Abstract
Fifty-five bacteriocinogenic lactic acid bacteria (LAB) isolated from seven different sources. Eight isolates were found to produce pediocin PA-1 like bacteriocin as detected by pedB gene PCR and dot-blot hybridization. The culture filtrate (CF) activity of these isolates exhibited strong antilisterial, antibacterial activity against tested food-borne pathogens and LAB. The identification and genetic diversity among the selected LAB was performed by conventional morphological and molecular tools like RFLP, RAPD, and 16S rDNA gene sequencing. The isolates were identified as, 1 each of Pediococcus acidilactici Cb1, Lactobacillus plantarum Acr2, and Streptococcus equinus AC1, 2 were of P. pentosaceus Cb4 and R38, and other 3 were Enterococcus faecium Acr4, BL1, V3. Partial characterization of the bacteriocins revealed that the peptide was heat-stable, active at acidic to alkaline pH, inactivated by proteolytic enzymes, and had molecular weight around 4.6 kDa and shared the properties of class IIa pediocin-family. The bacteriocin production at different temperatures, pH, and salt concentrations was studied to investigate the optimal condition for application of these isolates as a starter culture or as a biopreservative in either acidic or non-acidic foods.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The pediocin PA-1/AcH (pediocin PA-1) represent a class IIa bacteriocin of low molecular weight, unmodified anti-listerial peptides with a consensus motif of YGNGVXC at their N-terminal end [12]. Among all the class IIa bacteriocins, pediocin PA-1 is widely distributed and is more potent in inhibiting the growth of several pathogens associated with food spoilage and food related health hazards and hence can be a potential food bio-preservative agent [14].
Pediocin PA-1 is a plasmid encoded bacteriocin initially characterized from the strains of P. acidilactici PAC 1.0 [8]. Subsequently, other species viz P. pentosaceus, P. parvulus and other genera viz. Lactobacillus plantarum and B. coagulans were reported for the production of same bacteriocin where in different environmental conditions are known to influence bacteriocin production [3, 5, 7, 10]. The gene organization and sequences of pediocin PA-1 operon were found to be highly conserved and resides on plasmid size that ranges from 9 to 14 kb [10]. These reports are in concurrent observation that distribution of pediocin PA-1 operon among different bacteria took place by integration into the native plasmids [7]. In order to study such transfer, there is a need for detection and characterization of large number of pediocin PA-1 producers from different sources.
Although pediocin PA-1 producers are reported from different LAB, specific isolation of intergeneric and interspecific pediocin PA-1 like bacteriocin producers are not reported. Hence, in this study an attempt was made for the rapid detection of pediocin PA-1 like bacteriocin producers in different genera and species of LAB by using molecular tools. Influence of cultural conditions for the production of pediocin PA-1 like bacteriocin was also investigated.
Materials and Methods
Bacterial Strains and Maintenance
Standard pediocin PA-1 producers viz. Pediococcus acidilactici PAC1.0 [8], P. acidilactici K7 [6] and enterocin A producer Enterococcus faecium MTCC 5153 (MTCC, Chandigarh, India) were used in this study. All the above LAB cultures as well as P. acidilactici ΔK7, P. acidilactici ΔPAC1.0 (plasmid cured strains, obtained by novobiocin treatment), Leuconostoc mesenteroides NRRL B640 (NRRL, Peoria, USA) and the LAB isolates of the study were grown in de Man, Rogosa and Sharpe (MRS) broth or on MRS agar (Hi Media, Mumbai, India) at 37°C in static condition. The food-borne pathogenic indicator strains viz. Listeria monocytogenes ScottA, L. innocua FB 21, and L. murrayi FB 69 (obtained from Dr. AK Bhunia, Purdue University, USA); Aeromonas hydrophila NRRL B445; Yersinia entericolitica MTCC859 and Escherichia coli MTCC118, Staphylococcus aureus FR1722, Salmonella typhi FB231, and S. paratyphi FB254 (from Dr. E. Noterman, National Institute of Public health, Netherlands), were grown in Nutrient broth or BHI broth (Hi Media, Mumbai) at 37°C under shaking (200 rpm). The above mentioned strains were maintained at −40°C in lactobacilli MRS media and BHI or Nutrient media with 40% glycerol (v/v). Before being used, strains were propagated twice in their respective broth.
Isolation of Bacteriocinogenic LAB
The isolation of antilisterial bacteriocin producing LAB from fermented vegetable sources like carrot, cucumber, beans, and betel leaves was performed using ScottA as indicator described previously [6]. The other sources like fermented milk (curd) and chicken intestine sample were diluted and pour plated and observed for zone of inhibition against ScottA, and further characterized as described previously [6].
PCR Amplification of Pediocin PA-1 Genes
Total DNA from LAB was isolated as described by Mora et al. [11]. All the oligonucleotide primers were obtained from Sigma-Aldrich (Bangalore, India) and the PCR components were from Bangalore GeNei (Bangalore). The pedB gene was amplified by using primers, pedBF (5′GGTGATTTTATGAATAAGACTAAGTCG3′) and pedBR (5′CCCCTTTATCAGTACTATTGGCTAGGC3′) positioned at 3488–3514 and 3823–3849 as per the sequence of pSMB74 of P. acidilactici H (Accession number-U02482). The standard procedure for PCR amplification was followed as described by Sambrook and Russell [15] with annealing at 60°C. Similarly, pedAB gene was amplified as described earlier [6]. All the PCR amplicons were analyzed by 1.5% agarose (SRL, Mumbai, India) gel electrophoresis.
DNA Dot-Blot Hybridization
The PCR product of pedB gene obtained from P. acidilactici PAC1.0 was labelled with digoxigenin-dUTP using random primed DNA labeling kit (Roche Chemicals, Germany) and used as a probe for dot-blot analysis. Ten microlitre of total DNA (approximately 25–50 ng μl−1) of test culture was heat denatured, spotted on a Hybond Nylon membrane (Amersham International, UK) according to the method described earlier [15] and hybridized using probe. Hybridization and stringency washes were carried out at 42°C according to manufacturer instructions (Roche chemicals, Germany).
Phenotypic and Biochemical tests
The Gram-staining, catalase, fermentation of carbohydrate viz. 1% each of glucose, lactose, maltose, sucrose, mannitol, sorbitol, etc., gas production from glucose was performed as per standard microbiological methods. Growth of test cultures in MRS broth at different temperatures (10, 37, and 45°C) was evaluated upon incubation for 16 h. Similarly, growth in MRS broth containing 5 and 8% NaCl and at pH (4, 8, and 10) was also tested.
RAPD, RFLP, and 16S rDNA Gene Sequencing
Random Amplified Polymorphic DNA (RAPD) PCR of total DNA was carried out by primer M13 (5′GAGGGTGGCGGTTCT3′) in a 25 μl reaction volume as described earlier [17]. Digestion of 16S rDNA gene PCR product with HaeIII and AluI enzymes (Bangalore GeNei, Bangalore) for Restriction fragment length polymorphism (RFLP) analysis was performed. The primers and the PCR conditions used for amplification were followed as described earlier [13]. DNA sequences of 16S rDNA PCR product was sequenced at the sequencing facility of Vimta Labs (Hyderabad, India). The gene sequences obtained were analyzed using the BLAST search programme [1].
Antibacterial Activity Assay
The test cultures were grown in MRS broth at 37°C for 16 h under static condition. The cultures were centrifuged at 9000 rpm in 4°C for 15 min and the culture filtrate (CF) was collected, filtered through 0.4 μ filter (Millipore) and stored at 4°C until further use. The inhibitory effect of the CF was tested against food-borne pathogens and LAB cultures by spot-on-lawn assay [4].
Characteristics of Antimicrobial Compound
The CF of the test culture was subjected to treatment with different proteolytic enzymes such as proteinase K, papain, trypsin, pepsin (SRL) at a final concentration of 1 mg ml−1, reducing agents (conc. 10%) like β-mercaptoethanol (SRL) and Dithiothreitol (DTT) (SRL) were also used. Reaction mixture was incubated at 37°C for 2 h and residual activity was determined using ScottA, as described previously [13]. Similarly, stability of CF at different temperature and varying pH was also tested as above. The chloroform extracted bacteriocin from CF was subjected for bioassay by Tricine–SDS-PAGE [16] and overlaid with ScottA.
Bacteriocin Production at Different Temperatures, pH, and NaCl Concentration
MRS broth with pH of 4, 8, and 10 (adjusted by HCl or NaOH), as well as MRS with 4 and 8% sodium chloride (w/v) (SRL) was prepared and inoculated with 1% freshly grown test cultures and allowed growth for 16 h at static conditions at 37°C. Growth (OD 600 nm) and bacteriocin production of test cultures in MRS broth at different temperatures (15, 37, and 50°C) was also studied as described above. Antilisterial activity expressed as arbitrary unit per ml (AU ml−1) and defined as the highest dilution of test sample exhibiting the zone of inhibition against indicator ScottA.
Results and Discussion
Detection of Putative Pediocin PA-1 like Bacteriocin Producing LAB
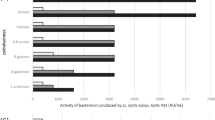
In order to study intergeneric and interspecific pediocin PA-1 producers, we have screened large number of antilisterial bacteriocin producing LAB. Among the screened sources, the LAB isolated from vegetables displayed strong antilisterial activity. From each representative source, the cultures with high activity were selected and subsequently tested for the presence of immunity protein of pediocin PA-1 (pedB) gene by PCR. Among 55, 8 cultures gave expected amplicons of 362 bp for pedB and 600 bp for pedAB genes. The results of CF activity against ScottA and pedB PCR analysis of the selected native isolates is shown in Fig. 1a, b. PCR results were additionally confirmed by dot-blot hybridization using pedB gene probe. As expected, cultures K7, BL1, Acr2, Acr4, Cb1, Cb4, V3, AC1, and R38 gave positive signal suggesting a conserved pediocin PA-1 gene in native isolates, whereas, E. faecium MTCC 5153 and ΔPAC1.0 did not react with the probe (data not shown). The detection of pediocin PA-1 by molecular tools was earlier reported in P. parvulus [3] and P. acidilactici [9].
Characteristics of Native LAB
All the selected isolates were Gram-positive, catalase negative and cocci in shape except the isolate Acr2, which was rod shaped. The gas production was observed only for Acr2 and V3 isolate. The isolate Acr2, was unable to grow at 45°C, 8% NaCl and at pH 10. Similarly AC1 isolate was unable to grow in 8% NaCl concentration, 10 and 45°C temperatures and also at pH 4 and 10. The isolates were able to ferment different carbohydrates tested, except AC1 which could not utilize lactose. The other isolates were able to grow at all the parameters used. These results suggested that, the isolates had distinct characteristic features.
Molecular Typing of Putative Pediocin PA-1 Producers
In order to differentiate the isolates among each other and also from native P. acidilactici K7, RAPD PCR was performed. RAPD showed five different banding profiles indicating, the selected isolates were different from each other. Similarly, RFLP also showed variability at their 16S rDNA gene (Fig. 2a, b). For species level identification, 16S rDNA gene sequencing followed by BLAST analysis was performed. DNA sequence homology in combination with the results of physiological and biochemical tests, the putative pediocin producers were identified as follows—Streptococcus equinus AC1, Pediococcus acidilactici Cb1, Pediococcus pentosaceus Cb4 and R38, Lactobacillus plantarum Acr2, Enterococcus faecium Acr4, BL1, and V3. The 16S rDNA gene sequences (~700 bp) have been deposited in the GenBank database under the Accession numbers GU222444–GU222450 for the LAB strains AC1, Cb1, Cb4, Acr2, V3, BL1, and Acr4, respectively. The bacteriocin producing LAB isolates reported in this study are deposited in the National Collection of Industrial Microorganisms (NCIM) at the National Chemical Laboratory, Pune, India.
Differentiation of the native pediocin PA-1 like bacteriocin producers by RAPD PCR (a) and RFLP of 16S rDNA gene digested with HaeIII and AluI (b). a Lane 1 AC1, 2 K7, 3 Cb1, 4 Cb4, 5 Acr2, 6 BL1, 7 V3, 8 Acr4, 9 R38, 10 PAC1.0 b Lane 1 K7, 2 PAC1.0, 3 Cb1, 4 Cb4, 5 BL1, 6 Acr2, 7 Acr4, 8 V3, 9 R38, 10 AC1, 11 MTCC 5153. M is a 10 Kb molecular marker (GeNei, Bangalore) in both the gels
Antibacterial Spectrum and Properties of Bacteriocin
All test isolates were studied for their ability to inhibit various food-borne pathogens as well as closely related LAB species like E. faecium 5153 and Leuconostoc mesenteroides NRRL B640. The tested isolates were able to inhibit all the Listeria spp., mutants of K7 and PAC 1.0 as well as Gram-negative Aeromonas and Yersinia sp, with an inhibition zone size of around 10–18 mm. Isolates St. equinus AC1, Lb. plantarum Acr2, E. faecium Acr4, and P. pentosaceus R38 were also able to inhibit Gram-positive Staphylococcus aureus. None of the isolates inhibited P. acidilactici K7, PAC 1.0, as well as Escherichia coli and Salmonella typhi. Inhibitory spectra of pediocin PA-1 to selected Gram-positive bacteria were earlier reported [12, 14]. The Gram-negative Aeromonas hydrophila B445 was similarly inhibited by pediocin SA1 [2]. In general, an antibacterial spectrum of Cb4, Acr2, Acr4, BL1, and V3 was found to be different than the pediocin PA-1 producing in P. acidilactici K7.
The protease sensitivity and inactivation by reducing agents suggested the proteinaceous nature and involvement of disulfide bridge, respectively, of the AMC. Optimum activity of CF for most of the isolate was found to be at pH 7–8 and temperature between 40 and 80°C. However, at pH 2–4 and temperature at 100 and 121°C, the activity was reduced to ~50%. Antilisterial activity was retained at even pH 10 and at 90°C suggesting heat stable and wide pH range AMC. Upon Tricine SDS-PAGE analysis, all isolates had the active peptide of 4.6 kDa (data not shown). The above reported observations for the selected isolates are similar to that described for pediocin PA-1 [14].
Effect of Cultural Conditions on Production of Bacteriocin
The bacteriocin production for all the cultures was more at 37°C when compared to 15 and 50°C. All the strains of E. faecium V3, Acr4, BL1 were able to grow and produce bacteriocin at all the temperatures, pH, and NaCl concentration, this could be due to the fact that E. faecium has wider adaptability to environment. The isolate P. acidilactici K7, Cb1, and Lb. plantarum Acr2 were able to grow at 15 and 37°C. The isolate St. equinus AC1 was unable to grow at 15 and 50°C, pH 4 and 10, and at 8% NaCl concentration. Similarly, the isolate Lb. plantarum Acr2 was unable to grow at pH 4 and 10 and did not produce bacteriocin (Fig. 3). The bacteriocin production is greatly influenced by the nutrients, temperature, pH, NaCl concentration [4]. The optimum condition for pediocin AcH, SA-1, and other class IIa bacteriocins was found to be at temperature 30–35°C, pH 5–7, and NaCl 1.5–3% [12]. Results obtained suggested that these native isolates could be used as a protective culture in acidic foods like pickles and yogurt, as they exhibited good growth and bacteriocin production at different cultural conditions.
In conclusion, we detected the presence of pediocin PA-1 gene cluster and PA-1 like bacteriocin properties in E. faecium and St. equinus besides P. pentosaceus and Lb. plantarum of vegetable and dairy origin. The molecular typing tools have proven to be useful in differentiation, characterization, and identification of LAB in spite of their high heterogeneity and phylogenetic inter-mixing. These cultures can be further used to study the pediocin PA-1 operon integration, revealing the possible mechanism of horizontal operon transfer as reported for B. coagulans I4 and Lb. plantarum 423 [7, 18]. We are presently investigating the flanking regions of the operon to discover the novel mobile genetic elements involved in such recombination events. Hence, Pediocin PA-1 produced by LAB other than P. acidilactici can be of industrial significance in different food systems because of their wider environmental adaptability.
References
Altschul SF, Maddan TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nuc Acids Res 25:3389–3402
Anastasiadou S, Papagianni M, Filiousis G, Ambrosiadis I, Koidis P (2008) Pediocin SA-1, an antimicrobial peptide from Pediococcus acidilactici NRRL B5627: production conditions, purification and characterization. Biores Technol 99:5384–5390
Bennik MHJ, Smid EJ, Gorris LGM (1997) Vegetable-associated Pediococcus parvulus produces pediocin PA-1. Appl Environ Microbiol 63:2074–2076
Biswas SR, Ray P, Johnson MC, Ray B (1991) Influence of growth conditions on the production of a bacteriocin, pediocin AcH, by Pediococcus acidilactici H. Appl Environ Microbiol 57:1265–1267
Ennahar S, Aoude-Werner D, Sorokine O, van Dorsselaer A, Bringel F, Hubert JC, Hasselmann C (1996) Production of pediocin AcH by Lactobacillus plantarum WHE92 isolated from cheese. Appl Environ Microbiol 62:4381–4387
Halami PM, Ramesh A, Chandrashekar A (2005) Fermenting cucumber, a potential source for the isolation of pediocin-like bacteriocin producers. World J Microbiol Biotech 21:1351–1358
Le Marrec C, Hyronimus B, Bressollier P, Verneuil B, Urdaci MC (2000) Biochemical and genetic characterization of coagulin, a new antilisterial bacteriocin in the pediocin family of bacteriocins, produced by Bacillus coagulans I4. Appl Environ Microbiol 66:5213–5220
Marrug JD, Gonzalez CF, Kunka BS, Ledeboer AM, Pucci MJ, Tooner MY, Walker SA, Zoetmulder LCM, Vandenbergh PA (1992) Cloning, expression, and nucleotide sequence of genes involved in production of pediocin PA-I, a bacteriocin from Pediococcus acidilactici PAC1.0. Appl Environ Microbiol 58:2360–2367
Martinez JM, Martinez MI, Herranz C, Suarez AM, Cintas LM, Fernandez MF, Rodriguez JM, Hernandez PE (2000) Use of genetic and immunological probes for pediocin PA-1 gene detection and quantification of bacteriocin production in Pediococcus acidilactici strains of Meat origin. Food Agric Immunol 12:299–310
Miller KW, Ray P, Steinmetz T, Hanekamp T, Ray B (2005) Gene organization and sequences of pediocin AcH/PA-1 production operons in Pediococcus and Lactobacillus plasmids. Lett Appl Microbiol 40:56–62
Mora D, Fortina MG, Parini C, Daffonchio D, Manachini PL (2000) Genomic sub-populations within the species Pediococcus acidilactici detected multilocus typing analysis: relationships between pediocin AcH/PA-1 producing and non-producing strains. Microbiol 146:2027–2038
Papagianni M, Anastasiadou S (2009) Pediocins: The bacteriocins of pediococci. Sources, production, properties and applications. Microb Cell Fact 8:1–16
Rai AK, Bhaskar N, Halami PM, Indirani K, Suresh PV, Mahendrakar NS (2009) Characterization and application of a native lactic acid bacterium isolated from tannery fleshings for fermentative bioconversion of tannery fleshings. Appl Microbiol Biotechnol 83:757–766
Rodriguez JM, Martinez MI, Kok J (2002) Pediocin PA-1, a wide-spectrum bacteriocin from lactic acid bacteria. Cr Rev Food Sci Nutr 42:91–121
Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual, 3rd edn. Cold Spring Harbor, New York
Schagger H, Von Jagow W (1987) Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for separation of proteins in the range from 1 to 100 kDa. Anal Biochem 166:368–379
Schillinger U, Yousif NMK, Sesar L, Franz CMAP (2003) Use of group-specific and RAPD-PCR analyses for rapid differentiation of Lactobacillus strains from probiotic yogurts. Curr Microbiol 47:453–456
Van Reenen CA, Van Zyl WH, Dicks LMT (2006) Expression of the immunity protein of plantaricin 423, produced by Lactobacillus plantarum 423, and analysis of the plasmid encoding the bacteriocin. Appl Environ Microbiol 72:7644–7651
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Devi, S.M., Halami, P.M. Detection and Characterization of Pediocin PA-1/AcH like Bacteriocin Producing Lactic Acid Bacteria. Curr Microbiol 63, 181–185 (2011). https://doi.org/10.1007/s00284-011-9963-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-011-9963-8