Abstract
Streptomyces coelicolor is the soil-dwelling bacterium with a complex life cycle and a strong ability to produce plenty of secondary metabolites which are strictly regulated by a variety of regulators. Amino acid alignment shows that the deduced protein of SCO2140 belongs to the family of Leucine-responsive regulatory proteins (Lrps). Disruption of SCO2140 significantly decreased the yields of actinorhodin and calcium-dependent antibiotics, and the complemented strain restored the antibiotic productions to the wild-type level. In contrast, overexpression of SCO2140 increased the actinorhodin production. In agreement with it, the transcriptions of actII-ORF4 and cdaR remarkably reduced in the SCO2140 disruption mutant. The aerial mycelium formation of the SCO2140 disruption mutant was clearly delayed in R2YE medium due to the decrease of ramS expression while its complemented strain could restore the normal formation of aerial mycelia. These results indicated that SCO2140 was involved in antibiotic biosynthesis and morphological differentiation of Streptomyces coelicolor A3(2).
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Streptomyces coelicolor A3(2) is the model strain of the Streptomyces genus. Twenty-nine biosynthetic gene clusters for secondary metabolites have been discovered in the genome of S. coelicolor A3(2) [1]. Up to now, 16 chemicals from the 15 clusters have been identified, which include the four distinct classes of antibiotics, the blue-pigmented polyketide antibiotic actinorhodin (ACT), the red oligopyrrole prodiginine antibiotic undecylprodigiosin (RED), the lipopeptide calcium-dependent antibiotic (CDA), and the yellow-pigmented type I polyketide (coelimycin P1) [2]. The regulation of secondary metabolic biosynthesis is a complex network, involving cluster-situated and global regulators, two-component systems, RNA regulatory elements, and some developmental genes (such as bldB, abaA, whiJ, and wblA) [3]. In S. coelicolor, actII-ORF4, redD, cdaR, and cpkO are the cluster-situated regulatory genes for ACT, RED, CDA, and coelimycin P1 biosynthesis, respectively [3].
The Leucine-responsive regulatory proteins (Lrps), also named FFRP or Lrp/AsnC, are widely distributed among prokaryotes and regulate a large number of genes’ expression [4]. Lrp functions as both an activator and a repressor in Escherichia coli, and affects the transcriptions of at least 10 % of all E. coli genes [5]. The Lrp-regulated genes are involved in amino acid metabolism and transport, the fimbriation, and the formation of virulence [6]. For instance, LrpA is vital to the long-term persistence of Mycobacterium tuberculosis and it may be a novel therapeutic target against M. tuberculosis [7]. The S. coelicolor genome contains 14 putative Lrp/AsnC-like protein encoding genes. Of them, bkdR encoding a transcriptional regulator of the branched-chain amino acid dehydrogenase complex affects the morphogenesis and antibiotic production in S. coelicolor [8]. However, little is known about the function of other Lrp/AsnC-like proteins in S. coelicolor. In this paper, the Lrp-like protein encoding gene SCO2140 was shown to be involved in the antibiotic production and morphological differentiation of S. coelicolor A3(2).
Materials and Methods
Bacterial Strains and Growth Conditions
Bacterial strains and plasmids used in this study are listed in Table S1. All media used for S. coelicolor M145 (the wild-type strain) and its derivatives were prepared according to Practical Streptomyces Genetics [9]. R5 liquid medium was used for the quantitative determination of antibiotic production as well as for RNA preparation of S. coelicolor. R2YE medium was used for the growth of S. coelicolor and observation of the aerial mycelium formation. Yeast extract–malt extract liquid medium (YEME) and 2× YT liquid medium were used for the growth of S. coelicolor. Mannitol soya flour medium (MS) was used for the sporulation and intergeneric conjugation of S. coelicolor. Difco Nutrient Agar (DNA) medium supplemented with 0.5 % (w/v) NaCl was used for the production of calcium-dependent antibiotic (CDA) and Staphylococcus aureus was used as the indicator of CDA production [10]. The Escherichia coli strains were cultured at 37 °C in Luria–Bertani (LB) medium supplemented with antibiotics (100 µg ml−1 for ampicillin, 100 µg ml−1 for kanamycin, 100 µg ml−1 for apramycin, 25 µg ml−1 for chloromycetin, and 15 µg ml−1 for tetracycline) when necessary for propagating plasmids.
Construction of the SCO2140 Disruption Mutant and Its Complementation
To construct the SCO2140 disruption mutant (SCO2140DM), a DNA fragment corresponding to the upstream region of SCO2140 (extending from the positions −655 to +39 with respect to the SCO2140 translation start codon) was amplified by using primers 2140UF/UR (Table S2). Then the amplified DNA fragment was inserted into the HindIII/BamHI sites of pUC119::Kan r [11], giving pD1. After digesting pD1 with HindIII and EcoRI, the fragment containing the upstream region of SCO2140 and the kanamycin resistance gene (Kan r) was inserted into the corresponding sites of pBluescript KS+ to generate pD2. The DNA fragment corresponding to the downstream region of SCO2140 (extending from the positions +226 to +1258 with respect to the SCO2140 translation start codon) was amplified by using primers 2140DF/DR (Table S2) and ligated into the EcoRI/XbaI sites of pD2 to give pD3. A 2.7 kb DNA fragment was isolated after digesting pD3 with HindIII and XbaI, and then it was inserted into the corresponding sites of pKC1132, giving pD2140. Subsequently, pD2140 was introduced into E. coli ET12567/pUZ8002 [9] and then it was transferred into the S. coelicolor wild-type strain (WT) by intergeneric conjugation. After growing on MS agar at 28 °C for 144 h, colonies were selected and transferred to the minimal medium with mannitol as carbon source (MMM) and supplemented with apramycin (10 µg ml−1) or kanamycin (10 µg ml−1). The kanamycin-resistant and apramycin sensitive strains were selected as SCO2140DM and confirmed by PCR using the primers 2140YF1/YR1 and 2140YF2/YR2 (Table S2).
To construct the complemented strain, the entire SCO2140 coding region was amplified using the primers 2140HF/HR (Table S2). Then the amplified DNA fragment was verified by DNA sequencing and inserted into the NdeI/BamHI sites of pIJ8600. The resulting plasmid pH2140 was introduced into SCO2140DM by conjugation to generate the SCO2140 complemented strain (SCO2140CM). To overexpress SCO2140, the entire SCO2140 coding region was amplified using the primers 2140HF1/HR1 (Table S2) and verified by DNA sequencing. Then the amplified DNA fragment was inserted into the EcoRV site of pIMEP [12]. The resulting plasmid pIMEP::SCO2140 was introduced into WT by conjugation to generate the SCO2140 overexpressed strain (SCO2140OE).
Determination of Antibiotic Productions
In order to assess the productions of ACT and RED, 1 × 109 spores of the S. coelicolor wild-type strain and its derivatives were cultured in 50 ml 2× YT liquid medium at 28 °C and shaked at 200 r/min for 10 h. Mycelia were collected and inoculated into 50 ml of R5 liquid medium at 28 °C and shaked at 200 r/min for 24, 48, 72, 96, 120, 144, and 168 h, respectively. Then the production of ACT was measured as described previously [9]. CDA production in DNA medium was assayed as described previously [10].
RNA Isolation and Real-Time RT-PCR
Total RNA was isolated from the S. coelicolor wild-type strain and SCO2140DM grown in R5 liquid medium or the R2YE medium for 24, 48, 72, 96, 120, and 144 h. The isolated RNA was further treated with DNase I (Promega) to remove the contaminated DNA, and then it was reverse transcribed into the complementary DNA by using PrimeScript RT reagent kit (TaKaRa). Real-time PCR was carried out in realplex2 Master cycler (Eppendorf, Germany) using Ultra SYBR Mixture (CWBIO). The condition is used as follows: 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s, 60 °C for 1 min. Three independent experiments were performed and three PCR replicates were used in parallel for each transcript. The hrdB was used as an internal control. The relative transcriptional levels of tested genes were normalized to hrdB and determined by using the \(- 2^{{\Delta \Delta C_{T} }}\) method [13]. The values were presented as fold change in comparison with the relative expression levels for each gene at the first test time point in the wild-type strain.
Expression and Purification of SCO2140 from E. coli
The SCO2140 coding region was amplified by using primers 2140OF/OR (Table S2) and verified by DNA sequencing. The amplified DNA fragment was inserted into the NdeI/XhoI sites of pET23b to give the plasmid pET23b::SCO2140. The plasmid was introduced into E. coli C43 [14] for gene expression. The E. coli C43 containing pET23b::SCO2140 was cultured at 37 °C in 200 ml of LB supplemented with kanamycin (100 µg ml−1) to an OD600 of 0.6. After isopropyl β-d-thiogalactopyranoside (IPTG) was added up to 0.1 mM, and the cultures were incubated at 37 °C for an additional 3 h. After centrifugation (6000×g for 5 min at 4 °C), the cells were collected and washed with binding buffer [20 mM Tris base, 500 mM NaCl, 5 mM imidazole, 5 % (v/v) glycerol (pH 7.9)], and re-suspended in 20 ml of the same buffer. After sonication on ice, the cell suspension was centrifugated (12,000×g for 20 min at 4 °C) and the supernatant was recovered. The supernatant was treated with Ni–NTA agarose chromatography (Novagen), and the purified SCO2140-His6 was separated from the whole-cell lysate. Protein purity was determined by Coomassie blue staining after electrophoresis on a 15 % (w/v) SDS–polyacrylamide gel. The purified protein was stored with 5 % (v/v) glycerol at −70 °C until used in the subsequent experiments.
Preparation of the Antibody and Western Blot Analysis
Approximately 2 mg of the purified SCO2140-His6 was mixed with Freund’s complete adjuvant and injected into two healthy mice for four times (once per week). Antisera were collected from the mice after 1 month and used as the polyclonal antibody against SCO2140-His6.
To detect the expression of SCO2140, WT, SCO2140DM, SCO2140CM, and SCO2140OE were cultured in R5 liquid medium at 28 °C for 24, 48, 72, 96, and 120 h. After collected, mycelia were ground in liquid nitrogen and sonicated on ice. The supernatant samples containing 50 µg proteins were separated by 15 % (w/v) SDS-PAGE and transferred to a polyvinylidene difluoride (PVDF) membrane (Roche). Then the proteins on the membrane were probed with anti-SCO2140 which were hybridized with the horseradish peroxidase-conjugated suitable secondary antibodies (Jackson), followed by detection with enhanced chemiluminescence (Pierce).
Electrophoretic Mobility Shift Assays (EMSAs)
DNA probes containing the promoter regions of actII-ORF4 and cdaR were generated by PCR using the primers act-PF/act-PR and cda-PF/cda-PR (Table S2). A mixture of the probes and varying quantities of SCO2140-His6 (0, 45, 180, 360, 450, 1800, 3600, 4500, and 6750 nM protein) was incubated at 25 °C for 20 min in the reaction buffer which contained 1 µg of poly-(dI-dC) (Sigma), 20 mM Tris–HCl (pH 7.5), 1 mM dithiothreitol (DTT), 10 mM MgCl2, 54 mM KCl, 0.5 mg ml−1 calf BSA, and 5 % (v/v) glycerol in a total volume of 20 µl. Then the mixture was separated on a 5 % (w/v) native polyacrylamide gel with a running buffer containing 45 mM Tris–HCl (pH 8.0), 45 mM boric acid, and 1 mM EDTA at 150 V for 1 h. Then the gel was stained with SYBR Gold Nucleic Acid Gel Stain (Invitrogen) for 30 min in 0.5× TBE buffer, and photographed under ultraviolet transillumination using Bio-Rad GelDoc XR.
Results and Discussion
SCO2140 Encodes a Novel Lrp/AsnC Family Protein
SCO2140 encodes a protein of 93 amino acids. In search of database, the deduced protein of SCO2140 is well conserved through Streptomyces species (Fig. S1a). SCO2140 shows 25 % identity and 48 % similarity over 84-aa region with Lrp from E. coli [15], 31 % identity and 44 % similarity over the 59-aa region with LrpA from M. tuberculosis [16]. However, sequence alignment showed that SCO2140 lacks the typical DNA binding domain (Fig. S1b), suggesting SCO2140 could not directly interact with DNA as other Lrps. As SCO2140 contains the AsnC-type C-terminal domain which is responsible for the ligand-response and protein–protein interaction, it is possible that SCO2140 plays a cooperative role via forming heterodimers with other transcriptional regulators.
Disruption of SCO2140 Reduced the Productions of ACT and CDA in S. coelicolor
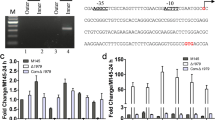
To study its function, SCO2140 was disrupted through homologous recombination (Fig. 1a). The expected mutants were selected randomly and confirmed by PCR. A distinctive band of 1377 bp was amplified from the SCO2140 disruption mutant (SCO2140DM) and a 587 bp band was obtained from WT as predicted while the 163 bp DNA fragment from the coding region of SCO2140 was amplified only from WT (Fig. 1b). These confirmed that SCO2140 was replaced by the kanamycin resistance cassette (Kan r). To further investigate its function, the complemented (SCO2140CM) and overexpressed (SCO2140OE) strains of SCO2140 were constructed. Western blotting showed the expression of SCO2140 significantly enhanced in SCO2140OE, compared with that of WT and SCO2140CM (Fig. 1c). Expectably, the expression of SCO2140 did not detect in SCO2140DM (Fig. 1c).
Construction of the SCO2140 disruption mutant (SCO2140DM) and Western blot analysis of SCO2140. a Strategy for construction of SCO2140DM via homologous recombination. WT, S. coelicolor M145; Kan r, kanamycin resistance gene. b Verification of SCO2140DM by PCR. PCR1 and PCR2 were performed with the gene-outside primers (2140YF1/YR1) and the gene-inside primers (2140YF2/YR2), respectively. NC negative control, Ladder 100 bp DNA ladder. c Expression of SCO2140. About 50 μg of total protein from WT, SCO2140OE, SCO2140CM, and SCO2140DM cultured in R5 medium was loaded for Western blot analysis with the anti-SCO2140 antibodies or the total protein shown by SDS–PAGE as the loading control. Data were representative of three independent experiments
In R5 liquid medium, ACT extracted from the mycelia of SCO2140DM was only about 40 % of that in WT (Fig. 2a). In agreement with it, ACT extracted from the medium of SCO2140DM also decreased (Fig. 2b). Combined with that SCO2140CM produced the same amount of ACT as that in WT, these results suggested that ACT decrease in SCO2140DM is due to the disruption of SCO2140. Meanwhile, the ACT extracted from the medium of SCO2140OE was nearly twofold of the wild-type level (Fig. 2b).
The productions of ACT and CDA. a ACT extracted from the mycelia of WT, SCO2140DM, SCO2140CM, and SCO2140OE, respectively. b ACT extracted from the cultured media of WT, SCO2140DM, SCO2140CM, and SCO2140OE, respectively. c The CDA productions of WT, SCO2140DM, and SCO2140CM. Staphylococcus aureus was used as the indicator strain in the presence or absence of Ca(NO3)2. Error bars represent the standard deviations from three independent experiments
When DNA medium was used, CDA production in SCO2140DM was also decreased. In SCO2140CM, the CDA production restored to the wild-type level (Fig. 2c). These results revealed that SCO2140 positively regulates the production of ACT and CDA in S. coelicolor. Unlike ACT production, the RED production was not affected by the disruption of SCO2140 (Fig. S2).
Disruption of SCO2140 Decreased the Transcriptions of actII-ORF4 and cdaR
As the cluster-situated regulatory genes, actII-ORF4 and cdaR positively regulate the production of ACT and CDA. The transcriptions of actII-ORF4 and cdaR were measured in SCO2140DM by real-time PCR. The transcriptional level of actII-ORF4 in SCO2140DM was only about 30 % of that in WT when its transcriptional level reached the highest level at 96 h (Fig. 3a). Like actII-ORF4, the transcriptional level of cdaR in SCO2140DM was only about 20 % of that in WT and it decreased rapidly after 24 h in both WT and SCO2140DM (Fig. 3b). These results suggested that disruption of SCO2140 reduced the production of ACT and CDA at least partially through decreasing the transcriptions of actII-ORF4 and cdaR. Since SCO2140 could not bind to the upstream regions of actII-ORF4 and cdaR (Fig. S3), SCO2140 may indirectly regulate antibiotic production or cooperate with other transcriptional regulators which involved in ACT and CDA production.
The transcriptions of actII-ORF4 and cdaR. a The transcriptional level of actII-ORF4 was detected in WT and SCO2140DM which were cultured in R5 medium. b The transcriptional level of cdaR was detected in WT and SCO2140DM which were cultured in DNA medium. The open columns represent the gene transcriptions in WT and the solid columns represent the gene transcriptions in SCO2140DM. Error bars represent the standard deviations from three independent experiments
Disruption of SCO2140 Affected the Morphological Differentiation of S. coelicolor
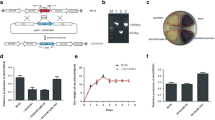
In R2YE medium, SCO2140DM showed rare aerial mycelium. Introduction of the entire wild-type SCO2140 in SCO2140DM restored the aerial mycelium formation, suggesting SCO2140 is required for the aerial mycelium formation in R2YE medium (Fig. 4a). Glucose is the main carbon source in R2YE medium, and the formation of aerial mycelium in this medium was related with the protein SapB [17]. Based on this consideration, the transcription of ramS which encodes SapB was detected in both WT and SCO2140DM. The transcriptional level of ramS decreased remarkably when SCO2140 was disrupted (Fig. 4b), indicating that SCO2140 positively regulates aerial mycelium formation through controlling the transcription of ramS.
Morphology analysis of the S. coelicolor M145 and its derivatives in R2YE media. a The phenotypes of WT, SCO2140DM, SCO2140CM, and SCO2140OE grown in R2YE medium for 3 days. b The transcriptional level of ramS was detected in WT and SCO2140DM which were cultured in R2YE medium. The open columns represent the gene transcriptions in WT and the solid columns represent the gene transcriptions in SCO2140DM. Error bars represent the standard deviations from three independent experiments
References
Nett M, Ikeda H, Moore BS (2009) Genomic basis for natural product biosynthesis diversity in the actinomycetes. Nat Prod Rep 26(11):1362–1384. doi:10.1039/b817069j
Gomez-Escribano JP, Bibb MJ (2012) Streptomyces coelicolor as an expression host for heterologous gene clusters. Methods Enzymol 517:279–300. doi:10.1016/B978-0-12-404634-4.00014-0
Liu G, Chater KF, Chandra G, Niu G, Tan H (2013) Molecular regulation of antibiotic biosynthesis in Streptomyces. Microbiol Mol Biol Rev 77(1):112–143. doi:10.1128/MMBR.00054-12
Pérez-Rueda E, Janga SC (2010) Identification and genomic analysis of transcription factors in archaeal genomes exemplifies their functional architecture and evolutionary origin. Mol Biol Evol 27(6):1449–1459. doi:10.1093/molbev/msq033
Tani TH, Khordusky A, Blumenthal RM, Brown PO, Matthews RG (2002) Adaptation to famine: a family of stationary-phase genes revealed by microarray analysis. Proc Natl Acad Sci USA 99(21):13471–13476. doi:10.1073/pnas.212510999
Peterson SN, Reich NO (2010) LRP: a nucleoid-associated protein with gene regulatory functions. In: Dame RT, Dorman CJ (eds) Bacterial Chromatin, 1st edn. Springer-Verlag, Berlin, pp 353–364
Parti R, Shrivastava R, Srivastava S, Subramanian A, Roy R, Srivastava B, Shrivastava R (2008) A transposon insertion mutant of Mycobacterium fortuitum attenuated in virulence and persistence in a murine infection model that is complemented by Rv3291c of Mycobacterium tuberculosis. Microb Pathog 45(5–6):370–376. doi:10.1016/j.micpath
Sprusansky O, Stirrett K, Skinner D, Denoya C, Westpheling J (2005) The bkdR gene of Streptomyces coelicolor is required for morphogenesis and antibiotic production and encodes a transcriptional regulator of a branched-chain amino acid dehydrogenase complex. J Bacteriol 187(2):664–671. doi:10.1128/JB.187.2.664-671.2005
Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA (2000) Practical Streptomyces genetics. The John Innes Foundation, Norwich
Anderson TB, Brian P, Champness WC (2001) Genetic and transcriptional analysis of absA, an antibiotic gene cluster-linked two-component system that regulates multiple antibiotics in Streptomyces coelicolor. Mol Microbiol 39(3):553–566. doi:10.1046/j.1365-2958.2001.02240.x
Yang H, Wang L, Xie Z, Tian Y, Liu G, Tan H (2007) The tyrosine degradation gene hppD is transcriptionally activated by HpdA and repressed by HpdR in Streptomyces coelicolor, while hpdA is negatively autoregulated and repressed by HpdR. Mol Microbiol 65(4):1064–1077. doi:10.1111/j.1365-2958.2007.05848.x
Wang SL, Fan KQ, Yang X, Lin ZX, Xu XP, Yang KQ (2008) CabC, an EF-hand calcium-binding protein, is involved in Ca2+-mediated regulation of spore germination and aerial hypha formation in Streptomyces coelicolor. J Bacteriol 190(11):4061–4068. doi:10.1128/JB.01954-07
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(−∆∆C(T)) method. Methods 25(4):402–408. doi:10.1006/meth.2001.1262
Miroux B, Walker JE (1996) Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J Mol Biol 260(3):289–298. doi:10.1006/jmbi.1996.0399
Calvo JM, Matthews RG (1994) The Leucine-responsive regulatory protein (Lrp), a global regulator of metabolism in Escherichia coli. Microbiol Rev 58(3):466–490
Reddy MC, Palaninathan SK, Shetty ND, Owen JL, Watson MD, Sacchettini JC (2007) High resolution crystal structures of Mycobacterium tuberculosis adenosine kinase—insights into the mechanism and specificity of this novel prokaryotic enzyme. J Biol Chem 282(37):27334–27342. doi:10.1074/jbc.M703290200
Capstick DS, Willey JM, Buttner MJ, Elliot MA (2007) SapB and the chaplins: connections between morphogenetic proteins in Streptomyces coelicolor. Mol Microbiol 64(3):602–613. doi:10.1111/j.1365-2958.2007.05674.x
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Yu, L., Pan, Y. & Liu, G. A Regulatory Gene SCO2140 is Involved in Antibiotic Production and Morphological Differentiation of Streptomyces coelicolor A3(2). Curr Microbiol 73, 196–201 (2016). https://doi.org/10.1007/s00284-016-1050-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-016-1050-8