Abstract
This study evaluated the effects of an increasing proportion of dietary grain on changes in bacterial populations in the goat ileum. Nine ruminally fistulated, castrated male goats were assigned to three diets in a completely randomized design. Goats were fed three different dietary treatments containing different proportions of corn grain (0, 25, and 50 %). The pH of the ileal contents and rumen fluid (P = 0.015) linearly decreased (P < 0.001), and the acetate, propionate, butyrate, and total volatile fatty acid in ileal contents increased (P < 0.05) with increases in dietary corn, and similar results were also observed in rumen fluid. The barcoded DNA pyrosequencing method was used to reveal 8 phyla, 70 genera, and 1,693 16S operational taxonomic units (OTUs). At the genus level, the proportions of Acetitomaculum, Enterococcus, Atopobium, unclassified Coriobacteriaceae, and unclassified Planctomycetaceae were linearly decreased (P < 0.05) with increases in corn grain. At the species level, high grain feeding linearly decreased the percentage of OTU8686 (unclassified Bacteria) (P = 0.004). To the best of our knowledge, this is the first study using barcoded DNA pyrosequencing method to survey the ileal microbiome of goats and the results suggest that increasing levels of dietary corn change the composition of the ileal bacterial community. These findings provide previously unknown information about the ileal microbiota of goats and a new understanding of the ileal microbial ecology, which may be useful in modulating the gut microbiome.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In the goat industry, a common practice is to feed meat goats with a high starch diet to improve meat production. However, because ruminants evolved as grazing herbivores whose diets had little if any starch, these animals have little need for intestinal amylases [30]. Consequently, if large amount of starch escapes from rumen and passes to the intestine, as can happen in grain-fed animals, bacterial overgrowth in the small intestine may occur [3, 30]. An increase in the numbers of bacteria in the small intestine can disrupt the microbial balance, and finally lead to systemic diseases such as enterotoxemia and peracute interstitial pneumonia [9, 30]. Therefore, it is important to maintain a well-balanced microflora in the small intestine of goat to optimize good health.
Although the microbiology of the rumen of goats has received considerable attention [5, 23, 27, 36]; to the best of our knowledge, the small intestine has not been well studied. This is surprising because the small intestine is the principal site of nutrient absorption. It is generally accepted that the rumen microflora has an impact on goat growth and health, and that the activities of the microflora can be manipulated by altering the diet [30, 36]. The effects of high grain feeding on the rumen of goats have been studied [36], but the role of dietary grain in relation to the small intestine microbial ecology is not very clear. Therefore, a full assessment is required of the possible variations that occur in the composition of small intestinal microbiology in response to high grain feeding.
In the past few years, a variety of different molecular fingerprinting techniques have been used to investigate various ruminants such as the domestic cow [43, 46], the reindeer [37, 38], the sheep [44, 45], the buffalo [7, 26, 48], and the Yak [14, 47]. However, studies applying molecular tools to describe the microbial diversity in the goat gut are scarce [34]. 454 pyrosequencing is a widely accepted sequencing method that has been used in the studies of microbial ecology in a variety of ecosystems, including the cecum of laying hens [4], the ileum of pigs [39, 40], and the rumen and hindgut of cattle [19, 33], and has provided a greater understanding of their microbial diversity. Therefore, in this article, we hypothesized that feeding high proportions of dietary grain could increase the ileum starch concentration, ultimately resulting in changes in the composition of ileal bacterial community, then, we used 454 pyrosequencing to analyze the bacterial communities of the goat ileum during high grain feeding by comparing the microbial communities in goats fed with diets containing different proportions of grain.
Materials and Methods
The experimental design and procedures were approved by the Animal Care and Use Committee of Nanjing Agricultural University following the requirements of the Regulations for the administration of Affairs Concerning Experimental Animals.
Animals and Diets
Nine male goats (Boer × Yangtze River Delta White) (25 ± 3 kg) were used in a completely randomized design. The experimental period was 21 days, with the first 14 days used for diet adaptation and the last 7 day used for sample collection. The animals were maintained in individual pens with free access to water and were fed twice daily at 08:00 or 20:00 h. The goats were challenged with graded amounts of rumen-fermentable carbohydrates in their diets by replacing of part of alfalfa hay and Aneurolepidium chinensis hay in a basic ration with 0, 25, or 50 % (DM basis) corn grain, to provide three different dietary treatments varying in their content of non-fiber carbohydrates. The amount of grain in the diet in the 50 % corn group was stepped up during the adaptation period. The diet was offered ad libitum to allow approximately 5 % feed refusals. All diets were formulated to meet or exceed the nutrient requirements of a 25 kg meat goat as per NRC (2007) guidelines (Table 1).
Sample Collection
Samples of rumen contents (50 mL) were obtained on days 1, 3, 5, and 7 of the measurement day shortly before the morning feeding (07: 45 h). All rumen contents samples were collected through the rumen cannula into a 140-mL plastic container. The rumen samples were strained through four layers of cheesecloth. The pH of the rumen fluid was measured immediately after collection by a mobile pH meter. Two milliliter of freshly prepared 25 % (250 mL/L) metaphosphoric acid was added to 8 mL of strained ruminal fluid. Samples were then centrifuged (17,000×g for 10 min), and supernatant fluid was stored at −20 °C for volatile fatty acid (VFA) determination [23].
On the measurement day 7, the goats were killed 6 h after the morning feeding, and the ileal contents were collected and mixed immediately. The pH was immediately measured using a mobile pH meter Three grams of mixed ileal content were stored at −80 °C. Another 10 g of mixed ileal content were stored at −20 °C for starch analysis. The remaining ileal content was prepared for VFA analysis by mixing 3 g of each sample with 1 mL of 25 % (w/v) metaphosphoric acid and 6 mL of water. Samples were then centrifuged (17,000×g for 10 min), and the supernatant was frozen at −20 °C until analysis of VFA, which was completed within 2 weeks after sample collection.
DNA Isolation
One gram of ileal content was used for DNA extraction. DNA was extracted by a bead-beating method using a mini-bead beater (Biospec Products, USA), followed by phenol–chloroform extraction [41]. The solution was precipitated with ethanol and the pellets were suspended in 50 μL Tris-EDTA buffer. The DNA samples were quantified using a Nanodrop spectrophotometer (Nyxor Biotech, Paris, France).
DNA Pyrosequencing
Universal 16S rRNA gene primers (Escherichia coli positions 8–533: E8F 5′-AGAGTTTGATCCTGGCTCAG-3′ and E533R 5′-TTACCGCGGCTGCTGGCAC-3′) were chosen for the amplification and subsequent pyrosequencing of the PCR products. The PCR mixture (final volume, 50 μL) contained 10 μL fivefold reaction buffer (TransStart™ FastPfu Buffer, TransGen Biotech), <100 ng of DNA, 0.4 μM each primer, 0.5 U Pfu polymerase (TransStart™ FastPfu DNA Polymerase, TransGen Biotech), and 2.5 mM dNTPs. Three independent PCRs were performed for each sample using a MG 96+Thermal Cycler (LongGene Scientific Instruments Co., Ltd). The PCR conditions were as follows: 95 °C for 2 min; 25 cycles of denaturation (95 °C; 0.5 min), annealing (52 °C; 0.5 min), and extension (72 °C; 0.5 min), followed by a final elongation (72 °C; 10 min). The DNA was quantified using a TBS-380 Mini-Fluorometer (Promega Corporation, CA, USA). The sequences of the partial 16S rRNA genes were determined using a Roche GS-FLX 454 pyrosequencer (Roche, Mannheim, Germany). Amplicons were sequenced according to the manufacturer’s instructions. The end fragments were blunted and tagged on both ends with one of nine ligation adaptors that contained a unique 10-bp sequence; these were recognized by the system software and the priming sequences.
Analysis of Pyrosequencing Data
Data preprocessing was performed upon software of MOTHUR program (v 1.24) [32]. The raw sequence was trimmed off the standard primers and barcodes. Sequences less than 200 bp in length and greater than 3 % low quality bases (quality score <27) were removed [33]. The chimeric sequences were also excluded using B2C2 [10]. These valid sequences were aligned to the bacterial SILVA database (SILVA 108) using a Needleman–Wunsch method [29]. The candidate sequences were screened, and preclustered to eliminate outliers; a distance matrix was generated from the resulting sequences. Sequences were clustered into OTUs using the furthest neighbor algorithm. Representative sequences from OTUs at a 0.03 distance were obtained and classified using the RDP’s Bayesian classifier [42]. For OTUs at 3 % distance, 1693 different phylotypes were detected among all the samples. Rarefaction analysis was performed by MOTHUR. From these, the Shannon diversities, and Ace and Chao1 richness estimations were calculated by MOTHUR. Aligned sequences were also used to generate a phylogenetic tree with FastTree [28] for beta-diversity (unweighted UniFrac) [21] metrics. Clustering was visualized using principal coordinates analyses (PCoA) for the unweighted UniFrac distances.
Data Analysis
The ruminal pH and VFA data were analyzed with the MIXED procedure of SPSS (SPSS v. 16, SPSS Inc., Chicago, IL) according to the following model: Y ijk = μ + D i + G j + T k + TD ik + e ijk , where Y ijk is the ith observation (the ruminal pH and specific VFA concentration in mmol/L) from the jth goat, μ is the overall mean, D is the fixed effect of diet, T k is the fixed effect of measurement time (k = 1–4 for days), TD ik is the fixed effect of diet by time interaction, G j is the random goat effect, e ijk is the residual error for the ith observation from the jth goat, residual terms are assumed to follow normal distributions. Measurements collected at different times on the same goat were considered as repeated measures in the ANOVA. The sums of squares were further partitioned by orthogonal polynomial contrast to study linear and quadratic effects of treatment.
The one-way ANOVA procedure of SPSS was used to analyze the ileal pH and VFA concentration. Nonparametric one-way ANOVA (Kruskal–Wallis) was performed to identify significant differences in the ileal microbiota composition between the control and grain treatment groups using GraphPad Prism (v 5.02). Linear and quadratic effects of increasing level of dietary corn in diets were tested using orthogonal contrasts. Significance was declared at P < 0.05 and a tendency was considered at P < 0.10.
Double dendrograms were constructed using the comparative functions and multivariate hierarchical clustering methods of NCSS 2007 (NCSS, Kaysville, Utah), on the basis of the abundances of the bacterial groups at genus level. Clustering was performed using the weighted pair linkage and Manhattan distance methods with no scaling.
Results
Effects of Increasing Levels of Corn Grain on the pH and VFA of Rumen and Ileal Content
The dietary treatment had a linear effect of lowering the pH of the rumen fluid (P = 0.015) and ileal contents (P < 0.001) (Table 2). The lowest ruminal pH (5.91) and ileal pH (6.58) were observed in goats fed the 50 % grain diet, and the highest ruminal pH (6.38) and ileal pH value (7.13) was obtained for the group fed no grain (Table 2). Increasing the amount of corn grain in the diets both linearly increased the concentration of acetate (P < 0.001) and total volatile fatty acid (TVFA) (P < 0.001) in ileal contents, as well as the percentage of starch (P < 0.001). A quadratic effect of dietary grain was detected for propionate (P < 0.001), butyrate (P < 0.001), isobutyric acid (P < 0.001), valerate (P < 0.001), and isovalerate (P < 0.001) in ileal contents. Similar results were also observed in corresponding individual VFA in rumen, except for a quadratic effect (P = 0.038) in acetate in rumen fluid.
General DNA Sequencing Observations
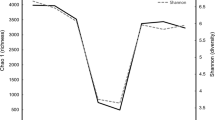
In total, 50174 reads were obtained for the 16S rRNA genes by pyrosequencing. After screening these gene sequences with our strict criteria (described in “Materials and Methods” section), 39,290 valid sequences were obtained, which accounted 78.1 % of their raw reads. The sequences were further analyzed by MOTHUR, and a total of 8 phyla were observed, distributed across all of the goats on the three diets (Fig. 1). In order of average abundance and with their respective ranges, these were: Firmicutes (96.6 %, 92.09–99.83 %), Actinobacteria (1.72 %, 0.11–3.19 %), Tenericutes (0.27 %, 0–0.68 %), Chloroflexi (0.09, 0–0.34 %), Planctomycetes (0.05 %, 0–0.14 %), Bacteroidetes (0.04 %, 0–0.13 %), Proteobacteria (0.03, 0–0.15 %), and Cyanobacteria (0.03 %, 0–0.09 %). Greater than 98.8 % of the total bacterial abundance was observed in these 8 phyla, with the remaining abundance represented by unclassified bacteria. Overall, at the genus level, the sequences were assigned to 70 different genera. Of these, the 0, 25, and 50 % grain group were represented by an average of 40, 37, and 32 genera, respectively (Table S1 in Electronic supplementary material). The number of high quality 16S OTUs recovered from each animal is listed in Table S1 in Electronic supplementary material. The average number of OTUs returned for each diet was: 0 % grain group, 381; 25 % grain group, 322; and 50 % grain group, 250.
Based on the OTUs assignment, the rarefaction curve was built to assess the sampling saturation (Fig. S1 in Electronic supplementary material). Rarefaction curves indicated that a high level of microbial diversity was obtained for subsequent analysis of dietary treatments. Furthermore, the number of observed OTUs (229–482) was much less than that calculated by estimators Ace (546–1,077). Good’s coverage revealed that the results represented the majority of bacterial 16S rRNA sequences present in each sample, with values ranging from 94.45 to 97.47 % (Table 3).
Influence of Increasing Levels of Corn Grain on Ileal Bacterial Phyla, Genera, and OTUs
As the proportion of dietary grain increased, the abundance of phylum Actinobacteria (P = 0.004) and Planctomycetes (P = 0.040) linearly decreased; there was a linear increase in the proportion of Firmicutes (P = 0.032) (Table 4). However, no significant differences were observed among the dietary treatments in the abundance of Bacteroidetes (P = 0.612), Chloroflexi (P = 0.412), Cyanobacteria (P = 0.296), Proteobacteria (P = 0.505), or Tenericutes (P = 0.646).
The influence of increased dietary grain on the ileal microbiota was observed by double hierarchal cluster analysis on the top 50 most abundant genera (≥97 % of the total number of bacterial genera observed), clustered by dietary treatment (Fig. 2). Among these genera, Solibacillus, unclassified Planococcaceae, Paenibacillus, unclassified Bacillus, unclassified Lachnospiraceae, and Bacillus occurred together in one cluster; the other 43 genera cohabited in another main cluster. Figure 3a shows the genera influenced (P < 0.10) or significantly affected (P < 0.05) by the increased dietary grain. In general, as the proportion of grain increased, the percentage of genera Acetitomaculum (P = 0.008), Enterococcus (P = 0.007), Atopobium (P = 0.007), unclassified Coriobacteriaceae (P = 0.013), and unclassified Planctomycetaceae (P = 0.037) linearly decreased. A quadratic effect of dietary grain was detected for the abundance of Aeriscardovia (P < 0.001), Ochrobactrum (P = 0.031), and Blautia (P < 0.001).
Double dendrogram of major bacterial genera found in the ileum of goats fed diets differing in grain content. The heat map indicates the relative percentage of each genera within each ileal sample. The distance between the samples, based on weighted pair linkage and Manhattan distance methods with no scaling, is shown at the top of the figure, along with a distance score. The 50 most abundant bacterial genera are provided on the y-axis with their associated distance scores. G-1-0 %, G-2-0 %, G-3-0 %, goats fed a 0 % corn grain diet; G-4-25 %, G-5-25 %, G-6-25 %, goats fed a 25 % corn grain diet; G-7-50 %, G-8-50 %, G-9-50 %, goats fed a 50 % corn grain diet
Influence of increasing corn grain on the microbiota of the goat ileum at level of genera (a) and species (b). Only the genera or OTUs that influenced (P < 0.10) or significantly affected (P < 0.05) by the increased dietary grain are presented. b G at the genera level, F at the family level, P at the phylum level
For OTUs at 3 % distance identity (i.e., species level), 1693 different phylotypes were detected among all of the samples (data not shown). Of these, a total of 955, 720, and 600 phylotypes were found in the 0, 25, and 50 % grain groups, respectively. Among the 1,693 OTUs detected in this study, only 6 OTUs were influenced or significantly affected by the increased dietary grain: as the proportion of grain increased (Fig. 3b), the percentages of OTU8686 (unclassified Bacteria) (P = 0.004) linearly decreased, while a tendency was noted for a linear decrease in the abundance of OTU1316 (Family: Planococcaceae) (P = 0.058). A quadratic effect of dietary grain was detected for the abundance of OTU622 (Family: Lachnospiraceaes) (P = 0.036), OTU8447 (Ruminococcus) (P = 0.035), OTU1885 (Solibacillus) (P = 0.002), and OTU8070 (Family: Lachnospiraceae) (P = 0.037).
Comparison of the bacterial communities by PCoA showed that goats fed the 50 % grain diet were separated from those receiving the 0 or 25 % grain diet (Fig. 4). In general, PCoA with unweighted UniFrac showed that PCA axis 1 was 23.66 % of the variation and PCA axis 2 was 13.69 % of the variation.
Discussion
In this study, we were able to see effects of the proportion of dietary corn on the ileal pH. The observed effect of supplemental fermentable carbohydrate on ileal pH in goats may be explained by the high starch content seen in the ileum of the high grain group. A previous study showed that, in cases of high grain feeding, the escape of large amounts of undigested feed from the rumen can result in extensive fermentation in the ileum and hindgut [13]. This fermentation results in increased acidity of the ileal contents. This study also showed the presence of high concentrations of acetate, propionate, butyrate, and TVFA in the rumen liquid in 50 % grain treatment compared with the control, with the values similar to these in other reports [23, 36]. In general, these results revealed that the supplementation of fermentable carbohydrate appeared to have a stimulatory effect on the fermentation in the rumen and ileum.
Deep sequencing of the ileal samples collected from the goats fed with the three diets provided a detailed view of the goat ileal microbiota. We detected members of 8 phyla of bacteria, with the majority of the pyrotags belonging to the Firmicutes and Actinobacteria, in agreement with previous reports in swine [16, 39]. These phyla have previously been shown to constitute the majority of gut-associated phylotypes in a variety of different mammalian species [5, 19, 20], which suggests that these phyla, especially the Firmicutes, play a critical role in the microbial ecology of the mammalian gut, including that of the goat. Other phyla including Chloroflexi, Bacteroidetes, Cyanobacteria, Proteobacteria, Planctomycetes, and Tenericutes represented a very small percentage of the sequences in the three groups, but the small proportion of sequences grouping with the phylum Bacteroidetes was of interest because members of the Bacteroidetes are often found to either be the most prevalent or second most prevalent in the rumen of the dairy cows or goats [5, 6, 17]. The reason for the small Bacteroidetes population observed in this study is unclear, but it is possible that the compositions of the microbiomes are different among the rumen, small intestine, and hindgut. Using comparative studies of T-RFLP fingerprints, Frey et al. [8] found that the microbial community structure changed as digesta passed from the rumen, into the small intestine and out of the animal in dairy cows. Recently, Isaacson and Kim [16] reviewed the intestinal microbiome of the pig and concluded that there were site-specific differences in the composition of microbial communities in jejunum, ileum, cecum, and colon in swine. At the phylum level, the dominant phyla were the Firmicutes or Bacteroidetes in compositions of the microbiomes of the colon and cecum. In the jejunum, bacteria in the phylum Firmicutes were the most dominant (>90 %). In the ileum, Firmicutes and Proteobacteria were the two dominant phyla [16]. Therefore, these results revealed that the site-specific differences in the composition of microbial communities may partly explain our finding that the Firmicutes were the most dominant phylum and that there is a small Bacteroidetes population in the composition of ileal community in goats. Perhaps quantification by real-time PCR of phylum Bacteroidetes in the ileum would resolve a greater number of Bacteroidetes in general.
As previously mentioned, the composition of microbial communities in rumen of a variety of ruminants including the domestic cow, the reindeer, sheep, the swamp buffalo, and the yak have been studied using molecular tools such as DGGE, RFLP, and clone library methods [34]. However, studies applying molecular tools to describe the microbial diversity in the goat gastrointestine are relatively scarce. Several reports showed that the predominant genus in the goat rumen were related to the genera Olsenella, Prevotella, Mogibacterium, Succiniclasticum, Selenomonas, Coprococcus, Butyrivibrio, Ruminococcus, and Oscillibacter [5, 27]. In contrast to the rumen, our finding showed that Solibacillus is the first predominant genera in the ileum of goats (Fig. 2). It is possible that this difference was caused by the site-specific differences in microbial composition in gastrointestinal tract as mentioned early, and the function difference in microbiology among the sites of gut. It is known that microbes in the rumen and hindgut are responsible for converting ingested feed into VFAs that serve as the major energy source for the ruminant host, and for providing a substantial portion of the host’s protein requirements [15]. On the contrary, culture-based studies have shown that microbial activity in the small intestine tends to be competitive with the host for energy and amino acids [12]. For example, bacterial utilization of glucose to produce lactic acid reduces the energy available to the host animal [31]. Normally, only small numbers of bacteria are found in the small intestine when compared with that found in the rumen and colon, and the abnormally large numbers of bacteria in the ileum may compete with the host for critical nutrients, alter host metabolism, directly damage the absorptive mucosa of the host, and produce gastrointestinal symptoms that reduce or alter food intake of the host.
The genus Solibacillus is a relatively unknown group of bacteria that was established by Krishnamurthi et al. [18]. Recent reports of 16S rRNA gene and ribosomal intergenic spacer analysis data indicated the presence of Solibacillus bacteria in the gut of adult sand fly [25] and the isolates from the potato leaf [24]. However, as only one species (Solibacillus silvestri) has been cultured [18], the physiological diversity of the members of this genera is unknown. As the isolated strain has activity for degrading N-acylhomoserine lactones (quorum-sensing signal molecules by many Gram-negative bacteria) and has the potential use in the biocontrol of plant diseases [24], the Solibacillus bacteria present in the goats may possibly perform useful functions, such as secretion of useful biocontrol agents that have quorum-quenching activities and provide protection from pathogens in the goat.
Feeding a high grain diet seemed to have a complex effect on the ileal microbiota. At the phylum level, the phyla Chloroflexi and Cyanobacteria were not detected in 50 % grain group, but were present in the 0 and 25 % groups. We also observed that the abundance of Actinobacteria was reduced by high grain feeding. At this point, the significance of the presence of Chloroflexi or Cyanobacteria in the low grain-feeding groups is unclear, and might merely indicate the presence of these species with unrelated physiologies. However, a general decrease in abundance of diverse bacterial groups was noted in the ileum of goats fed a grain diet compared with the forage group. Actinobacteria are regular, though infrequent, members of the digestive tract microbiota, and can represent up to 3 % of total rumen bacteria; however, information is considerably lacking regarding their ecology and biology [35]. So far, only a single Actinobacterium, Denitrobacterium detoxificans, has been cultivated from rumen contents [11], and the growth of this isolate was supported when 3-nitropropanol and 3-nitropropionate were supplied as electron acceptors [2]. The toxins 3-nitropropanol and 3-nitropropionate are constituents of many types of forage consumed by ruminants [22]. When absorbed into the bloodstream, 3-nitropropionate inhibits succinate dehydrogenase. On the other hand, while not toxic per se, 3-nitropropanol is converted to 3-nitropropionate by hepatic alcohol dehydrogenase [1]. In this study, we postulated that the lower proportion of members of the phylum Actinobacteria in goats fed with the high grain diet may indicate that these hosts have a low tolerance to the toxins. This finding therefore underlines the importance of continuing to research the links between bacterial genes and the tolerance of their hosts to toxic compounds.
At the genus level, a common set of taxa including Enterococcus, Atopobium, Acetitomaculum, Aeriscardovia, Blautia, Ochrobactrum, unclassified Coriobacteriaceae, and unclassified Planctomycetaceae seem to show a response to increasing grain in the diets. Some of these taxa have been identified in the rumen in other studies and show responses to a high grain diet, regardless of the differences in experimental protocols and animals (dairy cattle) [7, 19]. A more likely explanation for shifts in the microbial community structure in animals fed a high grain diet may be the abundance and digestibility of starch in the 25 or 50 % grain diet compared with the fibrous plant materials fed to the control group.
The ileum microbial population in the goat is ideally suited for the fermentation of sugars from fibrous plant materials. Goat species themselves do not produce the required fiber-degrading enzymes; instead, they harbor bacteria in their guts that can ferment the fiber. Thus, goat digestive physiology is dictated largely by the presence of fibrous materials in the ileum. If these animals are fed fiber-deficient diets, such as a high grain diet, their normal digestive processes can be disrupted, leading to the accumulation of fermentation acids and a resultant lowering of the ileal pH. These changes in the sources of sugar and starch, combined with shifts in pH, in turn alter the digestive habitat, resulting in an ileum bacterial community that can have an impact on VFA production and pathogen shedding [9, 30]. In this study, PCoA plot shows that the bacterial microbiome from 0 % grain group is similar to those of 25 % grain group, but is clearly distinct from the microbiome found in the 50 % grain group. In addition, this study revealed that the percentage of starch in the ileum from 50 % grain-fed animals was higher than that in the 0 % grain-fed goat. Therefore, the large amount of starch in the ileum contents due to high grain feeding caused more of the energy to be fermented by bacteria and absorbed as VFAs in the ileum, which prompted further changes in the composition of the ileum microbial community.
To the best of our knowledge, this is the first study using pyrosequencing method to survey the ileal microbiota and its differences among the goats fed high grain diets. Our analysis indicated that increasing levels of dietary corn increased microbial metabolites and altered the composition of the ileal bacterial community. This new information about the microbiota of the goat ileum may be useful in modulating the gut microbiome.
References
Alston TA, Mela L, Bright HJ (1977) 3-Nitropropionate, the toxic substance of Indigofera, is a suicide inactivator of succinate dehydrogenase. Proc Natl Acad Sci USA 74:3767–3771
Anderson R, Rasmussen M, Jensen N, Milton J, Allison R (2000) Denitrobacterium detoxificans gen. nov., sp. nov., a ruminal bacterium that respires on nitrocompounds. Int J Syst Evol Microbiol 50:633–638
Aslan V, Thamsborg SM, Jørgensen RJ, Basse A (1995) Induced acute ruminal acidosis in goats treated with yeast (Saccharomyces cerevisiae) and bicarbonate. Acta Vet Scand 36:65–77
Callaway TR, Dowd SE, Wolcott RD, Sun Y, McReynolds JL, Edrington TS, Byrd JA, Anderson RC, Krueger N, Nisbet DJ (2009) Evaluation of the bacterial diversity in cecal contents of laying hens fed various molting diets by using bacterial tag-encoded FLX amplicon pyrosequencing. Poult Sci 88:298–302
Cunha IS, Barreto CC, Costa OY, Bomfim MA, Castro AP, Kruger RH, Quirino BF (2011) Bacteria and Archaea community structure in the rumen microbiome of goats (Capra hircus) from the semiarid region of Brazil. Anaerobe 17:118–124
Fernando SC, Purvis HT 2nd, Najar FZ, Sukharnikov LO, Krehbiel CR, Nagaraja TG, Roe BA, Desilva U (2010) Rumen microbial population dynamics during adaptation to a high-grain diet. Appl Environ Microbiol 76:7482–7490
Franzolin R, St-Pierre B, Northwood K, Wright AD (2012) Analysis of rumen methanogen diversity in water buffaloes (Bubalus bubalis) under three different diets. Microb Ecol 64:131–139
Frey JC, Pell AN, Berthiaume R, Lapierre H, Lee S, Ha JK, Mendell JE, Angert ER (2010) Comparative studies of microbial populations in the rumen, duodenum, ileum and faeces of lactating dairy cows. J Appl Microbiol 108:1982–1993
Glock RD, DeGroot BD (1998) Sudden death of feedlot cattle. J Anim Sci 76(1):315–319
Gontcharova V, Youn E, Wolcott RD, Hollister EB, Gentry TJ, Dowd SE (2010) Black box chimera check (B2C2): a windows-based software for batch depletion of chimeras from bacterial 16S rRNA gene datasets. Open Microbiol J 11:47–52
Greening RC, Leedle JA (1989) Enrichment and isolation of Acetitomaculum ruminis, gen. nov., sp. nov.: acetogenic bacteria from the bovine rumen. Arch Microbiol 151:399–406
Hedde RD, Lindsey O (1986) Virginiamycin: a nutritional tool for swine production. Agri Pract 7:70
Hoover WH (1978) Digestion and absorption in the hindgut of ruminants. J Anim Sci 46:1789–1799
Huang XD, Tan HY, Long R, Liang JB, Wright AD (2012) Comparison of methanogen diversity of yak (Bos grunniens) and cattle (Bos taurus) from the Qinghai-Tibetan plateau, China. BMC Microbiol 19:237. doi:10.1186/1471-2180-12-237
Hungate RE (1966) The rumen and its microbes. Academic Press, New York, NY
Isaacson R, Kim HB (2012) The intestinal microbiome of the pig. Anim Health Res Rev 13:100–109
Khafipour E, Li S, Plaizier JC, Krause DO (2009) Rumen microbiome composition determined using two nutritional models of subacute ruminal acidosis. Appl Environ Microb 75:7115–7124
Krishnamurthi S, Chakrabarti T, Stackebrandt E (2009) Re-examination of the taxonomic position of Bacillus silvestris Rheims et al. 1999 and proposal to transfer it to Solibacillus gen. nov. as Solibacillus silvestris comb. nov. Int J Syst Evol Microbiol 59:1054–1058
Lee HJ, Jung JY, Oh YK, Lee SS, Madsen EL, Jeon CO (2012) Comparative survey of rumen microbial communities and metabolites across one caprine and three bovine groups, using bar-coded pyrosequencing and ¹H nuclear magnetic resonance spectroscopy. Appl Environ Microbiol 78:5983–5993
Ley RE, Hamady M, Lozupone C, Turnbaugh PJ, Ramey RR, Bircher JS, Schlegel ML, Tucker TA, Schrenzel MD, Knight R, Gordon JI (2008) Evolution of mammals and their gut microbes. Science 320:1647–1651
Lozupone C, Knight R (2005) UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol 71:8228–8235
Majak W (1992) Further enhancement of 3-nitropropanol detoxication by ruminal bacteria in cattle. Can J Anim Sci 72:863–870
Mao SY, Zhang G, Zhu WY (2008) Effect of disodium fumarate on ruminal metabolism and rumen bacterial communities in goat as revealed by denaturing gradient gel electrophoresis analysis of 16S ribosomal DNA. Anim Feed Sci Technol 140:293–306
Morohoshi T, Tominaga Y, Someya N, Ikeda T (2012) Complete genome sequence and characterization of the N-acylhomoserine lactone-degrading gene of the potato leaf-associated Solibacillus silvestris. J Biosci Bioeng 113:20–25
Mukhopadhyay J, Braig HR, Rowton ED, Ghosh K (2012) Naturally occurring culturable aerobic gut flora of adult Phlebotomus papatasi, vector of Leishmania major in the Old World. PLoS ONE 7:e35748
Pandya PR, Singh KM, Parnerkar S, Tripathi AK, Mehta HH, Rank DN, Kothari RK, Joshi CG (2010) Bacterial diversity in the rumen of Indian Surti buffalo (Bubalus bubalis), assessed by 16S rDNA analysis. J Appl Genet 51:395–402
Patel JKM, Jhala MK, Soni P, Shabir N, Pandya PR, Singh KM, Rank DN, Joshi CG (2011) Molecular characterization and diversity of rumen bacterial flora in Indian goat by 16S rDNA sequencing. VetScan 6:77–82
Price MN, Dehal PS, Arkin AP (2009) FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol 26:1641–1650
Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, Glöckner FO (2007) SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res 35:7188–7196
Russell JB, Rychlik JL (2001) Factors that alter rumen microbial ecology. Science 292:1119–1122
Saunders DR, Sillery J (1982) Effect of lactate and H+ on structure and function of rat intestine. Implications for the pathogenesis of fermentative diarrhea. Dig Dis Sci 27:33–41
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF (2009) Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541
Shanks OC, Kelty CA, Archibeque S, Jenkins M, Newton RJ, McLellan SL, Huse SM, Sogin ML (2011) Community structures of fecal bacteria in cattle from different animal feeding operations. Appl Environ Microbiol 77:2992–3001
Sirohi SK, Singh N, Dagar SS, Puniya AK (2012) Molecular tools for deciphering the microbial community structure and diversity in rumen ecosystem. Appl Microbiol Biotechnol 95:1135–1154
Suľák M, Sikorová L, Jankuvová J, Javorský P, Pristaš P (2012) Variability of Actinobacteria, a minor component of rumen microflora. Folia Microbiol (Praha) 57:351–353
Sun YZ, Mao SY, Zhu WY (2010) Rumen chemical and bacterial changes during stepwise adaptation to a high concentrate diet in goats. Animal 4:210–217
Sundset MA, Praesteng KE, Cann IK, Mathiesen SD, Mackie RI (2007) Novel rumen bacterial diversity in two geographically separated sub-species of reindeer. Microb Ecol 54:424–438
Sundset MA, Edwards JE, Cheng YF, Senosiain RS, Fraile MN, Northwood KS, Praesteng KE, Glad T, Mathiesen SD, Wright AD (2009) Rumen microbial diversity in Svalbard reindeer, with particular emphasis on methanogenic archaea. FEMS Microbiol Ecol 70:553–562
Vahjen W, Pieper R, Zentek J (2010) Bar-coded pyrosequencing of 16S rRNA gene amplicons reveals changes in ileal porcine bacterial communities due to high dietary zinc intake. Appl Environ Microbiol 76(19):6689–6691
Vahjen W, Pieper R, Zentek J (2011) Increased dietary zinc oxide changes the bacterial core and enterobacterial composition in the ileum of piglets. J Anim Sci 89:2430–2439
Van Kessel MA, Dutilh BE, Neveling K, Kwint MP, Veltman JA, Flik G, Jetten MS, Klaren PH, Op den Camp HJ (2011) Pyrosequencing of 16S rRNA gene amplicons to study the microbiota in the gastrointestinal tract of carp (Cyprinus carpio L.). AMB Express 1:41–49
Wang Q, Garrity GM, Tiedje JM, Cole JR (2007) Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73(16):5261–5267
Williams WL, Tedeschi LO, Kononoff PJ, Callaway TR, Dowd SE, Karges K, Gibson ML (2010) Evaluation of in vitro gas production and rumen bacterial populations fermenting corn milling (co)products. J Dairy Sci 93:4735–4743
Wright AD, Williams AJ, Winder B, Christophersen CT, Rodgers SL, Smith KD (2004) Molecular diversity of rumen methanogens from sheep in Western Australia. Appl Environ Microbiol 70:1263–1270
Wright AD, Toovey AF, Pimm CL (2006) Molecular identification of methanogenic archaea from sheep in Queensland, Australia reveal more uncultured novel archaea. Anaerobe 12:134–139
Wright AD, Auckland CH, Lynn DH (2007) Molecular diversity of methanogens in feedlot cattle from Ontario and Prince Edward Island, Canada. Appl Environ Microbiol 73:4206–4210
Yang LY, Chen J, Cheng XL, Xi DM, Yang SL, Deng WD, Mao HM (2010) Phylogenetic analysis of 16S rRNA gene sequences reveals rumen bacterial diversity in Yaks (Bos grunniens). Mol Biol Rep 37:553–562
Yang S, Ma S, Chen J, Mao H, He Y, Xi D, Yang L, He T, Deng W (2010) Bacterial diversity in the rumen of Gayals (Bos frontalis), Swamp buffaloes (Bubalus bubalis) and Holstein cow as revealed by cloned 16S rRNA gene sequences. Mol Biol Rep 37:2063–2073
Acknowledgments
This study was supported by Natural Science Foundation of China (NSFC) (31172228 and 30810103909), Fundamental Research Funds for the Central Universities (KYZ201114) and R&D Speical Fund for Public Welfare in Agriculture of China (200903003).
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Mao, S., Huo, W. & Zhu, W. Use of Pyrosequencing to Characterize the Microbiota in the Ileum of Goats Fed with Increasing Proportion of Dietary Grain. Curr Microbiol 67, 341–350 (2013). https://doi.org/10.1007/s00284-013-0371-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-013-0371-0