Abstract
On the basis of the DNA–DNA hybridization patterns and phenotypic characteristics, Fusobacterium nucleatum was classified into five subspecies. Previous studies have suggested that F. nucleatum subsp. vincentii is genetically similar to F. nucleatum subsp. fusiforme. The aim of this study was to investigate the possibility of classifying these two subspecies into a single subspecies by phylogenetic analysis using a single sequence (24,715 bp) concatenated 22 housekeeping genes of eight F. nucleatum strains including type strains of five F. nucleatum subspecies. The phylogenetic analysis indicated that F. nucleatum subsp. vincentii and F. nucleatum subsp. fusiforme were clustered in the same group and each strain of other F. nucleatum subspecies were also separated into the same cluster. These results suggested that F. nucleatum subsp. fusiforme and F. nucleatum subsp. vincentii can be classified into a single subspecies. F. nucleatum subsp. vincentii was early published name; therefore, F. nucleatum subsp. fusiforme Gharbia and Shah 1992 can be regarded as a later synonym of F. nucleatum subsp. vincentii Dzink et al. 1990.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Fusobacterium nucleatum is Gram-negative, non-motile, obligately anaerobic rod bacteria, which is frequently isolated from the oral cavity and may play an important role in periodontal disease [1, 2]. Based on the PAGE pattern of the whole-cell proteins and DNA homology [3] or electrophoretic patterns of glutamate dehydrogenase and 2-oxoglutarate reductase and DNA–DNA hybridization patterns [4], F. nucleatum was classified into five subspecies (nucleatum, polymorphum, vincentii, animalis, and fusiforme). Previous studies have suggested that F. nucleatum subsp. vincentii is genetically similar to F. nucleatum subsp. fusiforme [5, 6]. Therefore, this study was designed to investigate the possibility of classifying these two subspecies into a single subspecies by phylogenetic analysis of 22 housekeeping genes: rpoB (DNA-dependent RNA polymerase beta-subunit), atpC (F0F1-type ATP synthase gamma subunit), exoA (exodeoxyribonuclease III), pfkA (6-phosphofructokinase), pyrB (aspartate carbamoyltransferase), gapA (glyceraldehyde 3-phosphate dehydrogenase), pgk (phosphoglycerate kinase), gpmA (phosphoglycerate mutase), pckA (phosphoenolpyruvate carboxykinase), ldh (l-lactate dehydrogenase), ung (uracil-DNA glycosylase), tpi (triosephosphate isomerase), oadA (oxaloacetate decarboxylase alpha subunit), ddl (d-alanine-d-alanine ligase), murA (UDP-N-acetylglucosamine 1-carboxyvinyltransferase), eno (phosphopyruvate hydratase), dnaB (DNA helicase), pgi (glucose-6-phosphate isomerase), recA (recombinase A), gyrA (DNA gyrase subunit A), gyrB (DNA gyrase subunit B), and groEL (molecular chaperone GroEL) for phylogenetic analysis of the five F. nucleatum subspecies type strains. Table 1 showed the GenBank accession numbers and analyzed nucleotide location of the 22 housekeeping genes.
A 24,715-bp single sequence was generated by integrating individual genes in the mentioned order. Phylogenetic analysis was conducted using MEGA version 5 [7]. Phylogenetic trees were constructed using the neighbor-joining method and bootstrap analysis was carried out using 1,000 replications. Sequence similarities were calculated using MegAlign program (Lasergene™ 8.0, DNAStar, Inc., Madison, WI, USA).
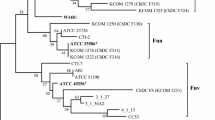
The phylogenetic tree showed that all of the F. nucleatum strains were clearly separated into four distinct clusters with 100 % bootstrap value (Fig. 1a). The genome sequence of eight strains used in this study has been deposited in GenBank. According to a dendrogram generated by the National Center for Biotechnology Information (NCBI; http://www.ncbi.nlm.nih.gov/genome?term=Fusobacterium%20nucleatum; NIH, Bethesda, MD, USA), F. nucleatum subsp. vincentii ATCC 49256T and F. nucleatum subsp. fusiforme ATCC 51190T were clustered in the same group and each strain of other F. nucleatum subspecies were also separated into the same cluster. Our data is consistent with that of NCBI except for the similarity between the four clusters composed of each F. nucleatum subspecies strain, including the one consisting of F. nucleatum subsp. vincentii ATCC 49256T and F. nucleatum subsp. fusiforme ATCC 51190T. The sequence similarity of the concatenated 22 housekeeping genes between F. nucleatum subsp. vincentii ATCC 49256T and F. nucleatum subsp. fusiforme ATCC 51190T was 98.5 % (Fig. 1b). The sequence similarities between the strains of the others F. nucleatum subspecies ranged from 98.3 to 99.5 %. The phylogenetic tree also delineated the F. nucleatum groups from Fusobacterium periodonticum (Fig. 1a). The sequences similarities between F. nucleatum strains and F. periodonticum ATCC 33369T ranged from 88.9 to 90.1 % (mean 89.4 %) (Fig. 1b). This is in agreement with our previous report, in that F. nucleatum subsp. vincentii and F. nucleatum subsp. fusiforme could not be distinguished from one another based on the analyses of the sequences of 16S rRNA gene, rpoB, and the zinc protease gene [6]. These findings suggest that these two subspecies can be classified into a single subspecies. F. nucleatum subsp. vincentii was early published name; therefore, F. nucleatum subsp. fusiforme Gharbia and Shah 1992 can be considered as a later synonym of F. nucleatum subsp. vincentii Dzink et al. 1990.
a Phylogenetic tree based on a single sequence (24,715 bp) by concatenating 22 housekeeping genes of eight Fusobacterium nucleatum strains, including five type strains of F. nucleatum subspecies. The resulting tree topology was evaluated by bootstrap analyses of the neighbor-joining tree based on 1,000 replications using MEGA version 5 [7]. b Percentage identity among the eight strains of F. nucleatum and F. polymorphum ATCC 33369T. Sequence similarities were calculated using the MegAlign program (DNAStar Lasergene™ 8.0, DNAStar, Inc.)
References
Albandar JM, Brown LJ, Löe H (1997) Putative periodontal pathogens in subgingival plaque of young adults with and without early-onset periodontitis. J Periodontol 68(10):973–981
Gharbia SE, Shah HN (1990) Heterogeneity within Fusobacterium nucleatum, proposal of four subspecies. Lett Appl Microbiol 10(2):105–108. doi:10.1111/j.1472-765X.1990.tb00276.x
Dzink JL, Sheenan MT, Socransky SS (1990) Proposal of three subspecies of Fusobacterium nucleatum Knorr 1922: Fusobacterium nucleatum subsp. nucleatum subsp. nov., comb. nov.; Fusobacterium nucleatum subsp. polymorphum subsp. nov., nom. rev., comb. nov.; and Fusobacterium nucleatum subsp. vincentii subsp. nov., nom. rev., comb. nov. Int J Syst Bacteriol 40(1):74–78
Gharbia SE, Shah HN (1992) Fusobacterium nucleatum subsp. fusiforme subsp. nov. and Fusobacterium nucleatum subsp. animalis subsp. nov. as additional subspecies within Fusobacterium nucleatum. Int J Syst Bacteriol 42(2):296–298
Gharbia SE, Shah HN, Lawson PA, Haapasalo M (1990) Distribution and frequency of Fusobacterium nucleatum subspecies in the human oral cavity. Oral Microbiol Immunol 5(6):324–327
Kim HS, Lee DS, Chang YH, Kim MJ, Koh S, Kim J, Seong JH, Song SK, Shin HS, Son JB, Jung MY, Park SN, Yoo SY, Cho KW, Kim DK, Moon S, Kim D, Choi Y, Kim BO, Jang HS, Kim CS, Kim C, Choe SJ, Kook JK (2010) Application of rpoB and zinc protease gene for use in molecular discrimination of Fusobacterium nucleatum subspecies. J Clin Microbiol 48(2):545–553
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA 5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony method. Mol Biol Evol 28(10):2731–2739
Acknowledgments
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (2012-003324) and in part by a grant from the KRIBB Research Initiative program.
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Kook, JK., Park, SN., Lim, Y.K. et al. Fusobacterium nucleatum subsp. fusiforme Gharbia and Shah 1992 is a Later Synonym of Fusobacterium nucleatum subsp. vincentii Dzink et al. 1990. Curr Microbiol 66, 414–417 (2013). https://doi.org/10.1007/s00284-012-0289-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-012-0289-y