Abstract
Many studies have demonstrated that the properties of enzymes expressed in eukaryotes can be affected by the position and extent of glycosylation on enzyme. In this study, two potential glycosylation sites (the 8th and the 58th asparagine) were identified and the effect of propeptide glycosylation on Rhizomucor miehei lipase (RML) expressed in Pichia pastoris was investigated. To better understand the effect of glycosylation on the activity of RML, three mutants (M1, generated by N8A; M2, generated by N58A; and M3, generated by N8A and N58A) were designed to generate deglycosylated enzymes. The results showed that deglycosylated RML exhibited a twofold higher activity compared to the wild type. However, it was also found that glycosylation on the propeptide was important for the removal of the propeptide by Kex2 protease and secretion of the enzyme. Thus, our study provided a further understanding into the role of glycosylation on enzyme function.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Lipases are biotechnologically important enzymes capable of catalyzing a wide array of esters with wide substrate specificity [22]. Rhizomucor miehei lipase (RML, Genebank accession No. A34959) is a particularly important enzyme in the food industry due to its ability to hydrolyze 1,3-position C(O)–O bonds in triglyceride esters [15], and is generated with 24 amino acid residues of signal peptide and 70 amino acid residues of propeptide ahead of its functional region [4]. Up until now, there have been many reports aimed at optimizing and demonstrating the functionality of various hosts for the large-scale production of RML in an industrial setting [8, 9, 20, 22]. Pichia pastoris has been found to be an ideal host for the secretory expression of RML and other genes from fungi due to its simple purification process.
It is reported that two forms of native RML from R. miehei, RML-A and RML-B, exist in fermentation broth. RML-A is glycosylated and exhibits a higher activity toward olive oil than the non-glycosylated RML-B [4]. However, little is known about the glycosylation of RML, such as the position or extent of glycosylation. In our previous study, only one form of RML was expressed using Escherichia coli as a host [20], while two forms of RML were found when using P. pastoris as a host. It is well known that E. coli lacks a post-translational modification system, so these results led us to speculate that one of two forms of RML expressed in P. pastoris was also glycosylated. In addition, two potential glycosylation sites were found in the propeptide of RML through Glycomod (http://web.expasy.org/glycomod/). It has been well documented that the glycosylation of proteins can enhance enzymatic activity depending on the position and extent of glycosylation on the enzyme [1, 13, 16, 18, 19]. It is also known that the propeptide of RML acts as an intramolecular chaperone [6, 7, 17] and is essential for the expression of RML [20]. In a previous study of carboxypeptidase Y (CPY) from Saccharomyces cerevisiae, it was shown that pro-CPY was less stable toward denaturation compared to mature CPY [21]. In turn, these findings inspired us to explore whether glycosylation exists in the propeptide of RML and how glycosylation affects the function of RML.
In this study, the wild type (WT) and three deglycosylated mutants (M1, M2, and M3; M1, deglycosylated mutant 1 generated by N8A; M2, deglycosylated mutant 2 generated by N58A; and M3, deglycosylated mutant 3 generated by N8A and N58A) of RML were expressed in P. pastoris. Then, a Kex2 protease recognition site was introduced between the propeptide and the functional portion of the enzyme to explore whether the glycosylation of propeptide could affect the cleavage of propeptide.
Materials and Methods
Materials
pET30a-pro-RML plasmid was constructed in our lab as described previously [20]. E. coli strain Top 10, the pPIC9K vector, and P. pastoris strain GS115 were purchased from Invitrogen (USA). Olive oil, T4 DNA ligase, restriction enzymes, and the BCA Protein Assay kit were purchased from Sangon Biotechnology (Shanghai, China). The filter used to concentrate the crude protein in the medium was purchased from Millipore. DNA polymerase was purchased from TaKaRa (Japan). The enzyme PNGaseF was purchased from New England Biolabs (Beijing). Plasmids were transformed into yeast by using an Eppendorf Electroporator 2510 (Germany). Column chromatography for enzyme purification was performed using the AKTA system (GE Healthcare) equipped with a Ni–NTA column (Bio-Science AB, Sweden). YPD medium (1 % yeast extract, 2 % peptone, and 2 % dextrose) was used for the growth of P. pastoris GS115, and all transformants were selected on MD plates (1.34 % yeast nitrogen base, 4 × 10−5 % biotin, and 2 % dextrose). BMGY (1 % yeast extract, 2 % peptone, 1.34 % yeast nitrogen base, 4 × 10−5 % biotin, 100 mmol/l potassium phosphate, pH 6, and 1 % glycerol) and BMMY (BMGY with 0.5 % methanol instead of 1 % glycerol) were used to express the recombinant P. pastoris.
Expression of RML
The maturation region of the RML gene (MR-RML) was amplified from pET30a-pro-RML by using the primers noproF and pasR (Table 1). RML, along with its propeptide, was amplified by using the primers pasF and pasR (Table 1). The PCR reaction mixtures contained 10 μl 5× primeSTAR buffer (Mg2+ plus), 4 μl dNTP mixture (2.5 mM), 1 μl of both the forward (noproF) and reverse (pasR) primers (10 μM each), 1.25 U PrimeSTAR HS DNA Polymerase, 1 ng template, and ddH2O in a total volume of 50 μl. The thermal cycling parameters were 98 °C for 10 s, 55 °C for 15 s, and 72 °C for 1 min for a total of 30 cycles. MR-RML and Pro-RML were inserted into the EcoRI-NotI sites of pPIC9K to generate pPIC9K-MR-RML and pPIC9K-WT, respectively. Then, the resulting plasmids were linearized with SalI and transformed into the P. pastoris by electroporation at 1.5 kV in a 0.2-cm-gap electroporation cuvette using an Eppendorf Eporator 2510. The transformants of P. pastoris GS115 were screened by an MD plate. Colonies were picked into YPD medium and incubated overnight. The 1 ml overnight culture was transferred into 50 ml BMGY medium for large-scale cultivation until OD600 = 2–5. The supernatant was discarded by centrifugation at 2,000 rpm for 5 min, and the cells were resuspended at OD600 = 1 in 50 ml BMMY medium. Methanol was added every 24 h. After shaking in flask for 144 h, the supernatant was collected.
Purification of RML and Deglycosylation by PNGaseF
The supernatant was filtered via a membrane (Φ45 μm), and then purified by a Ni2+-charged 5 ml HisTrap column using the binding buffer (20 mM sodium phosphate, 0.5 M NaCl, 20 mM imidazole, pH 7.4) and elution buffer (20 mM sodium phosphate, 0.5 M NaCl, 0.5 M imidazole, pH 7.4). The concentration of the enzyme was determined by a BCA protein kit. The enzyme was deglycosylated by PNGaseF. The molecular weights of the enzyme before deglycosylation and after deglycosylation were determined by SDS polyacrylamide gel electrophoresis (Fig. 2).
Lipase Activity Assay
The activity of RML was determined by titration with olive oil as the substrate [20]. One unit of RML activity was defined as the amount of liberated fatty acid from olive oil per minute per milligram of purified protein.
Constructions of Deglycosylated Mutants
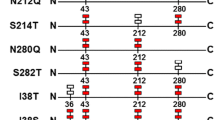
The deglycosylated RML mutant 1 (M1, Fig. 1) was generated by N8A with the primers M8F and pasR (Table 1). The deglycosylated mutant 2 (M2, Fig. 1) was generated by N58A. The primers pasF, pasR, M58R, and M58F (Table 1) were used in the overlap extension PCR. Both M1 and M2 were amplified with the WT as a template. The deglycosylated mutant 3 (M3, Fig. 1) was generated by N8A and N58A using M2 as a template, and M8F and pasR (Table 1) were used as primers.
Mutants constructed in the work. a Deglycosylated mutants. WT, wild type; M1, deglycosylated mutant 1 generated by N8A; M2, deglycosylated mutant 2 generated by N58A; M3, deglycosylated mutant 3 generated by N8A and N58A; b Introducing a Kex2 protease recognition site between the propeptide and mature portion of RML
Introducing a Kex2 Protease Recognition Site Between the Propeptide and Mature Portion of RML
The propeptide of RML could be removed completely by Kex2 protease expressed in yeast, which cleaves specifically at Lys–Arg sequences [5]. To remove the propeptide and obtain the mature part of RML, two mutants were generated by designing a Kex2 protease recognition site with the primers KexF and KexR (Table 1). The WT and M3 were used as the template to generate WT (Kex2) and M3 (Kex2), respectively (Fig. 1).
Results and Discussion
Expression of RML in P. pastoris and Deglycosylation by PNGaseF
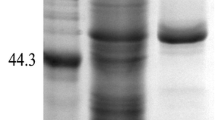
When the maturation region of the RML gene (MR-RML) was inserted into P. pastoris, no RML was expressed, which yielded the same result as the E. coli group in our previous study [20]. These results demonstrated that the propeptide of RML played a crucial role in protein expression. Our findings were also consistent with the theory that the propeptide could lower the activation energy barrier of protein folding [2, 7, 17]. However, when RML with the propeptide (WT) was cloned into pPIC9K, the purified enzyme of wide-type RML gave two major bands (Fig. 2, lane 1) at about 66 kDa and about 30 kDa. The molecular weight of RML without propeptide is 30 KDa, and the molecular weight of RML with the propeptide is about 37 kDa, which is much less than the 66 kDa obtained. Thus, we speculated that the smaller 30 kDa band was the mature RML without the propeptide while the larger band was the glycosylated form of RML with the propeptide. The enzyme produced by WT treated with PNGaseF gave two bands at 37 and 30 kDa (Fig. 2, lane 2). The band in 37 kDa was consistent with molecular weight of RML plus its propeptide. This suggested that RML was glycosylated when expressed in P. pastoris, and that the glycosylation site of RML was in the propeptide. PNGaseF treatment of M3 did not cause further shifts in mobility, which can be explained by the lack of N-glycosylation sites in the mature RML (data not shown).
Effect of Deglycosylation on Enzyme Activity
To explore whether the glycosylation of RML could affect enzyme activity, three mutants (M1, M2, and M3) were generated by mutating two asparagine residues in predicted N-glycosylation sites within the propeptide domain to alanine residues. Alanine residues were chosen to avoid generating new hydrogen bonds or charge interactions. The SDS-PAGE gel electrophoresis showed that the 8th residue of asparagine was glycosylated more severely than the 58th residue of asparagine (Fig. 3). M1 presented two bands at 40 and 30 kDa (Fig. 3, lane 2), while M2 showed two bands at about 63 and 30 kDa (Fig. 3, lane 3). When both the 8th site and 58th site were deglycosylated (M3), the mutant produced two bands at 37 and 30 kDa (Fig. 3, lane 4), which was the same as the WT treated with PNGaseF. The results indicated that the oligosaccharide chain covalently linked to the asparagine residue of the WT was removed. Furthermore, the enzyme activities of the three mutants were increased due to the deglycosylation of the propeptide in RML (Fig. 4). This result indicated that the individual removal of a glycosylation site in the propeptide of an enzyme can enhance its activity. There might be two possible reasons for this: first, the site of the glycosylation did not exist in the active center, and thus it cannot help enhance enzyme activity; second, the glycosylation has influences over the occupancy, structure, and folding of protein [14], so the fungi used might have produced high mannose-type N-glycans up to 100 sugars [3] to cover the active center, which resulted in a negative effect on the activity.
SDS-PAGE assay for the expression of the wild type and mutants. M, protein marker; lane 1, wild type; lane 2, M1 with mutation N8A; lane 3, M2 with mutation N58A; lane 4, M3 with mutation N8A and N58A. A glycosylated RML on its propeptide; B deglycosylated RML (N8A); C deglycosylated RML (N58A); D deglycosylated RML (N8A and N58A); E the mature region of RML
In Fig. 3, the proportion of the enzyme without the propeptide was gradually reduced in the total enzyme, which showed that the deglycosylation of the propeptide had a negative effect on the truncation of the propeptide. In addition, we speculate that the lower activity of the WT (Fig. 3, lane 1) was caused by glycosylation. Although other reports have investigated glycoproteins expressed in mammalian cells [12], the glycosylation of the lipase family had been seldom deliberated. Furthermore, the effect of propeptide glycosylation on enzyme activity has not been fully ascertained, though the majority of reported glycosylation events exhibited positive effects on enzyme activity. Brigitte and Huge successfully separated RML-A and RML-B in Mucor miehei fermentation, in which RML-B was formed by partial deglycosylation of RML-A, and found that the specific activity of RML-B toward olive oil was far less than that of RML-A (1,900 U/mg protein compared to 3,500 U/mg protein) [10]. In our work, the deglycosylated enzyme (M3) exhibited nearly the same activity (3,679 ± 300 U/mg protein) compared to the native enzyme RML-A. A comparison of results between the two studies demonstrated that the effect of deglycosylation was different for different hosts. In addition, the concentration of M1 and M2 in their supernatants was nearly the same as that of the WT, while M3 was less than that of the WT (Fig. 5a), as shown in the SDS-PAGE assay (Fig. 5b). These results indicated that deglycosylation on the propeptide site could decrease the secretion level of the recombinant protein.
The amount of the crude proteins in the supernatants. a Concentration of crude protein produced by WT, M1, M2, and M3 b SDS-PAGE for the crude protein in the medium; M, protein marker; lane 1, wild type; lane 2, M1 with mutation N8A; lane 3, M2 with mutation N58A; lane 4, M3 with mutation N8A and N58A. A Glycosylated RML on its propeptide; B Deglycosylated RML (N8A); C deglycosylated RML (N58A); D deglycosylated RML (N8A and N58A); E the mature region of RML
Effect of Glycosylation on Propeptide Removal
To further study the mature portion of RML, a Kex2 protease recognition site was introduced between the propeptide and the mature portion. The propeptide was successfully removed in the WT (Fig. 6, lane 1), and the specific activity of the mature portion in the WT was the same as the enzyme without N-glycosylation (M3, Fig. 4). This result indicated that the enzyme with and without the propeptide could produce the same enzyme activity. Furthermore, this also indicated that RML with glycosylation in its propeptide presented a lower enzyme activity than RML without glycosylation in its propeptide.
However, when a Kex2 protease recognition site was introduced in M3, the propeptide could not be removed completely (Fig. 6, lane 2). This suggested that the glycosylation could affect the efficiency of Kex2 protease, and that the glycosylation in the propeptide could contribute to the removal of the propeptide. The activities of the WT (Kex2) and M3 (Kex2) were about 3,600 U/mg protein (Fig. 4), which was the same as M3 and approximately twice that of WT.
There can also be two forms of glycosylation present, N-glycosylation on asparagine and O-glycosylation on serine or threonine, though O-glycosylation have not been studied in detail [11]. One of the functions of glycosylation is the stabilization of the protein conformation [12]. In our study, it was found that the glycosylation of RML could help remove the propeptide of RML in order to form mature RML, which exhibited a higher activity compared to RML containing its oligosaccharide chain. In M1, M2, and M3, the propeptide was not removed easily with the reduction of the oligosaccharide in the propeptide, which indicated that the glycosylations on the propeptide could facilitate the formation of mature RML. This finding inspired us to demonstrate that the glycosylation of the propeptide can help form a more stable structure of RML for improved function and extracellular secretion.
Conclusion
This work presented the first research on the effect of propeptide deglycosylation in RML expressed in P. pastoris. The results demonstrated that deglycosylation of the propeptide could greatly enhance enzymatic activity, though glycosylation of the propeptide is required for the removal of propeptide by Kex2 protease. This result also led to the hypothesis that the glycosylation of propeptides in other lipases might also have an effect on their activity. Finally, this work provides potential insights into furthering other properties of the lipase such as thermal stability and pH stability. Ultimately, we hope the concept of propeptide deglycosylation will pave the way for future development of new enzymatic modification strategies.
References
Aertgeerts K, Ye S, Shi L, Prasad SG, Witmer D, Chi E, Sang BC, Wijnands RA, Webb DR, Swanson RV (2009) N-linked glycosylation of dipeptidyl peptidase IV (CD26): effects on enzyme activity, homodimer formation, and adenosine deaminase binding. Protein Sci 13(1):145–154
Baker D, Sohl JL, Agard DA (1992) A protein-folding reaction under kinetic control. Nature 356(6366):263–265
Bobrowicz P, Davidson RC, Li H, Potgieter TI, Nett JH, Hamilton SR, Stadheim TA, Miele RG, Bobrowicz B, Mitchell T (2004) Engineering of an artificial glycosylation pathway blocked in core oligosaccharide assembly in the yeast Pichia pastoris: production of complex humanized glycoproteins with terminal galactose. Glycobiology 14(9):757–766
Boel E, Huge-Jensen B, Christensen M, Thim L, Fiil NP (1988) Rhizomucor miehei triglyceride lipase is synthesized as a precursor. Lipids 23(7):701–706
Brenner C, Fuller RS (1992) Structural and enzymatic characterization of a purified prohormone-processing enzyme: secreted, soluble Kex2 protease. Proc Natl Acad Sci 89(3):922–926
Chen YJ, Inouye M (2008) The intramolecular chaperone-mediated protein folding. Curr Opin Struct Biol 18(6):765–770
Eder J, Rheinnecker M, Fersht AR (1993) Folding of subtilisin BPN’: characterization of a folding intermediate. Biochemistry 32(1):18–26
Han ZL, Han SY, Zheng SP, Lin Y (2009) Enhancing thermostability of a Rhizomucor miehei lipase by engineering a disulfide bond and displaying on the yeast cell surface. Appl Microbiol Biotechnol 85(1):117–126
Huge-Jensen B, Andreasen F, Christensen T, Christensen M, Thim L, Boel E (1989) Rhizomucor miehei triglyceride lipase is processed and secreted from transformed Aspergillus oryzae. Lipids 24(9):781–785
Huge-Jensen B, Galluzzo DR, Jensen RG (1987) Partial purification and characterization of free and immobilized lipases from Mucor miehei. Lipids 22(8):559–565
Kukuruzinska M, Bergh M, Jackson B (1987) Protein glycosylation in yeast. Annu Rev Biochem 56(1):915–944
Lis H, Sharon N (2005) Protein glycosylation. Eur J Biochem 218(1):1–27
Nagai K, Ihara Y, Wada Y, Taniguchi N (1997) N-glycosylation is requisite for the enzyme activity and Golgi retention of N-acetylglucosaminyltransferase III. Glycobiology 7(6):769–776
Petrescu A-J, Milac A-L, Petrescu SM, Dwek RA, Wormald MR (2004) Statistical analysis of the protein environment of N-glycosylation sites: implications for occupancy, structure, and folding. Glycobiology 14(2):103–114
Rodrigues RC, Fernandez-Lafuente R (2010) Lipase from Rhizomucor miehei as an industrial biocatalyst in chemical process. J Mol Catal B 64(1):1–22
Seitz O (2000) Glycopeptide synthesis and the effects of glycosylation on protein structure and activity. ChemBioChem 1(4):214–246
Shinde U, Inouye M (1995) Folding pathway mediated by an intramolecular chaperone: characterization of the structural changes in pro-subtilisin E coincident with autoprocessing. J Mol Biol 252(1):25–30
Sinclair AM, Elliott S (2005) Glycoengineering: the effect of glycosylation on the properties of therapeutic proteins. J Pharm Sci 94(8):1626–1635
Tang SJ, Shaw JF, Sun KH, Sun GH, Chang TY, Lin CK, Lo YC, Lee GC (2001) Recombinant expression and characterization of the Candida rugosa lip4 lipase in Pichia pastoris: comparison of glycosylation, activity, and stability. Arch Biochem Biophys 387(1):93–98
Wang J, Wang D, Wang B, Mei Z-h, Liu J, Yu H-W (2012) Enhanced activity of Rhizomucor miehei lipase by directed evolution with simultaneous evolution of the propeptide. Appl Microbiol Biotechnol 96(2):443–450
Winther JR, Sørensen P (1991) Propeptide of carboxypeptidase Y provides a chaperone-like function as well as inhibition of the enzymatic activity. Proc Natl Acad Sci 88(20):9330–9334
Zhang WG, Han SY, Wei DZ, Lin Y, Wang XN (2008) Functional display of Rhizomucor miehei lipase on surface of Saccharomyces cerevisiae with higher activity and its practical properties. J Chem Technol Biotechnol 83(3):329–335
Acknowledgments
This work was financially supported by the Natural Science Foundation of China (21176215/21176102) and Outstanding Young Scholar of Zhejiang Province (R4110092) and the Program for Zhejiang Leading Team of S&T Innovation (2011R50007). We are grateful for the editors and reviewers and thank all the members of Prof. Yu’group.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Liu, Y., Xie, W. & Yu, H. Enhanced Activity of Rhizomucor miehei Lipase by Deglycosylation of Its Propeptide in Pichia pastoris . Curr Microbiol 68, 186–191 (2014). https://doi.org/10.1007/s00284-013-0460-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-013-0460-0