Abstract
To study the prevalence and isoforms of the pathogenicity island ETT2 among pathogenic Escherichia coli, as well as to determine the relationship between the ETT2 locus and other virulence factors, PCR amplifications target to the 35 ETT2-associated genes were established and used to investigate the presence of the ETT2 locus in 168 E. coli isolates from weaned piglets with edema and/or diarrhea or dairy cows with mastitis. The results showed that the ETT2 locus could be identified in the pathogenic E. coli isolates from colibacillosis in pigs and in the ones from mastitis in cows, but the presence of ETT2 among the isolates of porcine origin were significantly higher (85.87%) than that (47.37%) of bovine origin. Furthermore, 11 ETT2 isoforms were found in this research, including an intact form and 10 deletion types. The intact ETT2 was the prevalent form among the pathogenic E. coli isolates of porcine origin, and highly associated with the presence of shigatoxin type 2e (Stx2e), while the great majority isolates of bovine origin just carried various deletion types, and no distinct association with other virulence factors, e.g., the presence/absence of LT1, ST2, Cnf2, Tra, HPI, Hly, and F17a fimbriae.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Although toxin(s) and adhesin(s) are the predominant virulence factors [2–4, 6, 11, 16, 17], pathogenic Escherichia coli of animal origin always contain several mobile genetic elements such as plasmids, pathogenicity islands (PAI), or bacteriophages encoding a network of various virulence traits [14]. Thereinto, PAIs are a distinct class of genomic islands, which are acquired by horizontal gene transfer. These islands always encode accessory functions such as additional metabolic activities, antibiotic resistance, or properties involved in microbial pathogenesis, fitness, and/or symbiosis [8].
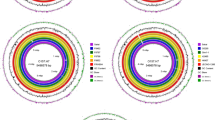
Among the PAIs, the E. coli type three secretion system 2 (ETT2) island, approximately 29.9 kb in size and localized adjacent to the tRNA locus glyU, was discovered through the analysis of genome sequences of enterohemorrhagic E. coli [10, 12–14]. As shown in Fig. 1 [15], there are at least 35 genes coding by the intact ETT2 locus, including yqe, yge, etr, epr, epa, eiv, etc., which were similar to the sequence of several Salmonella pathogenicity islands [1, 7, 9]. Except for the intact form, the ETT2 gene cluster was also found to be present in part in the E. coli strains [13, 14]. Compared to the parental E. coli strains, the ETT2 or eivA-deleted mutants exhibit defects in invasion and intracellular survival, suggesting its involvement in the pathogenesis of E. coli infection [18]. However, the ETT2-encoded proteins and their functions in pathogenesis or influenced on the virulence network of E. coli infection remain to be detailed defined as yet.
To investigate the prevalence and virulent association of the PAI ETT2 among E. coli isolates of different animal origins, in this study PCRs targeting 35 ETT2-associated genes were established and used to analyze the prevalence and isoforms of the ETT2 among 168 E. coli isolates from piglets with edema and/or diarrhea or dairy cows with mastitis. In addition, the association of the ETT2 with other virulence factors was also investigated.
Materials and Methods
Bacterial Isolates
Ninety-two E. coli isolates were isolated from weaned piglets with edema and/or diarrhea, including twenty-two enterotoxigenic E. coli (ETEC), forty-six shigatoxin-producing E. coli (STEC), and twenty-four STEC/ETEC [2, 3]. Seventy-six pathogenic E. coli isolates were from dairy cows with clinical or sub-clinical mastitis, which contain heat-labile toxin type 1 (LT1), heat-stable enterotoxin type 2 (ST2), cytotoxic necrotizing factor type 2 (Cnf2), transfer surface exclusion protein (Tra), hemolysin (Hly), F17a fimbriae, and high-pathogenicity island (HPI) [5]. Four ETT2+ (S443025, S451524, S452021, and S462234) and one ETT− (TG1) E. coli strains (Yangzhou University) were used as the positive and negative controls, respectively.
Primer Design for ETT2 Locus
Thirty-five sets of PCR primers (Table 1) were designed according to ETT2 ECs3703–ECs3737 gene sequences in GenBank using the PrimerSelect program in Lasergene software (DNASTAR, Inc), and synthesized by Sangon Biological Engineering Technology and Service Co. Ltd. (Shanghai, China). The two primers of each set were mixed together (each 50 mmol/l) before adding to the PCR mixture, respectively.
Bacterial Genomic DNA Extraction
Both the E. coli isolates and reference strains were grown overnight at 37°C on Luria–Bertani agar plates and single colonies were suspended in 50 μl deionized water. After boiling for 10 min, chilling on ice for 5 min and centrifugation at 10,000×g for 5 min, the supernatants were used as the DNA templates for PCR amplification.
PCR Detection of ETT2-Associated Genes in Pathogenic E. coli
Fifty microliter PCR reactions contained 5 μl of 10× PCR buffer (Mg2+ plus), 5 IU of Taq polymerase, 4 μl of dNTP mixture (2.5 mmol/l of each, TaKaRa), 1 μl of primer set, 2 μl of DNA template, and deionized water 37 μl. The 30-cycle PCRs were performed in 200 μl microcentrifuge tubes on the Applied Biosystems 2720 Thermal Cycler (America) using cycling parameters in Table 2. The PCR products were analyzed by 1% agarose gel electrophoresis along with DL2000 DNA markers.
Before amplification of ETT2-associated genes in 168 pathogenic E. coli isolates, the specificity of the PCR was tested using the genomic DNA template from ETT2+ (S443025, S451524, S452021, or S462234) or ETT− (TG1) E. coli as the positive or negative controls, respectively.
Sequence Analysis
To confirm the creditability of the PCR amplification, the PCR products of 35 ETT2-associated genes from the reference E. coli strain S443025 were subcloned into pGEM®-T vector (Promega) and transformed into E. coli DH5α, respectively. All the fragments in the recombinant plasmids were sequenced and compared with the published sequences (NC_002695).
Results
PCR Specificities for Amplification of ETT2-Associated Genes
The PCR was first performed using the DNA template from each of five reference E. coli strains. Agarose gel electrophoresis showed that 35 PCR products of expected sizes could be amplified from S443025 and S451524 strains, which harbor the intact ETT2 locus. For strain S452021 (with deletion from ECs3730 to ECs3735) or S462234 (with deletion from ECs3728 to ECs3735), 29 or 27 specific PCR products could be amplified. For TG1 strain (ETT2−), however, no single PCR product could be detected. To further evaluate the creditability of the PCR methods, the 35 DNA fragments of ETT2-associated genes of strain S443025 were sequenced, and the data showed that all the 35 PCR regions of ECs3703–ECs3737 genes are absolutely accorded with the expected size (Table 1) and share high homologies with the published sequence (NC_002695), respectively (Table 3).
Prevalence of ETT2 in Pathogenic E. coli Isolates
After 30 cycles of PCR amplification, agarose gel electrophoresis showed that the detection rate, including intact ETT2 or its deletion mutants, was 85.87% in the pathogenic E. coli isolates of porcine origin, which was significantly higher than that (47.37%) in the E. coli isolates of bovine origin (Table 4). Among different pathotypes of E. coli of porcine origin, prevalence of the ETT2 was 77.27, 89.13, and 83.33% in ETEC, STEC, and STEC/ETEC isolates, respectively.
Isoforms of ETT2 in Pathogenic E. coli Isolates
The gene-specific PCR amplification showed that the ETT2 in 168 E. coli isolates could be divided into 11 different isoforms, including the intact type containing the whole 35 genes (designated as type A) and 10 gene deletion mutants (designated as types B, C, D, E, F, G, H, I, J, and K). The schematic structures of 11 ETT2 genotypes are shown in Fig. 2. Type A was the complete form of ETT2 island with 35 genes; types B, C, D, E, and F were absence of 6, 7, 7, 8, and 9 genes in the ETT2 locus right (Ecs3737) regions, respectively; type G was absence of 8 genes in the right and 8 genes in the left (Ecs3703) region; type H was absence of 3 genes in the right, 8 genes in the middle, and 8 genes in the left region; form I was absence of 3 genes in the right and 13 genes in the middle region; and type J was absence of 3 genes in the right, 2 genes in the middle, and 8 genes in the left region; while type K was lost the great majority of genes of the locus.
For the pathogenic E. coli isolates from pig samples, five different ETT2 types could be detected, among which the types A (49.37%), B (12.66%), E (30.38%), F (5.06%), and G (2.53) were more prevalent form. For ETEC isolates, five different ETT2 types could be detected, among which the type A (15.79%), B (5.26%), E (47.37%), F (21.05%), and G (10.53%) were more prevalent. For STEC isolates, only two ETT2 types were detected, including the genotypes A (82.50%) and E (17.50%). For STEC/ETEC isolates, three different ETT2 types could be detected, including types A (15.00%), B (45.00%), and E (40.00%). For the pathogenic E. coli isolates from cow samples, eight ETT2 types (B, C, D, E, H, I, J, and K) were found and six of them (C, D, H, I, J, and K) were not found in the E. coli isolates of porcine origin. Among these the type D was the most prevalent (63.89%) and only two isolates contained types B or E, while there is no single isolates carried types A, F, and G.
Association of ETT2 with Other Virulence Factors
The virulence properties of the 168 bacteria under investigation are summarized in Table 4. Among the 92 E. coli isolates of porcine origin, 92.31% ETT2 type A isolates were Stx2e-positive, 100 or 90.00% type B isolates were ST1- or Stx2e-positive, and 60.87, 43.48, or 21.74% type E isolates were Stx2e-, ST2- or ST1-positive. Among the 76 E. coli of bovine origin, however, no distinct association was found between the ETT2 and other virulence factors, including LT1, ST2, Cnf2, Tra, HPI, Hly, and F17a.
Discussions
E. coli strains often acquire new complex pathogenic phenotypes by the acquisition of pathogenicity islands, which contain virulence genes clustered on the chromosome and which are acquired en bloc by horizontal gene transfer [8]. The type III secretion system (T3SS) is an important virulence factor used by several gram-negative bacteria to deliver effector proteins which subvert host cellular processes [10, 13]. ETT2 is an additional T3SS and has been found in many E. coli strains [10, 12–14], but its in vivo role is not known.
To investigate the prevalence of the ETT2 in pathogenic E. coli isolates of different animal origins, the PCR targeting to 35 individual genes of the ETT2 locus were established and were submitted to detection of the presence of ETT2 in 168 E. coli isolates. The experimental data suggest that the ETT2, including the intact form or 10 different gene deletion mutants, was significantly more prevalent in 92 E. coli isolates from weaned piglets with edema and/or diarrhea than that in 76 E. coli isolates from cows with mastitis. In addition, the intact ETT2 was the dominant genotype in the pathogenic E. coli isolates of porcine origin with a closer association with shigatoxin type 2e, while the deletion mutants were more prevalent in the E. coli isolates of bovine origin without distinct association with other virulence factors. Although the detailed reason(s) remains to be defined, it is possible that such difference could be due to different tissue origins (intestine vs. mammary gland) for bacterial isolation. This claim remains to be verified by further investigation.
Among 10 ETT2 deletion mutants, most of them had 6–9 gene deletions in their right regions, which encode the proteins responsible for bacterial invasion and/or intracellular survival [18]. These data indicate that ETT2 is a pathogenicity island not only for E. coli isolates of human origin [18], but also for those of animal origins, which will be further investigated in our future studies using gene deletion technique.
References
Blanc-Potard AB, Solomon F, Kayser J, Groisman EA (1999) The SPI-3 pathogenicity island of Salmonella enterica. J Bacteriol 1999(181):998–1004
Cheng D, Sun H, Xu J, Gao S (2005) Prevalence of fimbial colonization factors F18ab and F18ac in Escherichia coli isolates from weaned piglets with edema and/or diarrhea in China. Vet Microbiol 110:35–39
Cheng D, Sun H, Xu J, Gao S (2006) PCR detection of virulence factor genes in Escherichia coli isolates from weaned piglets with edema disease and/or diarrhea in China. Vet Microbiol 115:320–328
Cheng DR, Zhu SY, Ding WW, Chen XL, Gao XP, Sun HC (2010) Evaluation of the efficacy of maternal vaccination by using dominant pathogenic Escherichia coli isolates to control neonatal diarrhea in individual swine farm. Pure Appl Microbiol 4:487–495
Cheng DR, Zhu SY, Yin ZH, Ding WW, Mu ZX, Su ZR, Sun HC (2010) Prevalence of bacterial infection responsible for bovine mastitis. Afr J Microbiol Res 4:1110–1116
Fekete PZ, Gerardin J, Jacquemin E, Mainil JG, Nagy B (2002) Replicon typing of F18 fimbriae encoding plasmids of enterotoxigenic and verotoxigenic Escherichia coli strains from porcine postweaning diarrhoea and oedema disease. Vet Microbiol 85:275–284
Galán JE (1996) Molecular genetic bases of Salmonella entry into host cells. Mol Microbiol 20:263–271
Hacker J, Kaper JB (2000) Pathogenicity islands and the evolution of microbes. Annu Rev Microbiol 54:641–679
Hansen-Wester I, Hensel M (2001) Salmonella pathogenicity islands encoding type III secretion systems. Microbes Infect 3:549–559
Hayashi T, Makino K, Ohnishi M, Kurokawa K, Ishii K, Yokoyama K, Han CG, Ohtsubo E, Nakayama K, Murata T, Tanaka M, Tobe T, Iida T, Takami H, Honda T, Sasakawa C, Ogasawara N, Yasunaga T, Kuhara S, Shiba T, Hattori M, Shinagawa H (2001) Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res 8:11–22
Nagy B, Fekete PZ (1999) Enterotoxigenic Escherichia coli (ETEC) in farm animals. Vet Res 30:259–284
Osawa K, Shibata M, Nishiyama Y, Kurokawa M, Yamamoto G, Kinoshita S, Kataoka N (2006) Identification of the ETT2 locus in human diarrheagenic Escherichia coli by multiplex PCR. J Infect Chemother 12:157–159
Perna NT, Plunkett G III, Burland V, Mau B, Glasner JD, Rose DJ, Mayhew GF, Evans PS, Gregor J, Kirkpatrick HA, Pósfai G, Hackett J, Klink S, Boutin A, Shao Y, Miller L, Grotbeck EJ, Davis NW, Lim A, Dimalanta ET, Potamousis KD, Apodaca J, Anantharaman TS, Lin J, Yen G, Schwartz DC, Welch RA, Blattner FR (2001) Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529–533
Prager R, Bauerfeind R, Tietze E, Behrend J, Fruth A, Tschäpe H (2004) Prevalence and deletion types of the pathogenicity island ETT2 among Escherichia coli strains from oedema disease and colibacillosis in pigs. Vet Microbiol 99:287–294
Ren CP, Chaudhuri RR, Fivian A, Bailey CM, Antonio M, Barnes WM, Pallen MJ (2004) The ETT2 gene cluster, encoding a second type III secretion system from Escherichia coli, is present in the majority of strains but has undergone widespread mutational attrition. J Bacteriol 186:3547–3560
Rippinger P, Bertschinger HU, Imberechts H, Nagy B, Sorg I, Stamm M, Wild P, Wittig W (1995) Designations F18ab and F18ac for the related fimbrial types F107, 2134P and 8813 of Escherichia coli isolated from porcine postweaning diarrhoea and from oedema disease. Vet Microbiol 45:281–295
Salajka E, Salajkova Z, Alexa P, Hornich M (1992) Colonization factor different from K88, K99, F41 and 987P in enterotoxigenic Escherichia coli strains isolated from postweaning diarrhoea in pigs. Vet Microbiol 32:163–175
Yao Y, Xie Y, Perace D, Zhong Y, Lu J, Tao J, Guo X, Kim KS (2009) The type III secretion system is involved in the invasion and intracellular survival of Escherichia coli K1 in human brain microvascular endothelial cells. FEMS Microbiol Lett 300:18–24
Acknowledgments
This study was supported by Jiangsu Province Science and Technology Support Program (Agriculture) (Grant No. BE2010381), Natural Science Foundation of Jiangsu Province (Grant No. BK2009738), and a Project Funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions. We would like to give our thanks to all the staff of the veterinary microbiology laboratory of Yangzhou University for their help with some experiments.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Cheng, D., Zhu, S., Su, Z. et al. Prevalence and Isoforms of the Pathogenicity Island ETT2 Among Escherichia coli Isolates from Colibacillosis in Pigs and Mastitis in Cows. Curr Microbiol 64, 43–49 (2012). https://doi.org/10.1007/s00284-011-0032-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-011-0032-0