Abstract
Characterization, direct sequencing of the PCR amplicon and phylogenetic relationship was done to discover a novel Vip protein genes of the Bt isolates, to improve the prospects for insect control, more Vip proteins should be sought out and researched to predict their insecticidal activity. Characterization was based on direct sequencing of PCR amplicon using primers specific to vip3A gene was presented here. 12 out of 18 isolates screened were positive for vip gene-specific primers. Homology search for the partial sequences using BLAST showed that 11 isolates had high similarity to vip3Aa gene and only one fragment with vip3Ae gene (25–100% at nucleotide and amino acid level). Phylogenetic analysis showed that the gene sequences were responsible for geographic separation for divergence within vip genes, consistent with the evaluation of distinct bacterial population. Despite the geographical distances, strains harbouring vip genes have originated from common ancestors may significantly contribute to control resistant insect pests. Some strains have evolved to be quite distinct and others remain as members of closely related groups. The reported method is a powerful tool to find novel Vip3A proteins from large-scale Bt strains which is effective in terms of time and cost. Further the Vip proteins produced by different strains of B. thuringiensis are unique in terms of the sequence divergence and hence may also differ in their insecticidal activities.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The various crystal protein toxins (Cry) produced by Bt have been successfully used in pest management programmes both in agriculture and public health. In the recent years, there is a renewed interest in isolating potent Bt strains with increased host spectrum and also a source of isolation of novel genes. Bt has been isolated from different sources viz. fresh water, saw dust, cured tobacco leaves, rice bran, stored products, compost, phylloplane, marine sediments and ancient glacial ice [1–8]. The host range of Bt includes lepidoptera, coleopteran, diptera, acarina, protozoa, hymenoptera, trematode and nematodes [9]. Intensive, large-scale screening programmes have yielded new genes such as vegetative insecticidal proteins (vip3A) [10] are a group of insecticidal proteins and represent the second generation of insecticidal trans-genes that will complement the novel δ-endotoxins in future. Vip proteins have been reported to have insecticidal properties, providing a potent broad spectrum of insect control. Vip proteins fall into three families viz. Vip1, Vip2 and Vip3, where Vip1 and Vip2 proteins are the two components of a binary toxin that exhibits toxicity to coleopterans [11]. Host range of Vip3 proteins includes several major lepidopteran pests [12, 31].
vip3A gene homologues in approximately 15% of the Bacillus strains, which had shown activity against lepidopteran insect larvae, including black cutworm (Agrotis ipsilon), fall armyworm (Spodoptera frugiperda), beet armyworm (Spodoptera exigua), tobacco budworm (Heliothis virescens) and corm earworm (Helicoverpa zea) [11]. Histopathological observations showed that Vip3A ingestion by susceptible insects such as the black cutworm (Agrotis ipsilon) and fall armyworm (Spodoptera frugiperda) caused gut paralysis at concentrations as low as 4 ng/cm2 of diet spread on leaves, resulting in larval death at concentrations above 40 ng/cm2 [30]. The importance of Vip3A protein for the insecticidal activity of B. thuringiensis was determined by deleting the vip3A gene from strain HDl. Compared to HD1, the strain HD1Deltavip3A, with deleted vip3A showed only one-fourth toxicity to Agrotis ipsilon larvae and <10% toxicity to Spodoptera exigua larvae [29].
Unlike the crystal protein toxins (Cry) Vip3 proteins are secreted during vegetative growth and have no homology to Cry or cytolytic toxins (Cyt). Like Cry toxins, Vip3 proteins must be activated by proteases prior to recognition at the surface of the midgut epithelium of specific 80 and 100 kDa membrane proteins different from those recognized by Cry proteins [13]. Vip3 proteins are suitable candidates in Bt resistance management as these proteins recognize different receptors as compared to Cry proteins. However, the potential Vip proteins remains untapped to date, there are approximately 82 kinds of vegetative insecticidal protein genes that have been identified and cloned. These genes can be classified into three groups, eight subgroups, 25 classes and 82 subclasses according to the encoded amino acid sequence similarity (http://www.lifesci.sussex.ac.uk/home/Neil_Crickmore/Bt/vip.html/). The search for novel vip sequences might prove to be useful for increasing the target range of future insecticidal transgenic plants while allowing resistance management by associating vip genes with the functionally different cry genes. As the environment is diverse, hence the insecticidal proteins are also diverse showing differential insecticidal activities. Therefore, it is necessary to screen more Bt isolates to clone and characterize vip genes and their variants. Hence, in this study the local isolates of Bt were screened with vip3A gene-specific primers to identify isolates harbouring these genes to further clone and characterize full length of the same.
Materials and Methods
Bacterial Strains and DNA Preparations
Eighteen native lepidopteron active Bt isolates were cultured on Luria agar at 30°C for 72 h and stored at 4°C until further use. Single colony of the above Bt isolates was cultured in Luria broth at 30°C for 24 h and centrifuged at 9,000×g, 10 min at 4°C. The pellet was washed with double distilled water at 9,000×g, 10 min at 4°C and total DNA was isolated and used for PCR analysis [Published data 14].
Oligonucleotide Primers and PCR Analysis
For the detection of vip genes primers specific to vip3A gene viz. vip3A forward 5′-GCC CAT GGA CAA GAA TAA T-3′ and vip3A reverse 5′-GAA CTA GTT TCT GTA GCA A-3′ [15] with an expected product size of 584 bp was used. PCR was carried out in 25 μL total reaction volume, contained the following components template DNA—2 μl, Tris–HCl 10 mM pH 8.3; KCl—50 mM; MgCl2—2.5 mM—2.5 μl, dNTP mix—2.5 mM each Taq polymerase—0.5 U (5 U/μl) (Fermentas Life Sciences), forward and reverse primers—20 pmol of each and rest of the volume was made up with DNAase and RNAase-free water (Eppendrof). Amplification was done in a applied biosystem thermal cycler under the following conditions: initial denaturation −94°C for 3 min, followed by 40 cycles of denaturation—94°C for 30 s, annealing—47°C for 35 s, extension—72°C for 1 min and a final extension—72°C for 20 min. The PCR amplified products were analysed by 1.5% agarose gel in TBE buffer for 150 V/h and stained with ethidium bromide (10 μg/ml) and visualized in a transilluminator at 365 nm.
Sequencing and Analysis of vip Gene
The PCR amplified fragments were eluted using Perfect Prep® gel clean up kit (Eppendorf) according to the manufacture’s protocol. Direct sequencing of the above fragments was carried out using the forward primer of the gene-specific primers used in this study in an automated sequencer (ABI PRISM 310). Sequencing was repeated five times for each fragment and the sequences were aligned in a sequence alignment editor, ‘Bioedit’ to look for any introduced errors while sequencing, if any. Finally, one sequence was selected for each Bt isolate and homology search was carried out using BLAST X version 2.2.6 (http://www.ncbi.nlm.nih.gov) and were deposited with NCBI. As direct sequencing of PCR amplified fragments resulted in unequal lengths of partial sequences they were made of uniform length and compared with the first published vip3Aa sequence, L48811 [9] based on which vip3A gene-specific primers used in this study were designed. The sequence analysis was done after converting into FASTA format on internet (website: http://www.ncbi.nlm.nih.gov). Multiple alignments of nucleotide and deduced amino acid sequences were done using CLUSTAL W software (version 1.8). Both percent nucleotide identity and amino acid residue substitutions were studied on comparison with those different vip-like genes from various geographical regions. Phylogenetic analysis was performed using MEGA 4.1 (Beta 3) software. A rooted neighbour joining tree was constructed using the partial nucleotide and deduced amino acid sequences of the putative Bt isolates with other vip-like genes from different parts of the world. The other GenBank accession numbers used to construct tree AY074707, AY074708, AF399667, L48811, AY489126, AY743436, AY945939, L48812, EU294496, AJ971413, AF500478, DQ016969, EU107765, AY295778, DQ016968, AY547270, DQ241674, DQ250256, AY466020, AF399668, AY466018, AY466016, AY466015, AY466019, AY466017 and AF442384.
Nucleotide Sequence Accession Numbers
The sequence of the gene encoding the Vip3A protein has been deposited in the GenBank database (http://www.ncbi.nlm.nih.gov).
Results and Discussion
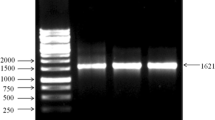
In contrast to insecticidal Cry proteins that have been extensively described and widely used in insect management programs for over 100 years [25], studies on the diversity and of Vip proteins are still in the early stage of discovery and application. Many instances of Bt with the cry gene have been reported, whereas only a few instances of Bt with vip genes had been found. The molecular and biological properties of VIPs established them as distinct class of insecticidal toxins that are different from Bt δ-endotoxin family. They represent second generation of insecticidal toxins that can be used to target important insect pests that are not susceptible to Bt δ-endotoxins [19]. To date, several PCR-based methods for cry gene identification have been developed [20–23], including specific primer PCR, multiplex PCR, exclusive PCR and PCR-RFLP. These methods directly detect known cry genes from B. thuringiensis [20, 21] as well as identify novel cry genes [23]. The NCBI accession numbers for the above sequences are EF679794 (IIHR-1), EF679795 (IIHR-2), EF679796 (IIHR-3), EF679797 (IIHR-4), EF679798 (IIHR-5), EF679800 (IIHR-7), EF679801 (IIHR-8), EF679802 (IIHR-9), EF679803 (IIHR-10), EF679805 (IIHR-12), EF679807 (IIHR-14) and EF679808 (IIHR-15). Sequence comparison of the above sequences with the reference L48811 showed that EF679803 had variations in 35 positions and EF679807 had a single nucleotide variation at 106th position (Fig. 1 and Supplementary data). Doss et al. [10] isolated vip3V gene employing primers used in this study, which corresponds to the coding region of the vip3Aa gene. Similarly by screening a large collection of Bacillus strains, vip3Ba1 identified using vip3A specific primers, which were initially designed to amplify the conserved regions in vip3A genes [14]. In this study, even though new gene was not detected it has yield vip3Aa and a related gene vip3Ae, potential of which could be studied after the isolation of full length of the same. The first report on the isolation of vip3A gene was by Estruch et al. [12, 13] and only a decade later vip3Ae gene was identified by Van Rie in the year 2005 (NCBI accession AJ872072) and Hernandez-Rodrignez et al. [16]. Recently, some of the vip3 genes have been employed to develop insect resistant transgenic plants. The number and diversity of vip genes available is abysmally low as compared to about 300 cry genes that have been cloned and characterized. Many of the known Vip toxins have insecticidal potential similar to that of Vip3Aa1. Therefore, enriching the diversity of available vip genes could broaden the spectrum of activity of the Vip3 family of proteins and facilitate the application for insect pest management [12]. Identification of new vip gene also increase the possibility of developing a broad spectrum vip genes, as it has been shown in the case of chimeric Vip3AcAa [17]. Hence, there is a potential of isolation and characterization of novel vip genes with improved toxicity and enhanced host spectrum. Further studies are needed to isolated full length of the different vip genes that are identified in this study in order to determine their toxicity and an effective tool for the management of Bt resistance as the mode of action of Vip proteins are found to be different to that of cry1Ab [18]. Finally, the results shows that the strategy used in this study can lead to the classification of known vip genes as well as the identification of novel vip genes from large scale of B. thuringiensis strains. The vip3Aa and vip3Ae gene proteins may be used to control insect pest or resistant insect pests by constructing genetically engineered strains or transgenic plants. The presence of vip3Aa1-insecticidal gene homologues in 15% of Bacillus strains analysed [11]. In a similar study, the presence of vip3Aa1-like genes in two out of 11 Bacillus thuringiensis kurstaki strains and absence in Bacillus thuringiensis israelensis strains [17]. Twenty-four serovars of Bacillus thuringiensis (Bt) were screened by polymerase chain reaction to detect the presence of vegetative insecticidal protein gene (vip)-like sequences by using vip3Aa1-specific primers. vip-like gene sequences were identified in eight serovars. These genes were cloned and sequenced. The deduced amino acid sequence of the vip3Aa14 gene from Bacillus thuringiensis tolworthi showed considerable differences as compared to those of Vips reported so far [32].
Positional differences in a nucleotide and b amino acids sequences of HD1 (vip3A) gene homologues and predicted Vip3A proteins in environmental isolates of Bacillus thuringiensis. The sequences of the vip3A homologues present in EF679794, EF679795, EF679796, EF679797, EF679798, EF679800, EF679801, EF679802, EF679803, EF679805, EF679807 and EF679808 and in Bacillus thuringiensis HD-1 in GenBank under accession numbers AF399667, respectively. The nucleotide and amino acid substitutions in the vip3A gene in those strains suggests the possibility that the observed mutations could be environmentally linked, though a much larger sampling size is required to support this hypothesis. The vip3A homologues of environmental isolates showed identities from 25 to 100% known vip3A genes at nucleotide and amino acid level
Analysis of insecticidal virulence factors in the Polish Bt collection, indicated that 57% of the vip3A-positive isolates (8 out of 14) also carried genes encoding insecticidal d-endotoxins potentially active against lepidopteran larvae, such as Cry1, Cry2 and/or Cry9. The same frequency of Bt isolates with both insecticidal toxins genes, vip3A and cry1I [24] in 125 strains surveyed. On one hand, these results may suggest a genetic linkage (e.g. carried by the same plasmid) between genes for insecticidal virulence factors, vip3A and cry1, in natural Bt collections. However, on the other hand, we cannot exclude the possibility that the co-occurrence of these genes in Bt is coincidental, due to a natural high frequency of cry1 than other genes encoding δ-endotoxins in Bt populations [2, 20, 26, 28]. Nevertheless, it is plausible that the co-occurrence of vip3A and cry toxin gene combinations significantly contributes to the successful occupation of new ecological niches and perhaps expanding the insecticidal host range and propagation of such Bt strains [27].
Phylogenetic analysis of this study showed that all vip gene isolates formed one distinct grouping (Fig. 2). Based on percent nucleotide identity of partial gene sequence and grouping in phylogenetic tree between vip-like genes, it can be inferred that the our isolates is closely related to its corresponding isolates suggested that comparison of gene sequence was responsible for geographic separation for divergence within vip-like genes/serotypes, consistent with the evaluation of distinct Bt isolates. Some isolates have evolved to be quite distinct and others remain members of closely related groups. These strains are candidates for application as commercialized biocontrol agent, therefore research on optimization of fermentation and formulation, greenhouse and field trials, and the bioassay of the selected isolates against other pests will be continued. Although clearly different, these genes will be submitted for consideration in the B. thuringiensis cry gene nomenclature only after cloning, full-length sequencing, and expression in a recombinant host strain to confirm the toxicity of the protein. Applying of molecular adaptation to nucleotide sequence data can help investigators identify specific amino acid substitutions for further experiments. In conclusions the identified, and characterized novel vip gene from Bt isolates, which can be used either alone or in gene pyramiding with other insecticidal protein genes for durable resistance against lepidopteran insect pests. Further studies on the mechanism of the Vip protein against its host may help us develop new effective insecticidal proteins and delay the insects’ resistance.
Neighbour-joining tree showing phylogenetic relationship amongst different Bacillus thuringiensis vip-like genes based on nucleotides. The NJ tree was constructed using CLUSTAL W with default parameters are indicated at the nodes. Sequences were obtained from the databases of the National Center for Biotechnology Information. The gene sequences were responsible for geographic separation for divergence within vip serotypes, consistent with the evaluation of distinct bacterial population. Despite the geographical distances, vip strains have originated from common ancestors. Some strains have evolved to be quite distinct and others remain as members of closely related groups
References
Asokan R, Puttaswamy (2007) Isolation and characterization of Bacillus thuringiensis Berliner from soil, leaf, seed dust and insect cadaver. J Biol Control 21:83–90
Ben-Dov E, Wang Q, Zaritsky A, Manasherob R, Barak Z, Schneider B, Khamraev A, Baizhanov M et al (1999) Multiplex PCR screening to detect cry9 in Bacillus thuringiensis strains. Appl Environ Microbiol 65:3714–3716
Bernhard K, Meadows M, Butt J, Ellis DJ, Roberts GM, Pauli S, Burges HD (1997) Natural isolates of Bacillus thuringiensis: worldwide distribution, characterization and activity against insect pests. J Invertebr Pathol 70:59–68
Bravo A, Sabaria S, Lopez L, Ontiveros H, Abarca C, Oritz A, Oritz M, Lina L, Villalobos JF, Pena G, Valdez NEM, Sobero M, Quintero R (1998) Characterisation of cry genes in Mexican Bacillus thuringiensis strain collection. Appl Environ Microbiol 64:4965–4972
Ceron J, Ortíz A, Quintero R, Guereca L, Bravo A (1995) Specific PCR primers directed to identify cryI and cryIII genes within a Bacillus thuringiensis strain collection. Appl Environ Microbiol 61:3826–3831
Cheryl EB, Leon Cour T, Annemie B, Roslyn M, Van Jeroen R, Raymond JA (2008) Unusually high frequency of genes encoding vegetative insecticidal proteins in an Australian Bacillus thuringiensis collection. Curr Microbiol 57:195–199
Christner BC, Thompson EM, Thompson LG, Reave JN (2003) Bacterial recovery from ancient glacial ice. Environ Microbiol 5:433–436
Crickmore N, Zeigler DR, Feitelson J, Schnepef E, Van Rie J, Lereclus D, Baum J, Dean DH (1998) Revision of the nomenclature for the Bacillus thuringiensis pesticidal crystal proteins. Microbiol Mol Biol Rev 62:813–870
Donovan WP, Donovan JC, Engleman JT (2001) Gene knockout demonstrates that vip3A contributes to the pathogenesis of Bacillus thuringiensis towards Agrotis ipsilon and Spodoptera exigua. J Invertebr Pathol 78(1):45–51
Doss VA, Kumar A, Jayakumar R, Sekar S (2004) Cloning and expression other insecticidal protein (vip3V) gene of Bacillus thuringiensis in Escherichia coli. Protein Exp Purif 26:82–88
Espinasse S, Chaufaux J, Buisson CH, Perchat S, Gohar M, Bourguet D, Sanchis V (2003) Occurrence and linkage between secreted insecticidal toxins in natural isolates of Bacillus thuringiensis. Curr Microbiol 47:501–507
Estruch JJ, Carozzi NB, Desai N, Duck NB, Warren GW, Koziel M (1997) Transgenic plants: an emerging approach to pest control. Nat Biotechnol 15:137–141
Estruch JJ, Warren GW, Mullins MA, Nye GJ, Craig JA, Koziel MG (1996) Vip3A, a novel Bacillus thuringiensis vegetative insecticidal protein with a wide spectrum of activities against lepidopteran insects. PNAS 93:5389–5395
Fang J, Xu X, Wang P, Zhao JZ, Shelton AM, Cheng J, Feng MG, Shen Z (2007) Characterization of chimeric Bacillus thuringiensis Vip3 toxins. Appl Environ Microbiol 73:956–961
Federici BA (2005) Insecticidal bacteria: an overwhelming success for invertebrate pathology. J Invertebr Pathol 89:30–38
Hernandez-Rodrignez CS, Boets A, Van Rie J, Ferrie J (2009) Screening and identification of vip genes in Bacillus thuringiensis strains. J Appl Microbiol 107(1):219–225
Ichimatsu T, Mizuki E, Nishimura K, Akao T, Asitoh H, Higuchi K, Ohba M (1998) Occurrence of Bacillus thuringiensis in fresh waters of Japan. Curr Microbiol 40:217–220
Juarez-Perez VM, Ferrandis MD, Frutos R (1997) PCR-based approach for detection of novel Bacillus thuringiensis cry genes. Appl Environ Microbiol 63:2997–3002
Jung YC, Kim US, Bok HS, Park YH, Opte CJ, Chung SY (1998) Characterization of Bacillus thuringiensis mutants and natural isolates by molecular methods. Can J Microbiol 44:403–410
Kaelin P, Gadani F (2000) Occurrence of Bacillus thuringiensis of cured tobacco leaves. Curr Microbiol 40:205–209
Kalman S, Kichne KL, Cooper N, Reynose MS, Yamamoto T (1995) Enhanced production of insecticidal proteins in Bacillus thuringiensis strains carrying an additional crystal protein gene in their chromosomes. Appl Environ Microbiol 61:3063–3068
Kati H, Sezen K, Demirbag Z (2007) Characterization of a highly pathogenic Bacillus thuringiensis strain isolated from common Cockchafer, Melolontha melolontha. Folia Microbiol 52:146–152
Lee MK, Walters FS, Hart H, Palekar N, Chen JS (2003) The mode of action of Bacillus thuringiensis vegetative insecticidal crystal protein Vip3A differs from that of Cry1Ab delta endotoxin. Appl Environ Microbiol 69:4648–4657
Maeda M, Mizuki E, Nakamura Y, Hatano T, Ohba M (2000) Recovery of Bacillus thuringiensis from marine sediments of Japan. Curr Microbiol 40:418–422
Martínez C, Caballero P (2002) Content of cry genes and insecticidal toxicity of Bacillus thuringiensis strains from terrestrial and aquatic habitats. J Appl Microbiol 92:745–752
Rang C, Gill P, Neisner N, Van Rie J, Frutos R (2005) Novel Vip3-related protein from Bacillus thuringiensis. Appl Environ Microbiol 71:6276–6281
Ritu B, Monika D, Siva KP, Borra J, Ajin DM, Singh AK, Polumetla AK (2005) Isolation, characterization and expression of a novel vegetative insecticidal protein gene of Bacillus thuringiensis. FEMS Microbiol Lett 243:467–472
Song F, Zhang J, Gu A, Wu Y, Han L, He K, Chen Z, Yao J et al (2003) Identification of cry1I-type genes from Bacillus thuringiensis strains and characterization of a novel cry1I-type gene. Appl Environ Microbiol 69:5207–5211
Swiecicka I (2008) Natural occurrence of Bacillus thuringiensis and Bacillus cereus in eukaryotic organisms: a case of symbiosis. Biocontrol Sci Technol 18:221–239
Swiecicka I, Mahillon J (2005) The clonal structure of Bacillus thuringiensis isolates from north-east Poland does not correlate with their cry gene diversity. Environ Microbiol 7:34–39
Wu JY, Zhao FQ, Bai J, Deng G, Qin S, Bao QY (2007) Evidence for positive darwinian selection of vip gene in Bacillus thuringiensis. J Genet Genomics 34:649–660
Yu CG, Mullins MA, Warren GW, Koziel MG, Estruch JJ (1997) The Bacillus thuringiensis vegetative insecticidal protein (vip) 3A lyses midgut epithelial cells of susceptible insects. Appl Environ Microbiol 63:532–536
Acknowledgments
Infrastructure facility and encouragement by The Director, Indian Institute of Horticultural Research (IIHR) is duly acknowledged. The authors are grateful to ICAR, New Delhi for funding this study under Network project on Application of Microbes in Agriculture and Allied Sectors (AMAAS).
Author information
Authors and Affiliations
Corresponding authors
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Asokan, R., Swamy, H.M.M. & Arora, D.K. Screening, Diversity and Partial Sequence Comparison of Vegetative Insecticidal Protein (vip3A) Genes in the Local Isolates of Bacillus thuringiensis Berliner. Curr Microbiol 64, 365–370 (2012). https://doi.org/10.1007/s00284-011-0078-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-011-0078-z