Abstract
For screening bilobalide (BB)-producing endophytic fungi from medicinal plant Ginkgo biloba, a total of 57 fungal isolates were isolated from the internal stem, root, leaf, and bark of the plant G. biloba. Fermentation processes using BB-producing fungi other than G. biloba may become a novel way to produce BB, which is a terpene trilactones exhibiting neuroprotective effects. In this study, a BB-producing endophytic fungal strain GZUYX13 was isolated from the leaves of G. biloba grown in the campus of Guizhou University, Guiyang city, Guizhou province, China. The strain produced BB when grown in potato dextrose liquid medium. The amount of BB produced by this endophytic fungus was quantified to be 106 μg/L via high-performance liquid chromatography (HPLC), substantially lower than that produced by the host tissue. The fungal BB which was analyzed by thin layer chromatography (TLC) and HPLC was proven to be identical to authentic BB. The strain GZUYX13 was identified as Pestalotiopsis uvicola via morphology and ITS rDNA phylogeny. To the best of our knowledge, this is the first report concerning the isolation and identification of endophytic BB-producing Pestalotiopsis spp. from the host plant, which further proved that endophytic fungi have the potential to produce bioactive compounds.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Ginkgo biloba L. is one of the most ancient plants on earth with fossil records dating back to more than 200 million years. It contains flavones and terpene trilactones and has widely and for a long time been used in China as an important and traditional medicine for various ailments [3]. As a class of main drug efficacy ingredients in G. biloba, terpene trilactones have many pharmacological activities such as acting as specific platelet-activating factor antagonists [5], selective antagonists of glycine receptors [21], and exhibiting neuroprotective effects [8]. Bilobalide (BB) was found to be the most active compound in previous studies on the consequences of hypoxia in hippocampal neurons [10] and endothelial cells [9] and on apoptotic cell death in neuronal cultures [1]. The anti-apoptotic effects of BB on the heart have not been researched much compared to those on the brain. However, BB has gained increasing attention as potential agents in the treatment of some brain disorders.
G. biloba is the only extracting source of BB currently used in herbal supplements and medicines. Although G. biloba actually has abundant BB content and rich resources, indiscriminate felling of trees, however, will destroy the ecological balance of nature. Thus, increasing efforts have been made to develop alternative means of BB production, such as using total chemical synthesis and ginkgo plant cell culture. However, thus far, the concentrations of BB in ginkgo cells and tissues in cell culture were even lower than that of natural plants [12], and the procedures were too complicated to make commercial-scale production of BB viable [14], although the complete chemical synthesis of BB has been accomplished.
In recent years, some studies on the metabolites from endophytic microorganisms residing in G. biloba have shown that Colletotrichum sp. could produce flavones which exhibited potent anti-cancer and antioxidant activities [22, 27], Alternaria No. 28 could produce cytotoxic metabolites [16], Chaetomium globosum ZY-22 could produce two polyhydroxylated steroids [15], and Fusarium sp. could produce ginkgolide B and other components [4, 26]. Thus, if a microbial source of BB is available, there is no need to harvest and extract the ginkgo plant for this drug. To our knowledge, this is the first report on BB isolated from endophytic Pestalotiopsis spp. associated with Ginkgo biloba. Therefore, this study aimed to isolate and study the endophytic BB-producing fungi in ginkgo plants collected from Guizhou province in China, and the potential of strain GZUYX13 for BB production was also evaluated.
Materials and Methods
Materials
Solvents used for chromatography were of high-performance liquid chromatography (HPLC) grade, while solvents used for extraction were of American Chemical Society grade. Authentic BB (≥98 % purity) was obtained from the National Institute for the Control of Pharmaceutical and Biological Products of China. All other chemicals were purchased from Guizhou Sciencelab Science & Technology Co., Ltd. Polymerase chain reaction (PCR) primers were synthesized by Shanghai Sangon Biologic Engineering Technology and Service Co., Ltd.
Samples of Ginkgo biloba were collected from Guizhou University campus, Guiyang city, Guizhou province, China, in November 2011. All samples were placed in polyethylene bags, immediately transported to the laboratory, and stored in a refrigerator at 4 °C.
Isolation of Endophytic Fungi from Ginkgo biloba
The collected samples were washed thoroughly under running water and then air-dried. The cleaned stems and roots were cut into pieces of about 1.5 cm length, and leaves and barks were cut into pieces of about 1 cm3 size. The cut stems, roots, leaves, and barks were surface-sterilized as follows: 75 % ethanol (v/v) for stems, roots and barks 3 min, leaves 1 min, and rinsed with sterile water five times. Subsequently, 0.1 % mercuric chlorine (v/v) was used to sterilize the tissues: stems and roots for 3 min, barks for 4 min, and leaves for 1 min. They were rinsed with sterile water five times and then dried on sterile filter papers. The sterilized stems and roots were opened longitudinally, and leaves and barks were cut to form the shape of a comb. The opened or cut plant materials were placed onto the potato dextrose agar (PDA) plates supplemented with 50 U/mL streptomycin and 30 U/mL penicillin. These plates were incubated at 28 °C for 3–15 days. At the first emergence of fungal growth from the plant tissue pieces, the hypha was transferred onto another PDA plate in time and incubated for 3–5 days. Strain GZUYX13 used in this study was 1 of the 57 endophytic fungi isolated from the Ginkgo biloba samples.
Fermentation and Extracts Preparation of Endophytic Fungi
The endophytic fungal isolates were inoculated, respectively, into 500 mL Erlenmeyer flasks containing 200 mL of potato dextrose liquid medium and cultured at 28 °C with 200 recycles/min for 10 days in a rotary shaker. The mycelia were harvested by centrifugation at 12,000×g for 10 min and frozen at −20 °C overnight. Dried mycelia were crushed and subjected to ultrasound-assisted extraction three times with acetic ether at room temperature for 30 min. All extracts were combined and dried under vacuum. The dry residues were dissolved in 1 mL of methanol (HPLC purity grade). The methanolic extracts were filtered through a 0.45 μm filter prior to chromatographic separation. BB crystals were dissolved in anhydrous methanol to be used as the standard solution.
Screening of Bilobalide-Producing Endophytic Fungi
Each extract was analyzed via TLC and HPLC. TLC analysis of the final extract as well as the standard solution of BB was developed in a solvent system (methylbenzene:ethyl acetate:acetone:methanol: at 10:5:5:0.6 v/v) by spotting on a 0.25 mm (10 × 20 cm) silica gel G plate. TLC analysis was performed at room temperature, and the TLC plates were dried naturally. After the solvent fully volatilized, the plate was fumigated in acetic anhydride for 15 min, and then heated at 140–160 °C for 30 min by stove. BB was detected under 365 nm ultraviolet light, which appeared as light-greenish spots. The BB spot was identified by comparison with authentic BB. HPLC was performed using a C18 column (5 μm, 4.6 × 250 mm) (Agilent, China). A 10 μL amount of each final extract was injected. The mobile phase was methanol:water (30:70, v/v) at a flow rate of 0.8 mL/min. The effluent was monitored by refractive index detection (RID). BB was quantified by comparing the peak area of the samples to that of the authentic BB.
Identification of Strain GZUYX13
After BB was determined in the culture of strain GZUYX13, the fungus was identified by morphological characteristics and ITS rDNA sequence analysis. The strain GZUYX13 was cultured on PDA plates at 28 °C, and the colonies’ morphologies and diameters were observed and recorded every day. A few mycelia were picked with a sterile needle directly from the surface of the cultures and placed onto a glass slide, and covered with a cover slip. The glass slide specimens were observed under a microscope, the morphology of conidia, conidiophores, and conidiogenous cells were observed.
The genomic DNA of strain GZUYX13 was extracted from fresh mycelia using the Fungal gDNA Kit GD2416 (Biomiga, CA, USA), according to manufacturers’ protocol. The fungal ITS rDNA fragments were amplified using the universal primers ITS1 (5′-TCCGTTGGTGAACCTGCGG-3′) and ITS4 (5′-TCCTCCG GTTATTGATATGC-3′) [23]. PCR mixture of 25 mL contained 12.5 μL 2× Taq PCR Master Mix (containing Taq DNA polymerase, buffer, MgCl2, and dNTPs), 1 μL DNA sample, 1 μL of each primer, and 9.5 μL ddH2O. The PCR conditions were as follows: initial pre-heating at 94 °C for 3 min, 30 cycles of 94 °C for 30 s, 58 °C for 30 s, 72 °C for 30 s, and a final extension at 72 °C for 10 min. The PCR products were separated on 1 % (w/v) agarose gel and purified using a DNA Gel Exaction Kit (AXYGEN, Suzhou, P.R. China). The resulting DNA was sequenced directly using the same primers (Shanghai Sangon Biologic Engineering Technology and Service Co., Ltd., Shanghai, P.R. China). The ITS sequence of the endophytic fungus was compared with the data available in NCBI using BLAST searches to obtain its allies. The resulting sequences were aligned using the Clustal X software [20] with gaps treated as missing data. The phylogenetic tree was constructed using the neighbor-joining method [17] and the Kimura two-parameter distance calculation in mega software version 3.1 [11]. The bootstrap was 1000 replications to assess the reliable level to the nods of the tree.
Results
Isolation of Endophytic Fungi
A total of 57 fungal strains were isolated from the healthy roots, stems, leaves, and barks of the medicinal plant G. biloba, and among them, 15 isolates from roots, 34 isolates from stems, 1 isolate from leaves, and 7 isolates from barks revealed the amount of endophytic fungi living inside the internal part of G. biloba. Based on colonial characteristics, the endophytic fungi were related to Alternaria spp., Aspergillus spp., Colletotrichum spp., Fusarium spp., Penicillium spp., Pestalotiopsis spp., Phomopsis spp., Trichoderma spp., Xylaria spp., and unidentified strains. These results confirm that host specificity and geographic structure affect biological diversity of endophytic fungi [7, 19].
Screening of Bilobalide-Producing Endophytic Fungi
The extracts of fungal cultures were examined for the presence of BB by TLC under UV illumination. The results showed that one of the fungal compounds from strain GZUYX13 exhibited the same Rf value (0.61) as authentic BB, indicating strain GZUYX13 containing BB (Fig. 1). The results of HPLC analyses further confirmed the presence of BB showing a retention time of 6.860 min, which was similar to authentic BB (6.859 min) (Fig. 2). The BB yield of strain GZUYX13 was about 106 μg/L or 75 μg/g (BB per dry wt of mycelium) when it was cultured in a 100 mL potato dextrose liquid medium at 28 °C with 200 recycles/min shaking for 10 days.
High-performance liquid chromatogram of authentic bilobalide (a) and fungal bilobalide (b). The mobile phase was methanol/water (30:70, v/v) with a flowrate of 0.8 mL/min. Registrations of peak and retention time were recorded by RID. Fungal sample showed a peak with retention time 6.860 min, which was found to be identical to authentic bilobalide
Phenotypic Characterization of Strain GZUYX13
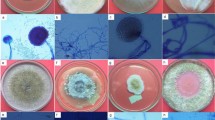
The endophytic fungus GZUYX13 was cultured on PDA at 28 °C for 7 days. The colonial morphological traits of the GZUYX13 isolate were as follows: white, cottony, and nearly round margins, then black fruiting body (conidiomata) in the central of colony; reverse of culture whitish to pale yellow (Fig. 3h, i). Conidiophores were indistinct, and conidiogenous cells were hyaline, simple, filiform, 4–12 μm long (Fig. 3f, g). Conidia: 18.3–22.8 × 4.9–6.1 μm, fusiform, straight to slightly curved, 4-septate, smooth, grayish brown; basal cell: conical, hyaline, and thin-walled, with three median cells, together being 11.0–13.4 μm long; second cell from base: 3.6–4.7 μm; third cell: 3.5–4.6 μm; and fourth cell 3.0–4.7 μm; and apical cell: hyaline, conic to triangular, with 2–3 apical appendages, arising from the apex of the apical cell, 4.9–6.1 μm long (Fig. 3a–e). The morphological characteristics of strain GZUYX13 were similar to those of Pestalotiopsis uvicola described by Bissett [2]. Based on the morphological characterization above, strain GZUYX13 was identified as P. uvicola.
ITS rDNA Sequence and Phylogenetic Analysis
To further determine the phylogeny of strain GZUYX13, the ITS rDNA fragments including 542 bp were amplified and sequenced. After homology searching against GenBank, the sequence was found to share 100 % similarity with P. uvicola (GenBank accession number AF409974). A phylogenetic relationship was established via phylogenetic analysis of ITS rDNA sequences of strain GZUYX13 and its allies in GenBank (Fig. 4). Strain GZUYX13 was identified as P. uvicola.
Discussion
This study isolated a BB-producing endophytic fungus GZUYX13 from the leaves of Ginkgo biloba collected from Guizhou University campus in Guizhou province in southwest China. Strain GZUYX13 was identified as P. uvicola on the basis of its morphology and ITS rDNA sequence. Previous studies have demonstrated that Pestalotiopsis spp. are distributed worldwide, and frequently obtained from numerous tree and crop plants. The genus Pestalotiopsis includes many species, with P. uvicola as one of the most common species. Pestalotiopsis has received considerable attention in recent years, not only because of its role as a plant pathogen but also as a commonly isolated endophyte which has been shown to produce a wide range of chemically novel diverse metabolites [13]. Pestalotiopsis spp. were reported to produce lots of bioactive secondary metabolites, including alkaloids, terpenoids, coumarins, chromones, quinones, semiquinones, peptides, xanthones, phenols, and other structural compounds [24, 25]. To the best of our knowledge, the endophytic fungus from Pestalotiopsis has never been reported to be capable of producing BB. The present study is the first report of BB-producing P. uvicola from Ginkgo biloba plants.
The discovery of BB-producing endophytic fungi associated with Ginkgo biloba is valuable for industrial application and for basic research. The consistent production of BB by P. uvicola GZUYX13 further supports the theory that during the long coevolution of endophytes and their host plants, endophytes adapted to their special microenvironments by genetic variation, including the uptake of some plant DNAs into their own genomes [6]. This could have led to the ability of certain endophytes to biosynthesize some phytochemicals originally associated with the host plant [18].
The amount of BB produced by P. uvicola GZUYX13 was 106 μg/L or 75 μg/g (BB per dry wt of mycelium) when it is cultured under the conditions described in this study. This suggests that P. uvicola GZUYX13 is a promising candidate for BB production. However, it is only a wild strain, and we believe that the BB yield of P. uvicola GZUYX13 will potentially be increased by strain improvement and optimization of the fermentation condition.
References
Ahlemeyer B, Mowes A, Krieglstein J (1999) Inhibition of serum deprivation- and staurosporine-induced neuronal apoptosis by Ginkgo biloba extract and some of its constituents. Eur J Pharmacol 367:423–430
Bissett J (1982) Pestalotiopsis menezesiana on greenhouse plantings of Cissus rhombifolia with notes on related fungi occurring on Vitaceae. Can J Bot 60:2570–2574
Chen P, Ozcan M, Harnly J (2007) Chromatographic fingerprint analysis for evaluation of Ginkgo biloba products. Anal Bioanal Chem 389:251–261
Cui YN, Yi DW, Bai XF, Sun BS, Zhao YQ, Zhang YX (2012) Ginkgolide B produced endophytic fungus (Fusarium oxysporum) isolated from Ginkgo biloba. Fitoterapia 83:913–920
Etienne A, Hecquet F, Soulard C, Spinnewyn B, Clostre F, Braquet P (1986) In vivo inhibition of plasma protein leakage and Salmonella enteritidis-induced mortality in the rat by a specific paf-acether antagonist: BN 52021. Agents Actions 54:368–370
Germaine K, Keogh E, Garcia-Cabellos G, Borremans B, Barac T, Dowling DN et al (2004) Colonisation of poplar trees by gfp expressing bacterial endophytes. FEMS Microbiol Ecol 48:109–118
Higgins KL, Arnold AE, Miadlikowska J, Sarvate SD, Lutzoni F (2007) Phylogenetic relationships, host affinity, and geographic structure of boreal and arctic endophytes from three major plant lineages. Mol Phylogenet Evol 42:543–555
Huang SH, Duke RK, Chebib M, Sasaki K, Wada K, Johnston GA (2003) Bilobalide, a sesquiterpene trilactone from Ginkgo biloba, is an antagonist at recombinant alpha1 beta2 gamma2 L GABA (A) receptors. Eur J Pharmacol 464:1–8
Janssens D, Michiels C, Delaive E, Eliaers F, Drieu K, Remacle J (1995) Protection of hypoxia-induced ATP decrease in endothelial cells by Ginkgo biloba extract and bilobalide. Biochem Pharmacol 50:991–999
Klein J, Chatterjee SS, Loffelholz K (1997) Phospholipid breakdown and choline release under hypoxic conditions: inhibition by bilobalide, a constituent of Ginkgo biloba. Brain Res 755:347–350
Kumar S, Tamura K, Nei M (2004) MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform 5:150–163
Laurain D, Tremouillaux-Guillerm J, Chenieux JC, van Beek TA (1997) Production of ginkgolide and bilobalide in transformed and gametophyte derived cell cultures of Ginkgo biloba. Phytochemistry 46:127–130
Maharachchikumbura SSN, Guo LD, Chukeatirote E, Bahkali AH, Hyde KD (2011) Pestalotiopsis—morphology, phylogeny, biochemistry and diversity. Fungal Diver 50:167–187
Michael TC, David KJ, Jeffrey LG (1993) Synthetic studies on the Ginkgolides: total synthesis of (±)-bilobalide. J Am Chem Soc 115:3146–3155
Qin JC, Gao JM, Zhang YM, Yang SX, Bai MS, Ma YT, Laatsch H (2009) Polyhydroxylated steroids from an endophytic fungus, Chaetomiumglobosum ZY-22 isolated from Ginkgo biloba. Steroids 74:786–790
Qin JC, Zhang YM, Hua L, Ma YT, Gao JM (2009) Cytotoxic metabolites produced by Alternaria No.28, an endophytic fungus isolated from Ginkgo biloba. Nat Prod Commun 4:1473–1476
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Stierle A, Strobel G, Stierle D (1993) Taxol and taxane production by Taxomyces andreanae, an endophytic fungus of pacific yew. Science 260:214–216
Strobel G, Daisy B (2003) Bioprospecting for microbial endophytes and their natural products. Microbiol Mol Biol Rev 67:491–502
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The Clustal_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
vanBeek TA (2005) Ginkgolides and bilobalide: their physical, chromatographic and spectroscopic properties. Bioorg Med Chem 13:5001–5012
Wang MX, Chen SL, Yan SZ, Huo J (2003) A preliminary study on the isolation, identification and media of an endophytic fungus producing flavones. J Nanjing Nor Univ (Nat Sci) 26:106–110
White TJ, Bruns T, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfland DH, Sninsky JJ, White TJ (eds) PCR protocols: a guide to methods and applications. Academic Press, New York
Xu J, Ebada SS, Proksch P (2010) Pestalotiopsis a highly creative genus: chemistry and bioactivity of secondary metabolites. Fungal Diver 44:15–31
Xu J, Yang XB, Lin Q (2014) Chemistry and biology of Pestalotiopsis-derived natural products. Fungal Diver 66:37–68
Yi DW, Zhang YX, He JY, Zhao WQ, Wu CF (2007) Fermentation metabolites of endophytic fungus isolated from Ginkgo biloba. J Microbiol 27:102–106
Zhang YJ, Wang JF, Huang YJ (2002) Screening for antitumor activities of endophytic fungi isolated from four species of gymnosperm. J Xiamen Univ (Nat Sci) 41:804–809
Acknowledgments
This work was funded by the talent introduction plan of the Guizhou University (Grant No. [2014]59), the joint funds plan of the Guizhou province (Grant No. LH [2014]7657), and the grants of the agricultural science and technology foundation of Guizhou province (Grant Nos. NY[2013]3042 and NY[2013]3044) from the Science and Technology Bureau of Guizhou University and the Science and Technology Department of Guizhou province, China. The authors would also like to thank the National Natural Science Foundation of China (Grant No. 31460011) for supporting and funding this research.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Qian, YX., Kang, JC., Luo, YK. et al. A Bilobalide-Producing Endophytic Fungus, Pestalotiopsis uvicola from Medicinal Plant Ginkgo biloba . Curr Microbiol 73, 280–286 (2016). https://doi.org/10.1007/s00284-016-1060-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-016-1060-6