Abstract
Limited knowledge currently exists regarding species diversity and antimicrobial activity of endophytic isolates of Preussia within Australia. This report describes endophytic Preussia species that were identified through molecular analysis of the internal transcribed spacer region. Screening for antimicrobial secondary metabolites was determined by testing crude ethyl acetate (EtOAc) extracts derived from fungal mycelia against a panel of ATCC type strains which included Bacillus cereus, Escherichia coli, Enterococcus faecalis, Pseudomonas aeruginosa, Serratia marcescens, methicillin-resistant Staphylococcus aureus (MRSA) and the opportunist yeast pathogen Candida albicans. Subsequently, high-performance liquid chromatography generated fractions of bioactive EtOAc extracts which were subject to confirmatory testing using the Clinical and Laboratory Standards Institute reference microdilution antimicrobial activity assay. A total of 18 Preussia were isolated from nine host plants with 6/18 having a <97 % sequence similarity to other known species in Genbank, suggesting that they are new species. In preliminary screening, 13/18 Preussia isolates revealed antimicrobial activity against at least one of the microbes tested, whilst 6/18 isolates, including 4/6 putative new species showed specific antimicrobial activity against MRSA and C. albicans. These results highlight the antimicrobial potential of Australian Preussia spp. and also the importance of Australian dry rainforests as an untapped repository of potentially significant bioactive compounds.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The genus Preussia is an environmentally diverse but poorly understood taxon. Members of the taxa are predominantly coprophilous, although they have been isolated from wood, soil, dead plant material and occasionally as endophytes [4, 5, 8, 13, 22, 30]. Morphological characteristics for the genus are cleistothecoid ascomata and dark brown phragmospores with elongated germinal slits [4, 5]. Differentiation of Preussia from the Sporormiella and Spororominula genera based on morphologic examination has been problematic, although supplementation of this data with molecular characterisation of nuclear ribosomal sequences has prompted a general acceptance that new species are now defined as Preussia [23]. Several species of Preussia have been documented in Australia and include P. africana, P. australis, P. minima, P. cylindrica and P. funiculata [8, 30]. Furthermore, several species have also been shown to produce bioactive secondary metabolites, particularly the preussomerins which display antimicrobial activity [14, 42, 43]. The spectre of emerging microbial resistance to last line antibiotics has increased the urgency for the discovery and development of new antibiotic formulations [3, 25, 28, 36]. Two strategies have developed to address this challenge. The first is based on combinatorial chemistry which aims at computational design of effective drugs [2, 15], whilst the second is for continued exploration of the natural biota for novel bioactive metabolites [10, 18, 29, 41]. Fungi have proved to be a rich source of antimicrobial compounds over many years [1, 18, 24, 31, 32, 37]. In this paper, we have explored a largely under-examined group of Preussia endophytes isolated from dry rainforests in south-east Queensland, Australia.
Materials and Methods
Sample Collection and Fungal Isolation
A total of 23 plants were sampled across three separate semi-evergreen vine thicket (dry rainforest) sites near Toowoomba in south-east Queensland, Australia. Plants were identified using Harden et al. [21]. Leaves were sampled in duplicate from living plants within each site and processed within 10 h of collection. Leaves were rinsed with water and surface-disinfected by immersion in ethanol and passing through a flame. For test samples, a sterile hole punch was used to remove eight tissue segments per leaf which were placed in Petri dishes containing potato dextrose agar (PDA) with 1.5 mg/ml sterile streptomycin and 1.5 mg/ml tetracycline and cultured at 23 °C under dark conditions.
Identification of Fungal Isolates
DNA was extracted from actively growing cultures using a DNeasy Plant Mini Kit™ (Qiagen, Doncaster, Victoria, Australia). PCR amplification was performed by adding 0.5 μl of the extracted fungal DNA to 14 μl sterile Milli-Q water, 2 μl 10× buffer (Scientifix, Cheltenham, Victoria, Australia), 2 μl 10 mM dNTP (Scientifix) and 0.5 μl of each of the universal fungal primers ITS1F [20] and ITS4 [44] and 0.5 μl of Hot Start DNA polymerase (Scientifix). PCR amplification was conducted at 35 cycles of 94 and 50 °C for 1 min each followed by 72 °C for 1 min and a concluding incubation period of 10 min at 72 °C. Amplicons were resolved by electrophoresis in a 2 % (w/v) agarose gel and observed under UV light. Following purification PCR products were sequenced at the Australian Genome Research Facility (AGRF; Brisbane, QLD, Australia). Sequences were analysed with the NCBI BLAST search program to establish closest sequence matches. Phylogenetic analysis was performed with MEGA version 5 [38]. Sequences were aligned using Clustal W [40] and a neighbour-joining tree [35] was constructed from the alignment file using the Maximum Composite Likelihood method [38] and bootstrapping of 1000 replicates [19]. Following molecular identification Preussia isolates were also grouped into morphotypes based on culture morphology and growth rate [6, 7].
Identification of Antimicrobial Activity
Preussia isolates were initially screened for antimicrobial activity using a modification of a method described by Amin et al. [1]. In brief, a 0.5 cm2 plug of fungal culture was placed in the centre of a Sensitest agar plate (Oxoid Australia, Adelaide, SA, Australia) and incubated at 23 °C. Once cultures were >2 cm in diameter, test microbes were streaked onto the plate beginning at the edge of the fungal colony and continuing outwards. The panel of microbes tested were: Bacillus cereus (ATCC 14579), Escherichia coli (ATCC 25922), Enteroccocus faecalis (ATCC 19433), Pseudomonas aeruginosa (ATCC 25668), Serratia marcescens (ATCC 14756) and methicillin-resistant Staphylococcus aureus (MRSA; ATCC 43300) and Candida albicans (ATCC 10231). Plates were then incubated overnight at 37 °C and examined for the presence of microbial growth inhibition zones of >1 mm adjacent to fungal hyphae. Following initial screening, biologically active Preussia sp. were cultured in 250 ml flasks containing malt extract broth (ME; Oxoid Australia) and incubated under static conditions at 23 °C until vigorous mycelial growth was observed. The temperature was then reduced to 20 °C in order to slow growth and increase secondary metabolite production. After 4 days of growth at 23 °C, a 1 ml mixture of autoclaved MRSA and C. albicans cells was aseptically transferred to the cultures to enhance production of antimicrobial compounds [1, 11]. Incubation was then continued for a further 3 weeks, following which the broth and mycelia were extracted using 500 ml of EtOAc. EtOAc extracts were then examined for specific activity against MRSA and C. albicans using the reference CLSI microtitre well antimicrobial assay [16]. Wells showing no microbial growth were sub cultured on appropriate agar medium to differentiate between microstatic or microcidal inhibition. Microbial sensitivity to the extracts was recorded as growth inhibition compared to negative (growth medium only) control. Antibacterial standards used were 4 μg/ml oxacillin, 6 μg/ml vancomycin, 3 μg/ml ciprofloxacin, 4 μg/ml tetracycline and 4 μg/ml amphotericin B as the anti-fungal standard.
Analytical HPLC Fractionation of EtOAc Extracts
1–5 mg of each fungal EtOAc extract was dissolved in 200 μl of DMSO of which 100 μl was injected onto an analytical C18 Onyx HPLC column (4.6 mm × 150 mm). Extracts were then fractionated using a methanol/water/0.1 % trifluoroacetic acid gradient, conditions of which have been reported elsewhere [12]. The total run time for each injection was 11 min with five fractions (5 × 1 min) collected between 2 and 7 min. Following fractionation of all eight bioactive extracts, a standard mixture consisting of methyl 4-hydroxy benzoate, ethyl 4-hydroxy benzoate, benzophenone and uracil (all in 0.125 mg/ml in DMSO) was injected as a positive control for the HPLC process. Following fractionation, solvents were evaporated to dryness, with each fraction being resuspended in 50 or 25 % EtOH, depending on solubility. Fractions were then retested for antimicrobial activity by the CLSI method as described above.
Results
Taxonomic Diversity of Preussia Isolates
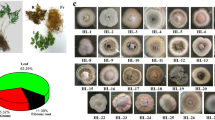
A total of 18 Preussia isolates were obtained from nine host plants (Table 1). Eight isolates were uncharacterised Preussia taxa having <97 % identity to known sequence when compared against GenBank using the BLAST search option. However, of the eight novel isolates ADM2.11 and NVM2.5 were found to have 98 % similarity over 360 bp and ELV3.11 and ELV3.2 were found to be 99 % similarity over 400 bp, reducing the number of potentially new taxa to six. Of the remaining ten isolates, two had identity to Preussia africana (99 % over 507–509 bp). Six isolates had identity to P. isomera and two had identity to two separate unnamed Preussia spp. (98–99 % over 514–426 bp). Phylogenetic analysis revealed seven separate clades with sufficient bootstrap support for all clades except clades 2 and 3 (Fig. 1). Clade 1 comprised isolates which had closest GenBank matches to P. isomera (EU551184.1), with the exception of CAH2.11 whose closest match was a different isolate of P. isomera (AY943053.1). Clade 1 also included the PAL2.3 isolate but this is most likely a separate Preussia species as it was only 95 % similar to the other members of the clade and had a velvety yellow appearance compared to the orange glossy colonies of the other isolates. When the sequences of clade seven were compared by the Blastn-2 alignment tool all three taxa were found to be less than 97 % similar suggesting they represent different species. Whilst two isolates (BSH2.9, GSM3.1) showed a sequence match of 99 % to P. africana, these were different submissions to GenBank and did not group together when phylogenetically analysed.
Antimicrobial Activity
During initial fungal colony screening, 13/18 isolates displayed antimicrobial activity against at least one of the microbe type species tested (Table 2). Subsequently, EtOAc extracts and HPLC-derived fractions were restricted to testing for specific activity against MRSA and C. albicans (Fig. 2, 3). The reason for this focus was that current therapeutic options against these two microbes are very limited and often toxic, particularly in the treatment of systemic infections, and all isolates had shown activity towards at least one of these (Fig. 2). The EtOAc extracts from 8/18 Preussia isolates showed antimicrobial activity against MRSA and/or C. albicans (Table 3). All HPLC fractions were re-tested for antimicrobial activity and revealed that the bioactive compounds eluted between 3 and 7 min from both known and presumptive new Preussia species (Table 3). Isolates which had similar bioactive profiles also exhibited similar HPLC traces (Online resource A).
HPLC Fractions of Preussia aff. africana Isolate
Preliminary chemical analysis of the bioactive HPLC fractions from isolate BSH2.9 (Preussia aff. africana) suggests that there are up to six new polyketide-derived compounds present in the later eluting fractions (Fig. 3). Chemical analysis consisted of proton nuclear magnetic resonance (NMR) spectroscopy and mass spectrometry (MS), followed by literature searching using SciFinder (http://www.cas.org/products/scifinder) and Dictionary of Natural Products [39]. Whilst the polyketide structure class is evident from the initial data analysis, additional chromatographic work is currently underway in order to purify larger quantities of the new compounds. This will enable structure determination studies to be undertaken via interpretation of 1D and 2D NMR data. Initial results indicate that HPLC fractions 2–4 contained secondary metabolites with antimicrobial activity against MRSA, but not C. albicans (Table 3).
Discussion
Species Diversity
The diversity of Preussia spp. present in dry rainforest plants of south-east Queensland is surprising given that only a small number of Preussia spp. have been previously described in Australia. Peterson et al. [30], for example, reported finding P. africana, P. australis, P. minima and P. isomera in the dung of Koalas whilst Bell [8] described P. cylindrica and P. funiculata, in a study of Australian coprophilous ascomycetes. Bell also identified nine Australian Sporormiella spp. all of which have corresponding sequences in GenBank, but none of these correlate with our samples. Samples ADL2.4, CAL2.1, CAH2.9, NVM2.6 and PAL2.1 all returned the same closest match in GenBank (EU551184.1/EU551184.2 Sporormiella isomera [syn. Preussia isomera]), displayed similar morphological characteristics and grouped together when phylogenetically analysed. From this, it may be concluded that they are different isolates of P. isomera. Given that sample PAL2.3 had a divergent ITS sequence and differing colony morphology, it is possible that this isolate is a related, but different Preussia species. Samples GSM3.1 and BSH2.9 both returned closest matches to P. africana; however, when the samples were phylogenetically analysed, they did not group together. Furthermore, when the samples were compared to each other, they only had a 92 % sequence similarity which would suggest that they are also different species. Moreover, GSM3.1 clustered more closely with P. lignicola. From this, it is possible that GSM3.1 is an isolate of P. lignicola whilst BSH2.9 is potentially P. africana. Further characterisation of the two unnamed species in GenBank, Preussia sp. (FJ210518) and Preussia sp. (FJ210521), together with the six new species obtained from this study will add to the understanding of species found in Australia for this fungal genus. Current convention is to use both molecular taxonomic and morphological characters for the description of new fungal taxa [27]. Preussia spp. are typically distinguished via differences in ascospore morphology [13]. We are endeavouring to induce sporulation in the fungal cultures and will combine morphological descriptions with the ITS data outlined here to name these new species. Phylogenetic analysis of the samples in this study and those in GenBank showed Sporomiella, Spororominula and Sporormia clustered within other known Preussia spp. Kruys and Wedin [23] argued for the merging of the genera Sporormiella, Spororominula and potentially Sporormia into the genus Preussia [23], and the results of this study would concur with this perspective.
Antimicrobial Activity in Secondary Metabolites
The search for novel classes of antimicrobial compounds is a critical strategy in addressing the emergence of antimicrobial resistance in medically important infections [9, 17]. For example, vancomycin is the antibiotic of choice for treating MRSA infections; however, the drug is renally toxic requiring constant dose monitoring and its effectiveness is being potentially compromised by the emergence of vancomycin-resistant enterococci [3, 41]. Similarly, amphotericin B, which is often the only effective antimycotic for treatment of systemic fungal infections (including C. albicans) is similarly limited due to severe infusion-related nephrotoxicity and numerous other side effects [25]. Consequently, we have explored the under exploited genus Preussia for potential drug development. Interest in examining endophytic species as a source of novel antimicrobial natural products has been heightened by reports which have shown that when endophyte-colonised plants are challenged by microbial pathogens they activate host defences more efficiently than non-symbiotic plants [29, 33, 34]. As indicated above, initial antimicrobial activity was demonstrated for 13/18 Preussia isolates. The activity of EtOAc extracts in eight of these isolates indicated that some of the bioactive compounds are lipid soluble and more specifically, HPLC fractions contained secondary metabolites that inhibited the growth of MRSA and/or C. albicans. Interestingly, isolates that had similar ITS regions showed differing bioactivity. In particular, isolate ADM2.11 was found to have a 99 % sequence similarity over 350 bp with NVM2.5. However, when these isolates were tested for bioactivity, sample ADM2.11 showed both antifungal and antibacterial activity whilst NVM2.5 did not. Variable Preussia isolate biochemistry has been described by Peterson et al. [30], who found that the two P. africana isolates produced such differing enzymatic activity that it was suggested they were potentially different species. However, differing antimicrobial activity among isolates of the same species has not been previously reported for the genus Preussia, although they have been reported for many other fungal species [26, 31]. The observance of several isolates that showed either a loss or gain of bioactivity at the HPLC stage is most likely due to concentration issues when testing fractions. Of particular interest was the bioactivity present in HPLC fractions from isolate BSH2.9 (Preussia aff. africana). Fractions collected at 3, 4 and 5 min each contained mixtures of compounds that inhibited MRSA. Thus far our chemical investigations have suggested the presence of at least six new compounds in these bioactive fractions. Additional HPLC work is currently underway in an attempt to purify all these new compounds.
References
Amin RM, Strobel GA, Vishnevetsky M, Ren Y, Geary B (2010) Biologically active endophytic Quambalaria sp. from Leptospermum junipae in Australia. Mycology 1(1):67–74
Aradi I, Erdi P (2006) Computational neuropharmacology: dynamical approaches in drug discovery. Trends Pharmacol Sci 27(5):240–243
Archibald L, Phillips L, Monnet D, McGowan JE Jr, Tenover F, Gaynes R (1997) Antimicrobial resistance in isolates from inpatients and outpatients in the United States: increasing importance of the intensive care unit. Clin Infect Dis 24(2):211–215
Arenal F, Platas G, Peláez F (2005) Preussia africana and Preussia isabellae, two new Preussia species based on morphological and molecular evidence. Fungal Divers 20:1–15
Arenal F, Platas G, Peláez F (2007) A new endophytic species of Preussia (Sporormiaceae) inferred from morphological observations and molecular phylogenetic analysis. Fungal Divers 25:1–17
Arnold A, Maynard Z, Gilbert G, Coley P, Kursar T (2002) Are tropical fungal endophytes hyperdiverse? Ecol Lett 3(4):267–274
Arnold A, Henk D, Eells R, Lutzoni F, Vilgalys R (2007) Diversity and phylogenetic affinities of foliar fungal endophytes in loblolly pine inferred by culturing and environmental PCR. Mycologia 99(2):185–193
Bell A (2005) An illustrated guide to the coprophilous Ascomycetes of Australia. Centraalbureau voor Schimmelcultures, Netherlands
Berdy J (2005) Bioactive microbial metabolites. J Antibiot 58(1):1–26
Bills G, Dombrowski A, Pelaez F, Polishook J, An Z, Watling R, Frankland J, Ainsworth A, Isaac S, Robinson C (2002) Recent and future discoveries of pharmacologically active metabolites from tropical fungi. In: Watling R, Frankland JC, Ainsworth AM, Isaac S, Robinson CH (eds) Tropical mycology: micromycetes, vol 2. CABI Publishing, Wallingford, pp 165–194
Boddington M (2009) Discovery and evaluation of novel antimicrobial agents against nosocomial pathogens. Master of science dissertation, University of Southern Queensland, Toowoomba
Camp D, Davis R, Campitelli M, Ebdon J, Quinn R (2012) Drug-like properties: guiding principles for the design of natural product libraries. J Nat Prod 75(1):72–81
Chang JH, Wang YZ (2009) The genera Sporormia and Preussia (Sporormiaceae, Pleosporales) in Taiwan. Nova Hedwigia 1(2):245–254
Chen X, Shi Q, Lin G, Guo S, Yang J (2009) Spirobisnaphthalene analogues from the endophytic fungus Preussia sp. J Nat Prod 72(9):1712–1715
Chen YP, Chen F (2008) Identifying targets for drug discovery using bioinformatics. Expert Opin Ther Targets 12(4):383–389
Clinical Laboratory Standards Institute (CLSI) (2006) Methods for dilution antimicrobial susceptibility test for bacteria that grow aerobically. Approved standard M7–A7. In: Clinical and Laboratory Standards Institute, Wayne, Pennsylvania
Coates A, Hu Y (2006) New strategies for antibacterial drug design: targeting non-multiplying latent bacteria. Drugs RD 7(3):133–151
Debbab A, Aly A, Edrada-Ebel R, Müller W, Mosaddak M, Hakiki A, Ebel R, Proksch P (2009) Bioactive secondary metabolites from the endophytic fungus Chaetomium sp. isolated from Salvia officinalis growing in Morocco. Biotechnol Agron Soc Environ 13(2):229–234
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Gardes M, Bruns T (1993) ITS primers with enhanced specificity for basidiomycetes—application to the identification of mycorrhizae and rusts. Mol Ecol 2(2):113–118
Harden GJ, McDonald WJF, Williams JB (2006) Rainforest trees and shrubs: a field guide to their identification in Victoria, New South Wales and subtropical Queensland using vegetative features. Gwen Harden Publishing, Nambucca Heads, NSW Australia
Kowalski DT (1968) Morphology and cytology of Preussia isomera. Bot Gaz 129:121–125
Kruys Å, Wedin M (2009) Phylogenetic relationships and an assessment of traditionally used taxonomic characters in the Sporormiaceae (Pleosporales, Dothideomycetes, Ascomycota), utilising multi-gene phylogenies. Syst Biodivers 7(4):465–478
Kumaran R, Muthumary J, Hur B (2008) Production of taxol from Phyllosticta spinarum, an endophytic fungus of Cupressus sp. Eng Life Sci 8:438–446
Laniado-LaborÃ-n R, Cabrales-Vargas MN (2009) Amphotericin B: side effects and toxicity. Rev Iberoam Micol 26(4):223–227
Larsen TO, Smedsgaard J, Nielsen KF, Hansen ME, Frisvad JC (2005) Phenotypic taxonomy and metabolite profiling in microbial drug discovery. Nat Prod Rep 22(6):672–695
Lebel T, Tonkin JE (2007) Australian species of Macowanites are sequestrate species of Russula (Russulaceae, Basidiomycota). Aust Syst Bot 20:355–381
Levy SB, Marshall B (2004) Antibacterial resistance worldwide: causes, challenges and responses. Nature Med 10:122–129
Lv Y, Zhang F, Chen J, Cui J, Xing Y, Li X, Guo S (2010) Diversity and antimicrobial activity of endophytic fungi associated with the alpine plant Saussurea involucrata. Biol Pharm Bull 33(8):1300–1306
Peterson R, Bradner J, Roberts T, Nevalainen K (2009) Fungi from koala (Phascolarctos cinereus) faeces exhibit a broad range of enzyme activities against recalcitrant substrates. Lett Appl Microbiol 48(2):218–225
Phongpaichit S, Rukachaisirikul V (2006) Antimicrobial activity in cultures of endophytic fungi isolated from Garcinia species. FEMS Immunol Med Microbiol 48(3):367–372
Raviraja N, Maria G, Sridhar K (2006) Antimicrobial evaluation of endophytic fungi inhabiting medicinal plants of the Western Ghats of India. Eng Life Sci 6(5):515–520
Redman RS, Freeman S, Clifton DR, Morrel J, Brown G, Rodriguez RJ (1999) Biochemical analysis of plant protection afforded by a nonpathogenic endophytic mutant of Colletotrichum magna. Plant Physiol 119(2):795–804
Rodriguez R, White J Jr, Arnold A, Redman R (2009) Fungal endophytes: diversity and functional roles. New Phytol 182(2):314–330
Saitou N, Nei M (1987) The neighbour-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Sakoulas G, Moellering RC Jr (2008) Increasing antibiotic resistance among methicillin-resistant Staphylococcus aureus strains. Clin Infect Dis 46(Supplement 5):S360–S367
Strobel G (2003) Endophytes as sources of bioactive products. Microbes Infect 5(6):535–544
Tamura K, Dudley J, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. doi:10.1093/molbev/msr121
Taylor, Francis Group (2012) Dictionary of Natural Products on DVD. CRC Press, London
Thompson J, Higgins D, Gibson T (1994) Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequencing weighting, position sequence gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Vaz AB, Mota RC, Bomfim MR, Vieira ML, Zani CL, Rosa CA, Rosa LH (2009) Antimicrobial activity of endophytic fungi associated with Orchidaceae in Brazil. Can J Microbiol 55(12):1381–1391
Vilella D, Sánchez M, Platas G, Salazar O, Genilloud O, Royo I, Cascales C, Martin I, Diez T, Silverman K (2000) Inhibitors of farnesylation of Ras from a microbial natural products screening program. J Ind Microbiol Biotechnol 25(6):315–327
Weber HA, Gloer JB (1991) The preussomerins: novel antifungal metabolites from the coprophilous fungus Preussia isomera Cain. J Org Chem 56(14):4355–4360
White T, Bruns T, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ (eds) PCR protocols, a guide to methods and applications. Academic Press, San Diego, pp 315–322
Acknowledgments
Rachel Mapperson is supported by an Australian Postgraduate award.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Mapperson, R.R., Kotiw, M., Davis, R.A. et al. The Diversity and Antimicrobial Activity of Preussia sp. Endophytes Isolated from Australian Dry Rainforests. Curr Microbiol 68, 30–37 (2014). https://doi.org/10.1007/s00284-013-0415-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-013-0415-5