Abstract
Successful clones of Acinetobacter baumannii cause a variety of nosocomial infections through serum resistance, biofilm formation, and antimicrobial resistance as virulence capabilities. Fifty clinical isolates of multidrug-resistant (MDR) A. baumannii were analyzed for clonal relatedness, serum resistance, biofilm formation, and in vivo assays. Furthermore, some virulence genes, sequence variation of ompA, and its expression were studied. The MLST (multilocus sequence typing) results showed that there were three sequence types among MDR isolates including ST2 (64%, 32/50), ST513 (30%, 15/50), and ST1 (6%, 3/50). The data showed that the clinical isolates recovered from sputum had mostly high biofilm-formation capacity, while isolates recovered from host interior fluids had high serum resistance. The results of PCR assays and in silico analysis represented patterns of virulence genes and even ompA sequence variations among MDR isolates which were clonally dependent. While quantitative reverse transcription-polymerase chain reaction (qRT-PCR) analysis showed that bacteremia-producing strains in C57/BL6 mice significantly overexpress ompA (P < 0.05) and have a direct relation with the level of IL-6 in bloodstream of mice. Moreover, the expressions of ompA among indistinguishable clones (ST2 or ST513) were clonally independent.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Acinetobacter baumannii has emerged as a challenge to the modern therapeutic system because of increasing infections and global spread of multidrug-resistant (MDR) strains [1]. In spite of its clinical importance, the mechanism of pathogenicity in A. baumannii remains uncertain [2]. One of the major mechanisms in the pathogenesis of A. baumannii is biofilm formation [3]. This bacterial strategy is associated with persistence of organism on indwelling medical devices [4]. Bap (biofilm-associated protein) is involved in biofilm formation [4]. AbOmpA plays multiple roles in bacterial pathogenesis, including adherence to host cells, induction of cell death, and serum resistance [5,6,7]. In A. baumannii, several proteins potentially play roles in adherence or biofilm formation, for example: Ata (Acinetobacter trimeric autotransporter) acts as adhesive material [8]; beta-lactamase PER-1 probably has correlated with the capacity to form biofilm and to adhere to epithelial cells [9]; filamentous hemagglutinin adhesion B (FhaB) is associated with biofilm formation [10], and phosphorylcholine (ChoP) interacts with host cells. [11].
Recently, it has been shown that A. baumannii harbors a wide repertoire of virulence factors, which are common to the genomes of most strains, but are differentially expressed according to each individual strain [6]. The purpose of this study was to perform molecular detection and in vitro and in vivo evaluations of virulence factors involved in serum resistance, biofilm formation, and cell attachment; additionally, clonal relatedness among MDR isolates of A. baumannii strains was analyzed.
Materials and Methods
Multilocus Sequence Typing (MLST)
In this study, 50 nonduplicate clinical MDR isolates of A. baumannii were considered randomly as previously described [12]. MLST was performed using Pasteur’s scheme as previously described http://pubmlst.org/abaumannii/. The nucleotide sequences and allele profiles of amplified genes (fusA, gltA, pyrG, recA, cpn60, rplB, and rpoB) were compared in http://pubmlst.org/abaumannii/ to identify the allele numbers and the sequence types (STs).
Serum-Resistance Assay
Serum resistance assay was performed based on a modified protocol as described in the study by Antunes et al. [13]. In brief, bacteria were cultured overnight on Muller Hinton agar. The next day, a single colony was inoculated in BHI broth until OD600nm reached values in the range of 0.08–0.13. Each strain of A. baumannii was separately suspended in 40% normal human serum (NHS) and heat-inactivated serum used as controls. Following 3-h incubation at 37 °C, bacterial isolates were serially diluted in PBS and cultured on Blood agar to determine colony counts (CFU/ml). A. baumannii strain AB-44 [14] and E. coli ATCC 25922 were used as highly serum-resistant and highly serum-susceptible strains, respectively. The CFU/ml of bacteria in heat-inactivated serums were divided into CFU/ml of bacteria in 40% NHS. The ratios of 0.6–1, < 0.6–0.4, and < 0.4 were considered as serum resistant (R), moderate (M), and susceptible (S), respectively.
Biofilm-Formation Test
Biofilm formation was determined using crystal violet method. In brief, bacterial suspensions in LB broth with 0.5% glucose were aliquoted in 96-well polystyrene microtiter plate and incubated at 37 °C for 72 h. After three washes with PBS, the crystal violet (0.1% W/V) bound to the adherent cells was re-solublized in ethanol/acetone 80:10 (V/V) and quantified in OD570nm using ELISA reader (BioTek Synergy4, Winooski, USA). The detailed method and biofilm capacities of the test strains were categorized as previously defined [15].
Detection of Virulence Genes
PCR assays were carried out for detection of six virulence genes including choP, ata, bap, fhaB, blaPER-1, and ompA using a set of specific primers and different annealing temperatures as shown in Table 1. The ompA genes in all isolates were also sequenced. The nucleotide sequences were compared with GenBank database using BLAST tool (https://blast.ncbi.nlm.nih.gov/Blast.cgi). Moreover, virulence genes on the selected genomes of various STs were detected using BLAST tool in ENSEMBL server (http://bacteria.ensembl.org/Multi/Tools/Blast?db=core).
ompA Expression
Eighteen isolates with strong serum-resistant capability were selected and analyzed for expression of the ompA gene. In brief, the bacteria were cultured in Luria–Bertani broth until OD reached 0.6 in 600 nm. The bacterial RNA was extracted using AccuZol kit (Bioneer, Seoul, Korea). Relative quantitative reverse-transcription polymerase chain reaction (qRT-PCR) was performed using a LightCycler® 96 (Roche Diagnostics, Mannheim, Germany) according to the protocol as previously described [16]. The primers and TaqMan probes were used as described in Table 1. The experiment was repeated in triplicate assay. The negative controls were TaqMan Real-Time PCR Master Mix (ThermoFisher, USA) without reverse-transcriptase that confirmed the absence of DNA contamination in the samples. Relative gene expression was determined using the 2−ΔΔCt method. The 16S rRNA gene was used as a reference gene for normalization of gene expression, and A. baumannii ATCC 19606 was used as a reference sample.
Spleen Colony Counts and Serum IL-6 Measurements
In this study, a murine model of disseminated peritoneal sepsis was used as described elsewhere [17]. In brief, bacteria were cultured overnight on Mueller-Hinton agar. The next day, a single colony was inoculated in BHI broth until OD600nm reached to 0.08–0.13. 100 μl of bacterial suspensions (including ~ 500 colony-forming units) was mixed with 100 μl of 10% (V/V) porcine stomach mucin (Sigma, USA). This suspension was injected intraperitoneally into 6–8 week-old C57BL/6 mice. The spleens aseptically were extracted followed by anesthetization of mice. The spleen suspensions were cultured on blood agar plates. After 24 h, the colonies were counted. A. baumannii ATCC 19606 was used as a reference strain. Moreover, for detection of bacteria in bloodstream, 100 µl heart’s blood was cultured on blood agar plates. The bacteremia of the selected strains was reported according to CFU/ml (Bacteremia index). Serum levels of IL-6 in mice were determined using IL-6 ELISA Ready-SET-Go kit (eBioscience, San Diego, Calif., USA) and measured using ELISA reader (BioTek Synergy4, Winooski, USA). Three mice were considered for each isolate. Moreover, spleen extractions, heart’s blood, and serum collections were obtained simultaneously 18 h post-intraperitoneal injections.
Statistical Analyses
The correlation between two ordinal variables was tested using Spearman’s rank correlation ρ test. Moreover, the in vivo results of strains were compared by One-Way ANOVA or unpaired t test. Statistical analyses and graphs were performed using GraphPad Prism version 6.01 (GraphPad Software Inc., San Diego California, USA). In all experiments, P < 0.05 was considered statistically significant.
Ethical Statement
This project was done based on ethical guidelines as previously approved by the Pasteur institute of Iran (Ethics No.: IR.PII.REC.1397.015).
Results
MLST Results
The sequences analysis revealed the presence of three sequence types among isolates including ST2 (64%, 32/50), ST513 (30%, 15/50), and ST1 (6%, 3/50) as shown in Table 3. The allele numbers of seven housekeeping genes among reported STs are as follows: ST1 (1–1–1–1–5–1–1), ST2 (2–2–2–2–2–2–2), and ST513 (56–3–55–2–9–4–14).
Serum Resistance and Biofilm Formation
The serum-resistance capacity among isolates was graded as 18% (9/50) sensitive, 28% (14/50) moderate, and 54% (27/50) resistant (Table 3). The biofilm-formation capacity among isolates was graded as 22% (11/50) weak, 66% (33/50) moderate, and 12% (6/50) strong (Table 3). No significant positive or negative correlation was observed in Spearman’s rank correlation (ρ) test between serum resistance and biofilm formation (P > 0.05).
Detection of Virulence Genes
PCR assays and in silico analysis revealed that patterns of virulence genes were strongly related to Pasteur’s ST as shown in Tables 2 and 3. The patterns of virulence genes including choP, ata, bap, fhaB, blaPER-1, and ompA types among ST2, ST513, and ST1 were (−, +, +, +, − and type I), (−, −, +, −, + and type II), and (+, −, +, −, − and type II), respectively. DNA sequences of ompA type I and type II were deposited in GenBank database under accession numbers KP271243 and KP271242, respectively. The naming of type I and II is contractual.
Spleen Colonization and Determination of Bacterial Loads in Bloodstream
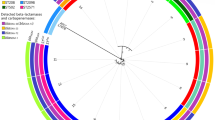
We determined the bacterial loads in spleen to estimate proliferation capacity of strains in mouse model. Moreover, the bacterial loads in bloodstream (calculated as ‘bacteremia index’, BI = CFU/ml of bacterial loads in bloodstream obtained 18 h’ post-injection) were measured for all mice [18]. The average of Log10 CFU/g ± SD of bacteria recovered from spleens of C57/BL6 mice is shown in Fig. 1. The range of Log10 CFU/g was from 4.10 (AB-40 strain) to 7.72 (AB-33 strain). One-Way ANOVA test showed that the Log10 CFU/g results of strains (except AB-40, AB-38, and AB-15) had significant differences with reference strain (P < 0.05). Moreover, the bacteremia was confirmed in 9/18 of strains (Table 3).
The mean of Log10 CFU/g ± SD among MDR isolates recovered from spleens of C57/BL6 mice. There is a wide range of Log10 CFU/g from 4.10 (AB40 strain) to 7.72 (AB33strain). The One-Way ANOVA test showed the Log10 CFU/g of MDR strains) had significant differences with reference strain (P-value < 0.05). A. baumannii ATCC 19606 was used as reference strain
Levels of IL-6 in Serum
It seems that the serum level of IL-6 can be considered as a serum marker characterizing the invasiveness of A. baumannii strains [19]. Therefore, we measured this proinflammatory cytokine in mice serum. We selected two groups of MDR isolates based on spreading in bloodstream in C57/BL mice. Group A including AB-38, AB-40, AB42, AB-47, AB-30, AB-1, AB-5, AB-45, and AB-43 (with spreading capability in bloodstream); and group B including AB-32, AB-12, AB-11, AB-15, AB-3, AB-19, AB-46, AB-41, and AB-33 (without spreading capability in bloodstream) are shown in Fig. 1. The average levels of IL-6 ± SD among groups A and B were 229.38 ± 24.22 and 145.15 ± 12.04 pg/ml, respectively. Unpaired t test showed that the amounts of IL-6 between groups A and B had a significant difference (P value < 0.05). See Fig. 2.
The amounts of IL-6 (pg/ml) in serum of C57/BL6 mice. There are two groups A (with capability of bacteremia in mice) and B (without capability of bacteremia in mice) among clinical MDR isolates. Unpaired t test showed that the levels of IL-6 between groups A and B had a significant difference (P-value < 0.05)
ompA Expression
The means of fold-changes in ompA expressions among MDR isolates have been shown in Table 3. The range of expression was 0.823–2.005. Unpaired t test results showed that groups A and B were statistically significant (P < 0.001). However, there was no statistically significant difference between ompA expression of ST2 and ST513 (P > 0.05).
Discussion
Acinetobacter baumannii causes a variety of nosocomial infections with a particular ability to survive in hostile environments and capture numerous antimicrobial resistance elements [20]. Data have shown that majority of outbreaks reported around the world are associated with MLST clonal complexes CC92/CC2 (Oxford/Pasteur), corresponding to Pasteur sequence types including ST2, ST47, ST414, ST415, and ST524 (Table 2). Our findings are consistent with previous results. Moreover, ST513 as a singleton and endemic clone in Middle East is closely related to ST323 (56–3–55–2–5–1–14) as previously described in Iran [21].
It can be argued that clinical isolates recovered from exterior fluids including urine, sputum, etc., need adhesion factors involved in attachment and biofilm formation. On the other hand, isolates recovered from interior fluids including blood, synovia fluid, and cerebrospinal fluid need serum-resistance capability to establish their infections. In our study, isolates recovered from sputum mostly showed a high capacity to form a biofilm. However, all isolates recovered from interior fluids showed a high serum-resistance capacity.
In the present study, in silico comparative analysis of bacterial genomes extracted from ENSEMBL database revealed the presence or the absence of choP, ata, bap, fhaB, and blaPER-1 in genomes as well as variations in ompA sequences. On the other hand, the rest of virulence genes including csuA/BABCDE, pgaABCD, pbpG, ptk, epsA, pld, entA, plc1, nfuA, CipA, lpsB, Omp33-36, omp22, carO, tuf, abeD, gacS, paaE, surA1, uspA, and recA were mostly present in all Acinetobacter spp. genomes except SDF as a nonpathogenic strain. Comparative in silico analysis of surface-exposed virulence factors of A. baumannii has shown that the known or putative virulence determinants were restricted to specific clonal lineages suggesting that these virulence determinants may be crucial for overcoming these successful clones [22]. Our PCR assays and in silico analysis revealed that the pattern of virulence genes was strongly related to Pasteur’s ST as shown in Tables 2 and 3. However, as was proved earlier, the virulence-associated phenotypes cannot be associated with clonal relatedness, or vice versa.
AbOmpA as a multifaceted protein was recently shown to be a major virulence factor [23]. Paired sequence alignment of type I and type II of AbOmpA showed there are sequence variations in extracellular loops (data not shown). It seems that these sequence variations render a different virulence capability among clinical isolates. qRT-PCR analysis showed that sepsis-producing strains overexpress ompA significantly with respect to the level of IL-6 in bloodstream of mice model. These results showed that high expression of ompA possibly plays an important role in the infectious capacity caused by A. baumannii. Moreover, it is noteworthy that the difference in ompA expressions observed between MDR isolates is clonally independent.
In sepsis murine model after the peritoneal injection, the acute inflammatory response is triggered, and subsequently, innate immune cells infiltrated in peritoneal cavity. The bacteria are phagocyted and transported to spleen for antigen presentation [24]. Our study showed that all 18 selected strains proliferated in the spleen of C57/BL6 mice. On the other hand, only 9/18 of bacteria were detected in bloodstream of mice model. These findings suggest that the bacteria possibly have different proliferation capabilities and bloodstream invasions. Interestingly, AB-38 and AB-40 strains were released in bloodstream efficiently, while they had low Log10 CFU/g scores (4.69 and 4.1, respectively). Moreover, the strains which caused bacteremia in mice had higher and significant levels of serum IL-6 compared to isolates without bacteremia in mice model. This finding showed that during spleen transportation of the strains which had strong invasiveness, the larger population of them evades from host immune system and released into bloodstream.
The present study showed that indistinguishable clones have the same pattern of virulence genes, while, the virulence traits and expressions of ompA were clonally independent. Recently, some studies have focused on transcriptome and expression of virulence apparatus to clarify different virulence capability among clinical isolates [25, 26]. However, drawing a complete picture of the host–pathogen interaction associated with A. baumannii remains to be explored and elucidated.
References
Durante-Mangoni E, Zarrilli R (2011) Global spread of drug-resistant Acinetobacter baumannii: molecular epidemiology and management of antimicrobial resistance. Future Microbiol 6:407–422
Harding CM, Hennon SW, Feldman MF (2018) Uncovering the mechanisms of Acinetobacter baumannii virulence. Nat Rev Microbiol 16:91
Algburi A, Comito N, Kashtanov D et al (2017) Control of biofilm formation: antibiotics and beyond. Appl Environ Microbiol 17:83
Brossard KA, Campagnari AA (2012) The Acinetobacter baumannii biofilm-associated protein plays a role in adherence to human epithelial cells. Infect Immun 80:228–233
Lee CR, Lee JH, Park M et al (2017) Biology of Acinetobacter baumannii: pathogenesis, antibiotic resistance mechanisms, and prospective treatment options. Front Cell Infect Microbiol 7:55
McConnell MJ, Actis L, Pachón J (2013) Acinetobacter baumannii: human infections, factors contributing to pathogenesis and animal models. FEMS Microbiol Rev 37:130–155
Kim SW, Choi CH, Moon DC et al (2009) Serum resistance of Acinetobacter baumannii through the binding of factor H to outer membrane proteins. FEMS Microbiol Lett 301:224–231
Bentancor LV, Camacho-Peiro A, Bozkurt-Guzel C et al (2012) Identification of Ata, a multifunctional trimeric autotransporter of Acinetobacter baumannii. J Bacteriol 194:3950–3960
Lee HW, Koh YM, Kim J et al (2008) Capacity of multidrug-resistant clinical isolates of Acinetobacter baumannii to form biofilm and adhere to epithelial cell surfaces. Clin Microbiol Infect 14:49–54
Darwish SA, Rasooli I, Mousavi SG (2017) Filamentous hemagglutinin adhesin FhaB limits A. baumannii biofilm formation. Front Biosci 9:266–275
Smani Y, Docobo-Pérez F, López-Rojas R et al (2012) Platelet-activating factor receptor initiates contact of Acinetobacter baumannii expressing phosphorylcholine with host cells. J Biol Chem 287:26901–26910
Mirshekar M, Shahcheraghi F, Azizi O et al (2018) Diversity of class 1 integrons, and disruption of carO and dacD by insertion sequences among Acinetobacter baumannii isolates in Tehran, Iran. Microb Drug Resist 24:359–366
Antunes LC, Imperi F, Carattoli A et al (2011) Deciphering the multifactorial nature of Acinetobacter baumannii pathogenicity. PLoS ONE 6:e22674
Badmasti F, Ajdary S, Bouzari S et al (2015) Immunological evaluation of OMV (PagL) + Bap (1-487aa) and AbOmpA (8-346aa) + Bap (1-487aa) as vaccine candidates against Acinetobacter baumannii sepsis infection. Mol Immunol 67:552–558
Badmasti F, Siadat SD, Bouzari S et al (2015) Molecular detection of genes related to biofilm formation in multidrug-resistant Acinetobacter baumannii isolated from clinical settings. J Med Microbiol 64:559–564
Azizi O, Shahcheraghi F, Salimizand H et al (2016) Molecular analysis and expression of bap gene in biofilm-forming multi-drug-resistant Acinetobacter baumannii. Rep Biochem Mol Biol 5:62–72
Harris G, KuoLee R, Xu HH et al (2017) Mouse models of Acinetobacter baumannii infection. Curr Protoc Microbiol 46(1):6G-3
Aziz RK, Kansal R, Abdeltawab NF et al (2007) Susceptibility to severe Streptococcal sepsis: use of a large set of isogenic mouse lines to study genetic and environmental factors. Genes Immun 8:404–415
Noto MJ, Boyd KL, Burns WJ et al (2015) Toll-like receptor 9 contributes to defense against Acinetobacter baumannii infection. Infect Immun 83:4134–4141
Kröger C, Kary SC, Schauer K et al (2016) Genetic regulation of virulence and antibiotic resistance in Acinetobacter baumannii. Genes 8:12
Hojabri Z, Pajand O, Bonura C et al (2014) Molecular epidemiology of Acinetobacter baumannii in Iran: endemic and epidemic spread of multiresistant isolates. J Antimicrob Chemother 69:2383–2387
Eijkelkamp BA, Stroeher UH, Hassan KA et al (2014) Comparative analysis of surface-exposed virulence factors of Acinetobacter baumannii. BMC Genom 15:1020
Mortensen BL, Skaar EP (2012) Host–microbe interactions that shape the pathogenesis of Acinetobacter baumannii infection. Cell Microbiol 14:1336–1344
Buras JA, Holzmann B, Sitkovsky M (2005) Model organisms: animal models of sepsis: setting the stage. Nat Rev Drug Discov 4:854
Kim J, Lee JY, Lee H et al (2017) Microbiological features and clinical impact of the type VI secretion system (T6SS) in Acinetobacter baumannii isolates causing bacteremia. Virulence 8:1378–1389
Wright MS, Jacobs MR, Bonomo RA et al (2017) Transcriptome remodeling of Acinetobacter baumannii during infection and treatment. MBio 8:e02193-16
Acknowledgments
The authors would like to thank the personnel in the Bacteriology Department of the Pasture Institute of Iran for their help. This research was supported by the Pasture Institute of Iran under Grant No. B-0137.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors have no conflicts of interest to declare.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Shirazi, A.S., Shafiei, M., Solgi, H. et al. Different Virulence Capabilities and ompA Expressions in ST2 and ST513 of Multidrug-Resistant Acinetobacter baumannii. Curr Microbiol 76, 723–731 (2019). https://doi.org/10.1007/s00284-019-01686-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-019-01686-9