, Battershill CN (2004) Diverse microbial communities inhabit Antarctic sponges. Environ Microbiol 6(3):288–300
Zak DR, Holmes WE, White DC, Peacock AD (2003) Plant diversity, soil microbial comuunities and ecosystem function: are there any links? Ecology 84:2042–2050
Su XL, Li YB, Yang B, Li Q (2018) Effects of plant diversity on soil microbial communities in subtropical forests. Chin J Ecol 37(8):2254–2261
Wang JW, Zhao CZ, Zhao LC, Wang XP, Li Q (2018) Response of root morphology and biomass of Phragmites australis to soil salinity in inland salt marsh. Acta Ecol Sin 38(13):4843–4851
Sandaa RA, Enger Torsvik V (1999) Abundance and diversity of archaea in heavy-metal-contaminated soils. Appl Environ Microbiol 65:3293–3297
Dave BP, Soni A (2013) Diversity of halophilic archaea at salt pans around Bhavnagar Coast, Gujarat. Proc Natl Acad Sci India B 83:225–232
Zafrilla B, Martínezespinosa RM, Alonso MA, Bonete MJ (2010) Biodiversity of archaea and floral of two inland saltern ecosystems in the Alto Vinalopó Valley, Spain. Aquat Biosyst 6(1):10
Green JL, Holmes AJ, Westoby M, Oliver I, Briscoe D, Dangerfield M (2004) Spatial scalingof microbial eukaryote diversity. Nature 432(7018):747–750
Da CM, Santos H, Galinski EA (1998) An overview of the role and diversity of compatible solutes in Bacteria and Archaea. Adv Biochem Eng/Biotechnol 61:117–153
Xu XW, Wu M, Totti D, Ababekley GB (2006) Cultivation of halophilic archaea diversity in Ebinur Lake and Yiwu Lake, Xinjiang. Biodiversity 14:359–362
Cui HL, Yang Y, Diliber T, Zhou PY, Liu SJ (2006) Study on the diversity of halophilic archaea cultured in two salt lakes in Xinjiang. J Microbiol 46:171–176
Torsten O, Felicitas P, Christa S (2002) Diversity of archaea in hypersaline environments characterized by molecular-phylogenetic and cultivation studies. Extremophiles 6(4):267–274
Pan H, Wang XY, Fang ZX, Wu Y (2018) Diversity and phylogenetic analysis of the archaea in volcanic sediments of the Wudalianchi region, China. Chin J Appl Environ Biol 24(05):1000–1008
Wang BJ, Liu SJ (2013) Advances in new technologies of environmental microbial culture. Microbiology 40(1):6–17
Bao SD (2000) Soil agrochemical analysis. China Agriculture Press, Beijing
Li X (2014) Microbial diversity of different saline-alkaline soil analyzing by PLFA in the Hetao area of inner Mongolia. Ecol Sci 33(03):488–494
Sun JJ, Yi JD, Jie YH, Yang YL, Su XW, Liu BD (2010) Study on microbial ecological characteristics of Tianjin coastal saline-alkali soil. J Nanjing For Univ 34:057–061
Cao YS, Fu SL, Zou XM, Cao HL, Shao YH, Zhou LX (2010) Soil microbial community composition under Eucalyptus plantations of different age in subtropical China. Eur J Soil Biol 46:128–135
Sardinha M, Müller T, Schmeisky H, Joergensen RG (2003) Microbial performance in soils along a salinity gradient under acidic conditions. Appl Soil Ecol 23:237–244
Li X, Zhang HH, Yue BB, Jin WW, Xu N, Zhu WX, Sun GY (2012) Effects of mulberry-soybean intercropping on microbial diversity of carbon metabolism in saline-alkaline soil. J Appl Ecol 23:1825–1831
Zhang F, Shen J, Li L, Liu X (2004) An overview of rhizosphere processes related with plant nutrition in major cropping systems in China. Plant Soil 260(1/2):89–99
Killham K (1994) Soil ecology. Cambridge University Press, Cambridge
Franzluebbers AJ (2010) Soil organic matter stratification ratio as an indicator of soil quality. Soil Tillage Res 66:95–106
Marschner P, Kandeler E, Marschner B (2003) Structure and function of the soil microbial community in a long-term fertilizer experiment. Soil Biol Biochem 35:453–461
Acknowledgements
This work was supported by the National Natural Science Foundation of China [Grant Number 31160026]. We declare that we have no financial and personal relationships with other people or organizations that can inappropriately influence our work. The authors would like to thank Gao Xiang, Director of the Ebinur Lake wetland National Nature Reserve administration, and Xu Wei, chief of the Ebinur Lake wetland bird island station in Xinjiang, for having permitted the collection of samples for this study. The authors would like to thank the editor and anonymous reviewers for their valuable comments and suggestions to improve the quality of this paper.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
He, S., Tan, J., Hu, W. et al. Diversity of Archaea and Its Correlation with Environmental Factors in the Ebinur Lake Wetland. Curr Microbiol 76, 1417–1424 (2019). https://doi.org/10.1007/s00284-019-01768-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-019-01768-8