Abstract
The release of extracellular DNA molecules (eDNA) contributes to various biological processes, such as biofilm formation, virulence, and stress tolerance. The quantity of eDNA released by bacteria is usually regulated by extracellular nucleases that are secreted by different systems. In this study, we show that high concentrations of eDNA inhibit the growth of two strains of Deinococcaceae, Deinococcus radiodurans, and Deinococcus radiopugnans, but have no effect on other selected organisms, such as Escherichia coli. In D. radiodurans, an extracellular nuclease was shown to be secreted through the Sec pathway. Disruption of one member of this pathway, SecD/F, inhibited cell growth, suggesting that the Sec pathway plays an important role in growth rate. However, the Sec pathway mutant exhibited a greater deficiency in growth rate compared with the extracellular nuclease mutant, indicating that the pathway not only secretes the extracellular nuclease, but has other unknown functions as well.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Both dying and living cells release extracellular DNA molecules (eDNA) all the time. Organisms often benefit from eDNA or its degradation products via their effects on various biological processes, such as biofilm formation, virulence, and stress tolerance [24, 30]. Extracellular nuclease-degraded eDNA molecules are also important nutrient sources for cells [4, 9]. However, if eDNA is not degraded immediately, damaged bases will be reincorporated into the genome, which can increase the mutation rate and lead to cell death [2]. In such instances, extracellular nucleases secreted by cells, as well as their corresponding secretion pathways, play important roles in regulating the degradation of eDNA.
Nearly one-third of cellular proteins are transported across the cytoplasmic membrane and function in the periplasm, outer membrane, or growth medium [25]. Diverse protein secretion systems, which result from the structural differences of the cell wall and cytoplasmic membrane, play roles in biofilm formation, virulence, and stress tolerance [18, 21, 22]. Several routes for protein export and secretion are known, including the general secretion (Sec) pathway and twin-arginine translocation (Tat) pathway. Among them, the Sec pathway was the first secretion pathway to be discovered, and it is conserved across the three domains of life [3, 6]. Proteins secreted through this pathway are immature precursors, and they are non-functional until they properly fold, which occurs after they cross the cytoplasmic membrane [16, 19]. Although there are some differences between Gram-negative and Gram-positive bacteria, all elucidated Sec pathways include similar elements, such as a signal peptide, signal peptidase, and the Sec transport machinery [10, 12].
Deinococcus radiodurans R1 is a red, non-pathogenic, aerobic, Gram-positive bacterium that exhibits strong resistance to ionizing and ultraviolet radiation, hydrogen peroxide, drying, and some chemical mutagens owing to its robust DNA repair and anti-oxidation abilities [2, 15, 17, 23]. After gamma radiation stress, D. radiodurans releases large amounts of damaged DNA fragments (about 1 kb in length) during the process of genome restoration [26]. An extracellular nuclease degrades these eDNA fragments immediately to avoid their reabsorption into the cells, thereby reducing the mutation rate of its genome.
Our previous studies suggested that dGMP, one of the final products of degraded DNA fragments, could elevate the bacterium’s oxidation resistance. Mutation of the only extracellular nuclease, DRB0067, obviously reduced H2O2 stress resistance [13]. Here, we show that DRB0067 was secreted through the Sec pathway, and deletion of the secD/F homolog dr1822 strongly represses cell growth, indicating that the Sec pathway plays an important role in the growth of D. radiodurans.
Results
High Levels of Extracellular DNA Repress the Growth of Deinococcaceae
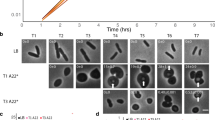
Our previous study indicated that a high level of eDNA inhibits the growth of D. radiodurans while having little effect on Escherichia coli. Further investigation suggested that the only extracellular nuclease of D. radiodurans, DRB0067, could relieve the inhibition [13]. Here, we selected five other species (D. radiopugnans, Acinetobacter baumannii, Enterococcus faecium, Lactococcus lactis, and Saccharomyces cerevisiae) and tested whether their growth was also affected by eDNA. As shown in Fig. 1, the growth of D. radiopugnans was obviously inhibited by eDNA, while the other four strains were barely affected. Combining with previous result, it seems that excessive eDNA fragments repress the growth of both D. radiodurans and D. radiopugnans.
Cell growth of five species in response to high levels of eDNA. Five other species (Deinococcus radiopugnans, Acinetobacter baumannii, Enterococcus faecium, Lactococcus lactis, and Saccharomyces cerevisiae) were added with 0, 1.5, and 3.0 mg/ml eDNA, respectively, to test their growth rates with the plate streaking method
The Extracellular Nuclease in D. radiodurans is Secreted by the Sec Pathway
Using the online program SignalP (http://www.cbs.dtu.dk/services/SignalP/), we analyzed the signal peptide of extracellular nuclease DRB0067 in D. radiodurans and predicted it to be a member of the Sec pathway. The genome of D. radiodurans was then searched for homologs of Sec pathway proteins, which revealed that DR0575, DR1822, DR2116, DR2048, and DR1825 are counterparts of SecA, SecD/F, SecY, SecE, and SecG, respectively, in E. coli. Usually, SecA acts as a central player in the Sec pathway, associates with the SecYEG channel, and transports preproteins through the cytoplasmic membrane. In some bacteria, SecD/F interacts with YajC to form a complex subunit, which links with SecYEG and stabilizes SecA in its conformation [5, 7, 8]. It has been proved in vivo that secA, secD, and secF are required for the secretion of proteins through the Sec pathway in E. coli [1, 20]. We firstly attempted to construct mutant strain that lacks dr0575, but failed, even though a variety of different gene deletion methods were employed, suggesting that SecA homolog is essential for the survival of D. radiodurans. Because SecYEG is also an essential complex in Sec pathway, we chose dr1822 for deletion and successfully obtained the corresponding mutant using the deletion–replacement method, indicating that SecD/F is dispensable for the survival of the bacteria.
To confirm that DRB0067 is secreted by the Sec pathway, we tested whether deletion of dr1822 decreased the secretion of DRB0067. As shown in Fig. 2, no nuclease activity was detected in the supernatant of the △dr1822 strain. However, the activity was fully restored when the mutant was complemented with the gene dr1822. The results demonstrate that inactivation of dr1822 strongly influenced the secretion of nuclease DRB0067, verifying the above prediction that the extracellular nuclease was secreted through the Sec pathway in D. radiodurans.
Measurement of extracellular nuclease activity. Extracellular nuclease activity of the wild-type strain (WT), the nuclease-deletion strain (Δdrb0067), dr1822 mutant strain (Δdr1822), and dr1822 complementary strain (Δdr1822 C) were measured. At various intervals (0, 15, 30, and 45 min), samples were removed and analyzed
Disruption of the SecD/F Inhibits the Growth of D. radiodurans
It has been proved that deletion of the extracellular nuclease gene drb0067 does not affect the growth of D. radiodurans. Nevertheless, the growth rate of the △drb0067 strain is greatly suppressed when excessive eDNA fragments exist [13]. Meanwhile, our above-mentioned data showed that DRB0067 was secreted by the Sec pathway. Thus, we measured the growth rate of the dr1822 mutant strain using the plate streaking method. Figure 3 shows that the △dr1822 strain exhibited a slower growth rate than the wild-type R1 and △drb0067 strains under non-eDNA-treated circumstances. However, the growth rate of the △dr1822 strain was much more dramatically inhibited than the wild-type strain and the △drb0067 strain in the presence of abundant eDNA. These results demonstrated that the inactivation of the Sec pathway slowed the growth of D. radiodurans, which is partly dependent on the deletion of drb0067 or dr1822.
Discussion
In the previous and present studies, we tested the effects of high levels of eDNA on six selected microorganisms, including one eukaryote (S. cerevisiae) and five prokaryotes (including the Gram-positive bacteria D. radiodurans, D. radiopugnans, and E. faecium, and Gram-negative bacteria E. coli, A. baumannii, and L. lactis). The results showed that excessive eDNA fragments strongly inhibit the growth of D. radiodurans and D. radiopugnans, but have little effect on the other selected microorganisms. Both D. radiodurans and D. radiopugnans belong to the Deinococcaceae, which possess relatively strong natural transformation and homologous recombination abilities, as well as extreme stress resistance [11].
Upon exposure to severe stresses, such as gamma radiation, the genome of D. radiodurans releases a large amount of damaged DNA fragments. During the subsequent genome recovery, these fragments can be reabsorbed by the cells as a result of their strong transformation ability, which would normally be expected to greatly increase the mutation frequency of the bacteria [14]. However, the genome of this unusual microorganism is consistently maintained in a high-fidelity state [29]. Thus, we investigated how the eDNA is degraded in the bacteria.
As was illustrated, the drb0067 mutant strain, which lacks the only extracellular nuclease gene, has the same growth rate as the wild-type strain under non-eDNA-treated conditions, whereas the growth of both strains decreased in the presence of excessive DNA fragments [13]. However, this was not the case for the dr1822 mutant strain, which exhibited a slower growth rate even under non-eDNA-treated condition, suggesting that the Sec pathway is essential for the growth of D. radiodurans. The mutant devoid of SecA homolog leads to the death of the bacteria, while disruption of SecD/F homolog decreases the growth rate either in the presence or absence of eDNA, indicating the relationship between growth and the Sec pathway.
Experimental Procedures
Bacterial Strains and Growth Conditions
Deinococcus radiodurans R1 (ATCC13939) was cultured in TGY medium (0.5 % tryptone, 0.1 % glucose, 0.3 % yeast extract; 1.5 % agar was added for solid medium) at 30 °C. Deinococcus radiopugnans was also cultured at 30 °C in a medium containing 3 % Peptone-B and 0.3 % tryptone. Acinetobacter baumannii was cultured in Luria-Bertani (LB) medium at 37 °C, while Enterococcus faecium, Lactococcus lactis, and Saccharomyces cerevisiae were cultured at 30 °C in brain–heart infusion (BHI) medium, MRS medium (invented by de Man, Rogosa and Sharp), and YPAD medium (2 % tryptone, 1 % yeast extract, 2 % glucose, and 0.1 mg/ml adenine sulfate), respectively. The final concentration of kanamycin added to the medium for the D. radiodurans mutant strains was 30 μg/ml.
Detection of Extracellular DNase Activity Using Methyl Green Agar Plate
Corresponding culture media were added with a certain quality of herring sperm DNA and sterilized at 121 °C for 15 min. When cooling to about 50 °C, 1 ml of 0.5 % methyl green solution was added to 100 ml of medium to form methyl green agar plates. For detection of extracellular DNase activity, a monoclonal strain was selected and cultured to an OD600 of 1.0. About 20 μl of culture was then dotted on the corresponding plate and incubated at 30 or 37 °C for several days before observing the surrounding colonies.
Growth Rate Measurement
The plate streaking method was used to determine the growth rate of bacterial strains. Various strains were streaked onto their corresponding plates and cultured at optimal temperatures for several days to observe their growth rates. Plates containing DNA were prepared as follows: herring sperm DNA was dissolved in water, filtered through a 0.22-μm membrane, and added to the solid medium.
Construction of the Mutants Δdr0575 and Δdr1822
These genes were knocked out with a deletion method that takes advantage of the strong homologous recombination ability of D. radiodurans [28]. To construct the Δdr1822 mutant, pairs of primers upstream and downstream dr1822 were used to clone DNA fragments upstream and downstream of the gene, using the wild-type R1 genome as a template for the PCRs. Then, the DNA fragments were digested with BamHI and HindIII, respectively, and ligated to a kanamycin-resistance cassette which was pre-digested with both BamHI and HindIII (Fig. 4a). Next, a tri-ligation of upstream, kanamycin-resistance, and downstream fragments was performed. The resulting fragment was then amplified and transformed into the wild-type D. radiodurans strain. The mutant Δdr1822 was screened on TGY plates containing 30 μg/ml kanamycin and verified by PCR and DNA sequencing (Fig. 4b). The complemented plasmid Δdr1822 C was constructed as described previously [27].
Measurement of Extracellular Nuclease Activity In Vitro
The wild-type R1 and mutant △dr1822 strains were cultured to an OD600 of about 2.0 and centrifuged for 10 min at 16,000 g. The supernatants were collected for the digestion reaction, using the plasmid pRADK as a substrate. MgCl2 was added to a final concentration of 10 mM, and the reaction was performed at 30 °C. At various intervals, samples were removed and analyzed by 1 % agarose electrophoresis.
References
Arkowitz RA, Wickner W (1994) SecD and SecF are required for the proton electrochemical gradient stimulation of preprotein translocation. EMBO J 13:954–963
Battista JR (1997) Against all odds: the survival strategies of Deinococcus radiodurans. Annu Rev Microbiol 51:203–224
Beckwith J (2013) The Sec-dependent pathway. Res Microbiol 164:497–504
Blokesch M, Schoolnik GK (2008) The extracellular nuclease Dns and its role in natural transformation of Vibrio cholerae. J Bacteriol 190:7232–7240
Chatzi KE, Sardis MF, Economou A, Karamanou S (2014) SecA-mediated targeting and translocation of secretory proteins. Biochim Biophys Acta 1843:1466–1474
Chatzi KE, Sardis MF, Karamanou S, Economou A (2013) Breaking on through to the other side: protein export through the bacterial Sec system. Biochem J 449:25–37
Economou A (2000) Bacterial protein translocase: a unique molecular machine with an army of substrates. FEBS Lett 476:18–21
Fekkes P, Driessen AJ (1999) Protein targeting to the bacterial cytoplasmic membrane. Microbiol Mol Biol Rev 63:161–173
Finkel SE, Kolter R (2001) DNA as a nutrient: novel role for bacterial competence gene homologs. J Bacteriol 183:6288–6293
Freudl R (1992) Protein secretion in gram-positive bacteria. J Biotechnol 23:231–240
Jung S, Joe M, Im S, Kim D, Lim S (2010) Comparison of the genomes of deinococcal species using oligonucleotide microarrays. J Microbiol Biotechnol 20:1637–1646
Kudva R, Denks K, Kuhn P, Vogt A, Muller M, Koch HG (2013) Protein translocation across the inner membrane of Gram-negative bacteria: the Sec and Tat dependent protein transport pathways. Res Microbiol 164:505–534
Li M, Sun H, Feng Q, Lu H, Zhao Y, Zhang H, Xu X, Jiao J, Wang L, Hua Y (2013) Extracellular dGMP enhances Deinococcus radiodurans tolerance to oxidative stress. PLoS One 8:e54420
Makarova KS, Aravind L, Wolf YI, Tatusov RL, Minton KW, Koonin EV, Daly MJ (2001) Genome of the extremely radiation-resistant bacterium Deinococcus radiodurans viewed from the perspective of comparative genomics. Microbiol Mol Biol Rev 65:44–79
Minton KW (1994) DNA repair in the extremely radioresistant bacterium Deinococcus radiodurans. Mol Microbiol 13:9–15
Mori H, Ito K (2001) The Sec protein-translocation pathway. Trends Microbiol 9:494–500
Moseley BE, Evans DM (1983) Isolation and properties of strains of Micrococcus (Deinococcus) radiodurans unable to excise ultraviolet light-induced pyrimidine dimers from DNA: evidence for two excision pathways. J Gen Microbiol 129:2437–2445
Ochsner UA, Snyder A, Vasil AI, Vasil ML (2002) Effects of the twin-arginine translocase on secretion of virulence factors, stress response, and pathogenesis. Proc Natl Acad Sci USA 99:8312–8317
Pohlschroder M, Hartmann E, Hand NJ, Dilks K, Haddad A (2005) Diversity and evolution of protein translocation. Annu Rev Microbiol 59:91–111
Qi HY, Bernstein HD (1999) SecA is required for the insertion of inner membrane proteins targeted by the Escherichia coli signal recognition particle. J Biol Chem 274:8993–8997
Quiblier C, Seidl K, Roschitzki B, Zinkernagel AS, Berger-Bachi B, Senn MM (2013) Secretome analysis defines the major role of SecDF in Staphylococcus aureus virulence. PLoS One 8:e63513
Renier S, Chagnot C, Deschamps J, Caccia N, Szlavik J, Joyce SA, Popowska M, Hill C, Knochel S, Briandet R, Hebraud M, Desvaux M (2014) Inactivation of the SecA2 protein export pathway in Listeria monocytogenes promotes cell aggregation, impacts biofilm architecture and induces biofilm formation in environmental condition. Environ Microbiol 16:1176–1192
Slade D, Radman M (2011) Oxidative stress resistance in Deinococcus radiodurans. Microbiology Mol Biol Rev 75:133–191
Svensson SL, Pryjma M, Gaynor EC (2014) Flagella-Mediated Adhesion and Extracellular DNA Release Contribute to Biofilm Formation and Stress Tolerance of Campylobacter jejuni. PLoS One 9:e106063
Tseng TT, Tyler BM, Setubal JC (2009) Protein secretion systems in bacterial-host associations, and their description in the Gene Ontology. BMC Microbiol 9(Suppl 1):S2
Vukovic-Nagy B, Fox BW, Fox M (1974) The release of a deoxyribonucleic acid fragment after x-irradiation of Micrococcus radiodurans. Int J Radiat Biol 25:329–337
Wang L, Xu G, Chen H, Zhao Y, Xu N, Tian B, Hua Y (2008) DrRRA: a novel response regulator essential for the extreme radioresistance of Deinococcus radiodurans. Mol Microbiol 67:1211–1222
Wang LY, Yin LF, Xu GZ, Li MF, Zhang H, Tian B, Hua YJ (2012) Cooperation of PprI and DrRRA in response to extreme ionizing radiation in Deinococcus radiodurans. Chin Sci Bull 57:98–104
Zahradka K, Slade D, Bailone A, Sommer S, Averbeck D, Petranovic M, Lindner AB, Radman M (2006) Reassembly of shattered chromosomes in Deinococcus radiodurans. Nature 443:569–573
Zhu L, Kuang Z, Wilson BA, Lau GW (2013) Competence-independent activity of pneumococcal enda mediates degradation of extracellular DNA and nets and is important for virulence. PLoS One 8:e70363
Acknowledgments
This work was supported by Grants from the National Natural Science Foundation of China (31210103904, 31370102), a major project for genetically modified organisms breeding from the Ministry of Agriculture of China (2014ZX08009-003), a special Fund for Agroscientific Research in the Public Interest (201103007), the Natural Science Foundation of Zhejiang Province (LY13C010001), the Public Project of Zhejiang Province (2014C33024), the Project for Experimental Technology of Zhejiang University, and the Project for Zhejiang Provincial Construction of technology innovation team (2010R50033).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Wang, L., Tan, H., Cheng, K. et al. Sec Pathway Influences the Growth of Deinococcus radiodurans . Curr Microbiol 70, 651–656 (2015). https://doi.org/10.1007/s00284-014-0767-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-014-0767-5