Abstract
Xanthomonas campestris pv. campestris (Pammel) Dowson (Xcc) causing black rot of crucifers is a serious disease in India and causes >50% crop losses in favorable environmental conditions. Pathogenic variability of Xcc, X. oryzae pv. oryzae (Xoo), and X. axonopodis pv. citri (Xac) were tested on 19 cultivars of cruciferae including seven Brassica spp. viz., B. campestris, B. carinata, B. juncea, B. napus, B. nigra, B. oleracea and B. rapa, and Raphanus sativus for two consecutive years viz., 2007–2008 and 2008–2009 under field conditions at Indian Agricultural Research Institute, New Delhi. Xcc (22 strains) and other species of Xanthomonas (2 strains), they formed three distinct groups of pathogenic variability i.e., Group 1, 2, and 3 under 50% minimum similarity coefficient. All strains of Xcc clustered under Groupl except Xcc-C20. The strains of Xcc further clustered in 6 subgroups viz., A, B, C, D, E, and F based on diseases reaction on host. Genetic variability of 22 strains of Xcc was studied by using Rep-PCR (REP-, BOX- and ERIC-PCR) and 10 strains for hrp (hypersensitive reaction and pathogenecity) gene sequence analysis. Xcc strains comprised in cluster 1, Xac under cluster 2, while Xoo formed separate cluster 3 based on >50% similarity coefficient. Cluster 1 was further divided into 8 subgroups viz., A, B, C, D, E, F, G, and H at 75% similarity coefficient. The hrpF gene sequence analysis also showed distinctness of Xcc strains from other Xanthomonads. In this study, genetic and pathogenic variability in Indian strains of Xcc were established, which will be of immense use in the development of resistant genotypes against this bacterial pathogen.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Black rot of crucifers caused by Xanthomonas campestris pv. campestris (Pammel) Dowson (Xcc) is the most important disease, substantially damage the crops by 10–50% in favorable environmental conditions. The pathogen infects a large number of cruciferous plants, including agriculturally important crops like cole crops (broccoli, cabbage, cauliflower, and knol khol), turnip, radish, oliferous brassica crops, ornamental plants, and weeds [1–3]. India has a wide range of climates ranging from temperate to tropical, where different crucifer crops are grown all over the country. Among crucifers, especially cole crops are grown round the year for fresh vegetable consumption. Initially, the black rot disease was reported on cabbage from Bombay, Maharashtra in India and now, it occurs across the country including the states of West Bengal, Uttar Pradesh, Himachal Pradesh, Rajsthan, Delhi, and Meghalaya [2]. Based on the interaction of various strains of Xcc with different Brassica species, a total of seven pathogenic races of Xcc were established [1, 4–7]. Xanthomonas campestris possesses interspecies variations on the level of pathovars, haplotypes, serogroups, and races [1, 8].
Various molecular techniques viz., RFLP patterns, repetitive sequence based PCR (rep-PCR), 16S rRNA gene analysis, hrp (hypersensitive reaction and pathogenecity) gene, and amplified fragment length polymorphism have been used to study the genetic variability in various bacteria [8–12]. Although, the PCR-based DNA-fingerprinting is a fast, reliable, and comparatively low cost method to study genetic diversity of bacteria, its effectiveness depends on primers chosen for analysis and quality of DNA. There are many highly conserved, repetitive DNA sequences, present in the genomes of Gram-negative bacteria, and that can be used for study of genetic diversity of bacteria employing PCR with different primers homologous to repetitive sequences named as rep-PCR. Three families of repetitive sequences (Rep) including repetitive extragenic palindromic (REP) sequences [13], enterobacterial repetitive intergenic consensus (ERIC) sequences, and BOX element [14] have been identified. No comprehensive information on resistance against black rot is available in Indian commercial lines of Brassica. Thus, the pathogenic and genetic variability within the strains of X. campestris pv. campestris must be determined to design effective control strategies and to develop resistant cultivars.
Hence, the present study was undertaken to find out the genetic and pathogenic variability in Xcc population of the country.
Materials and Methods
Bacterial Strain
The strains Xcc, Xoo, and Xac used in the present study were routinely grown on YDC and nutrient sucrose agar medium at 28°C (Table 1). Bacterial cultures were stored in mineral water at 4°C and ambient temperature.
Pathogenic Variability
The pathogenic variability was studied during winter season (November to March) in two consecutive years viz., 2007–2008 and 2008–2009 at Division of Plant Pathology, Indian Agricultural Research Institute, New Delhi under field conditions by scoring disease reaction on the leaves of crucifers as given in Table 2. A total of 19 cultivars/accessions of crucifereae family, comprising 18 cultivars of 7 economically important Brassica species viz. B. campestris, B. carinata, B. juncea, B. napus, B. nigra, B. oleracea, and B. rapa and one cultivar of Raphanus sativus grown in India were included for pathogenic variability. Bacterial cultures were grown in nutrient sucrose broth medium, adjusted to 0.1 optical densities at 600 nm, diluted to contain 108 CFU/ml. Forty-eight hours old culture of 22 strains of Xcc, Xoo (Xoo-4), and Xac (Xcc-C63) were inoculated on 35 days old plants of turnip, radish, and oliferous Brassica spp. and 30 days after transplanting of cole crops. The plants were inoculated by clipping the secondary veins at the margins with small scissors dipped in the bacterial suspension. Bacterial cultures were inoculated at 10 points per leaf and the three youngest leaves were inoculated per plant with three replications. The total number of inoculated points and number of points showing disease symptoms were recorded and the percentage of infected points were calculated. The severity of symptoms were assessed on a six- point scale of 0–9 based on the relative lesion size as 0 = no symptoms, 1 = small necrosis or chlorosis surrounding the infection point, 3 = typical small V-shaped lesion with black veins, 5 = typical lesion half way to the middle vein, 7 = typical lesion progressing to the middle vein, and 9 = lesion reaching the middle vein. The inoculated plants were grouped into four categories based on disease scores and percentage of inoculated points showing symptoms as resistant, partial resistant, susceptible, and highly susceptible as described by Vicente et al. [7]. Pathogenic variability was calculated by giving 1 for partial resistance, susceptible and highly susceptible cultivars, and 0 for resistance cultivars and a dendogram was prepared using NTSYS software (version 2.02e) program.
Genomic DNA Extraction from Bacteria
The strains of X. campestris pv. campestris and other test bacteria X. oryzae pv. oryzae and X. axonopodis pv citri were grown in nutrient broth at 28°C for 24 h. Total DNA from bacteria was extracted with guanidium thiocyanate as described by Pitcher et al. [15]. Eluted DNA was diluted by adding 100 μl 1× TE buffer for the use in PCR and stored at −20°C.
Genomic Fingerprinting and Analysis
Twenty-two strains of Xcc and one strain each of Xoo and Xac were fingerprinted using primers corresponding to prokaryotic enterobacterial repetitive intergenic consensus (ERIC) sequences, the BOX A subunit of the BOX element and repetitive extragenic palindromic (REP) sequences. Two independent amplifications were performed using the conditions as described earlier [2, 16] in thermo cycler (BIO-RAD, C1000™ Thermal cycler). Amplified PCR products were separated by horizontal gel electrophoresis in 1.5% agarose gel at 100 V in 0.5× Tris–acetate–EDTA buffer (TAE: 40 mM Tris, 20 mM acetic acid and 1 mM EDTA, pH 8.3) for 6 h. One kb of DNA ladder was included to normalize the banding pattern of all the three methods of rep-PCR (ERIC, BOX, and REP-PCR) profiles. Gels were stained in dilute ethidium bromide (2 μ g/ml), and DNA was visualized under UV light and photographed using the gel documentation system (BIO-RAD, GEL DOC™ XR+ with image Lab™ software). The amplified DNA fragments were scored for each isolate as 1 for band present and 0 for band absent. A similarity matrix was generated from the pooled binary data using SIMQUAL module for the NTSYSpc 2.02e computer software. The similarity coefficient was used to derive similarity among isolates. The similarity matrix, thus, generated was used for cluster analysis by unweighted pair group method of arithmetic average (UPGMA) using sequential, agglomerative, hierarchical, nested clustering module of NTSYSpc. The output data were graphically presented as a phylogenetic tree.
Hrp Gene Based Marker
Primer pair DLH 120 forward 5′-CCGTAGCACTTAGTGCAATG-3′ and DLH 125 reverse: 5′-GCATTTCCATCGGTCACGATTG-3′ with a predicted PCR product size 619 bp amplified the 3′ end of hrpF were used to confirm strains of Xcc [17]. PCR assays were performed in a thermal cycler. The amplifications were carried out in a final volume of 25 μl containing 1.5 mM MgCl2, 200 μM dNTPs (Promega), 1× PCR buffer, 1 unit Taq Polymerase, 500 nM each primer, and 20 ng DNA template. Each PCR experiment included a control without DNA. Reactions were run for 40 cycles each consisting of 40 s at 95°C, 40 s at 63°C, 40 s at 72°C with initial denaturation of 3 min at 95°C, and final extension of 5 min at 72°C. A 15 μl aliquot of each amplified PCR product was separated on a 1.0% agarose gel in 0.5% TBE, 80 V for 1.5 h, stained with ethidium bromide and visualized under a gel documentation system (BIO-RAD, GEL DOC™ XR+ with image Lab™ software).
Partial hrpF Gene Sequence of X. campestris pv. campestris
PCR product of hrpF gene amplified at 619 bp of strains Xcc-C1, Xcc-C7, Xcc-C10, Xcc-C11, Xcc-C12, Xcc-C19, Xcc-C20, and Xcc-C22 were directly used for sequencing, whereas, strains Xcc-C4 and Xcc-C5 of the same gene fragment hrpF gene were cloned in T-DNA hanging site of the pGEMT vector using competent cells of E. coli DH 5α for transformation. Plasmid DNA was extracted and purified using PCR purification kit (Promega kit) according to the recommendations of the manufacturer. Sequencing was performed by Applied Biosystem Machine-3130, Chromas Biotech, Bangalore, India. The evolutionary history was inferred using the neighbor-joining method. The optimal tree with the sum of branch length = 5.228 is shown. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (500 replicates) is shown above the branches [18]. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Maximum Composite Likelihood method [19] and are in the units of the number of base substitutions per site. Codon positions included were 1st + 2nd + 3rd + Noncoding. All positions containing gaps and missing data were eliminated from the dataset (Complete deletion option). There were total 388 positions in the final dataset. Phylogenetic analyses were conducted in MEGA4 [20].
Results
Natural occurrence of black rot disease was observed at research farms and farmer’s field. The isolates of Xcc were collected from five states viz., Delhi, Uttar Pradesh, Karnataka, Meghalaya, and Jharkhand belonging to different agro-climatic regions of India ranging from tropical to sub-tropical conditions. As such, 22 strains of Xcc were isolated from four cultivated species of Brassica viz. B. oleracea, B. juncea, B. rapa, and B. campestris and one species of Raphanus (R. sativus) at all the stages of plants starting from nursery seedling to seed producing plants (near to maturity) in 2006–2008 are presented in Table 1. All the isolates from cruciferous hosts were pathogenic to their respective hosts, and a visible symptom of the disease initiated after 6 days of inoculation in susceptible host by producing typical symptoms of black rot disease as V-shaped, yellow lesions with black veins and usually with necrotic centre. These isolates were considered to be Xcc on the basis of morphological and biochemical tests. Molecular marker based on hrpF gene sequence was used for further confirmation (Table 1) and it was found that all the Xcc strains collected from crucifers were amplified at 619 bp, whereas strains of Xoo and Xac were not amplified.
Pathogenic Variability
Xanthomonas campestris pv. campestris strains were inoculated on 19 cultivars of crucifereae family comprising 18 Brassica species viz., Brassica campestris, B. carinata, B. juncea, B. napus, B. nigra, B. oleracea, and B. rapa and one of R. sativus (Table 2) and they showed pathogenic variability to cause black rot disease. Among the Xcc strains, Xcc-C7 and Xcc-C19 were found highly virulent and 16 cultivars showed susceptible reaction, whereas Xcc-C20 strain was least virulent and infected only 5 cultivars of three Brassica spp. viz., B. campestris, B. carinata, and B. rapa. Among the species, B. campestris showed susceptibility to most of the strains of Xcc followed by B. oleracea . Wide range of pathogenic variability was found in Xcc and they were arbitrarily categorized into three distinct groups viz., Group 1, 2, and 3 at 50% minimum similarity coefficient. Out of 22 strains of Xcc, 21 strains were clustered in Group 1 except Xcc-C20, which was found least virulent and infected only five cultivars (Table 2). At 70% similarity coefficient, Group 1 further distinctly divided into five subgroups viz., A, B, C, D, and E; Group 2 having one subgroup F, whereas Group 3 has distinct subgroup G consisting of Xoo and Xac strains, which were non pathogenic to crucifers (Fig. 1), hence, the both strains fall under same cluster. The strains of Xcc, isolated from B. oleracea (cole crops) including cauliflower, cabbage, broccoli, and knol khol and R. sativus belonged to cluster subgroup A at 75% similarity coefficient. Further, strains Xcc-C10 and Xcc-C11 isolated from V-shaped symptoms on radish gave the similar type of disease reaction in all the cole crops. Xcc-C18 and Xcc-C19 isolated from B. rapa make a cluster of subgroup E with other strains of Xcc isolated from B. campestris (Xcc-C21) and B. juncea (Xcc-C17). Based on these grouping, 22 strains of Xcc were divided into six subgroups as Group A, B, C, D, E, and F (Table 2), Group A contained 13 strains of Xcc which were isolated from B. olearacea (cauliflower, cabbage, broccoli, and knol khol) and R. sativus. Group B and C contained Xcc-C12 and Xcc-C13, were isolated in 2006 and 2007 from the B. juncea host were formed different groups as B and C, respectively. The B. oleracea (cole crops), B. campestris, and B. rapa cultivars under study showed susceptible reaction against Group A strains of Xcc, while cultivars of B. juncea, B. nigra, and B. carinata showed resistant reaction. B. juncea (Pusa Jagannath) and B. carinata (Pusa Aditya) were found resistant against Xcc but knocked down only by 4 and 8 isolates, respectively, out of 22 strains of Xcc. B. oleracea cultivars showed resistant reaction against three strains of Xcc viz., Xcc-C17, Xcc-C18, and Xcc-C20 isolated from B. juncea, B. rapa and B. campestris, respectively. Strains of Xoo and Xac were tested on all the cultivars of crucifers and did not show symptoms of the disease.
Cluster analysis of pathogenic variability of 22 strains of Xanthomonas campestris pv. campestris and one strain each of X. oryxae pv. oryzae and X. axonopodis pv. citri based on pathogenicity reaction. The phylogenic tree was constructed by the UPGMA methods to the similarity matrix generated using Pearson’s correlation coefficient applied to the whole patterns. The minimum similarity coefficient of all isolates was used to define distinct groups, which are labeled numerically. Distinct group of pathogenic variability are labeled alphabetically
Genetic Variability
Data on DNA fingerprinting were generated from genomic DNA extracted from all the 22 strains of Xcc and one each of Xoo and Xac. DNA fragments of 300 to 10 kb were amplified and revealed a high level of genetic diversity among strains of Xcc. Maximum number of amplicons were found in ERIC-PCR (33 amplicons) followed by 30 amplicons in REP and 29 in BOX-PCR (Table 3). Amplification in each strain of Xanthomonas varied in all three methods of PCR as amplicons 13–21 in REP-PCR, 13–24 in ERIC, and 15–22 in BOX-PCR. In REP-PCR, maximum 21 fragments of amplicons were found in the Xcc-C10 isolated from host R. sativus from Ranchi (Jharkhand); in ERIC-PCR, 24 fragments of amplicons in Xcc-C16 isolated from B. juncea (Pusa Sag) from Delhi, while in BOX-PCR, 21 fragments of amplicons were found in four strains viz., Xcc-C10, Xcc-C13, Xcc-C19, and Xcc-C63 isolated from R. sativa, B. juncea (Indian mustard), B. rapa and citrus (lemon), respectively. Based on qualitative differences implication profiles and more 50% similarity coefficient, the strains were distinctly divided into three major groups, which separated Xanthomonas species as Group 1 comprises Xcc, Xac under Group 2 while Xoo under Group 3. Further cluster Group 1 contained all the 22 strains of Xcc and these strains were further clustered into 8 subgroups designating as A, B, C, D, E, F, G, and H at 75% similarity coefficient. It was interestingly noted that the strains of Xcc collected from cole crops (B. oleracea) were clustered into subgroup A. However, Xcc-C17 and Xcc-C18 collected from B. juncea (leafy mustard, Pusa Sag) and turnip (B. rapa) clustered subgroup B (Fig. 2), while Xcc-C19, Xcc-C22, Xcc-C10, Xcc-C12, Xcc-C16 of Xcc strains made distinct cluster subgroups C, D, E, G, and H, respectively. Xanthomonas species were clearly separated from each other like Xcc has distinct group from Xoo and Xac by using rep-PCR.
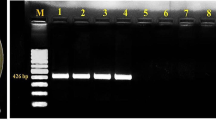
a Cluster analysis of Xanthmonas campestris pv. campestris: unweighted paired group mathematical average (UPGMA) dendrograms were generated using Pearson’s correlation coefficient. The significant of each branch indicated by the bootstrap percentage calculated for 1,000 subsets (only values greater than 50% are shown). The minimum similarity coefficient of all strains of X. campestris pv. campestris, X. oryzae pv. oryzae (Xoo-4) and X. axonopodis pv. citri (Xac-C63) was used to define distinct groups which are labeled numerically. Distinct groups of band based genotypes are labeled alphabetically. b Rep-PCR analysis of Xanthomonads, M 1 kb DNA ladder, 1 Xcc-C1, 2 Xcc-C2, 3 Xcc-C3, 4 Xcc-C4, 5 Xcc-C5, 6 Xcc-C6, 7 Xcc-C7, 8 Xcc-C8, 9 Xcc-C9, 10 Xcc-C10, 11 Xcc-C11, 12 Xcc-C12, 13 Xcc-C13, 14 Xcc-C14, 15 Xcc-C15, 16 Xcc-C17, 18 Xcc-C18, 19 Xcc-C19, 20 Xcc-C20, 21 Xcc-C21, 22 Xcc-C22, 23 Xoo-4, 24 Xac-C63
Specific primers were used from hrpF gene as earlier published by Berg et al. [17]. The PCR with the primers resulted in a single specific 619 bp band in the strains of Xcc. This band did not appear in the other Xanthomonas species (not shown) or strains of common plant associated bacteria, including Ralstonia solanacearum, Erwinia carotovora subsp. carotovora. The PCR fragment DLH120 and DLH125 amplified at 619 bp found nearly all 22 tested strains of Xcc and based on host and geographical distribution, 10 strains of these viz., Xcc-C1, Xcc-C4, Xcc-C5, Xcc-C7, Xcc-C10, Xcc-C11, Xcc-C12, Xcc-C19, Xcc-C20, Xcc-C22 were cloned, sequenced, and data were deposited to the NCBI Gene Bank under accession number HN176579, HN174139, HN174140, HN176583, HN176582, HN176576, HN176577, HN176580, HN176578, and HN176581, respectively. Phylogenetic sequevar analysis of hrpF gene demonstrated that Indian strains formed distinct cluster from the strains of Xcc from NCBI database of other countries. Out of 10 strains, 7 were clustered in Group 1 and 3 strains in Group 3. Highly similar sequence of Xanthomonas (Xcc strain B100, Xac strain 306, and X. fuscans subsp. fuscans) taken from NCBI gene bank were clustered in Group 2 and highly dissimilar sequence of Xanthomonas (X. axonopodis pv. glycinea strain 8ra and Xoo) and fungus Ajelodomyces dermatidis SLH14081 into Group 4 (Fig. 3). The strain Xcc-C1 isolated from cauliflower in Delhi, Xcc-C7 from cabbage in Bangalore (Karnataka), and Xcc-C20 from Indian mustard in Delhi fell into Group 3. In contrast, Xcc-C4 from cauliflower, Xcc-C5 from cabbage, Xcc-C11 from radish, Xcc-C19 from turnip, Xcc-C12 from Indian mustard in Delhi, Xcc-C10 from radish in Jharkhand, and Xcc-C22 from Indian mustard in Uttar Pradesh were clustered in Group 1 (Fig. 3).
Taxonomic analysis of partial hrp gene sequences of 10 strains of X. camapestris pv. campestris and six other strains of Xanthomonas and a fungus from database showing the resolution of distinct groups. The dendogram was generated by MEGA (version 4.0) software using the UPGMA mean applied to the distance matrix of nucleotide differences. Number at branch points indicates per cent bootstrap support for 500 iterations. The scale bar represents the average number of nucleotide substitutions among subgroup members at branch point
Discussion
The isolates of Xcc were collected from the different agro-climatic zones of India from various cultivars in cole crops (B. oleracea) like cauliflower, cabbage, broccoli and knol khol, turnip (B. rapa), radish (R. sativus), oliferous crops (B. campestris, and B. juncea). Three pathovars of Xanthomonas campestris viz., X. campestris. pv. campestris, pv. armoraceae and raphani infected the cruciferous plants and produce two distinguished types of symptoms commonly known as black rot and leaf spot. It is well established that the symptoms are dependent on environmental conditions, particularly host plants or specific gene [21]. In the present studies, only black rot producing strains of Xcc were considered to study genetic and pathogenic variability. However, the blight symptoms have also been reported and it was characterized by necrosis and sudden collapse of large areas of mesophyll in advanced stage causing blackening of veins [22]. They suggested that blight symptoms occur because of aggressive variants of Xcc. The initial investigations for resistance to Xcc were concentrated on B. oleracea, mainly because of the economic importance of this disease in cole crops (cauliflower, cabbage, Brussels sprouts, kale etc.). In recent studies, using defined races, race specific resistance in different accessions of B. oleracea, B. rapa and B. napus have been identified [23]. Jensen et al. [1] reported that races 4, 1, and 6 were the most common in cabbage in Nepal and blight like symptoms were frequently produced by strains of race 7 and by some strains of races 5 and 6 and also by a few strains of race 1 but no symptoms were produced by strain of race 4. In present case, all the strains of Xcc produced typical V-shaped symptoms of black rot disease. In present study, 19 cultivars of crucifers including seven species of Brassica (B. campestris, B. carinata, B. juncea, B. napus, B. nigra, B. oleracea and B. rapa) and R. sativus have been evaluated against Xcc, Xac, and Xoo. Screening was carried out in the field during winter seasons, where temperature ranged from 7.2 to 30.1°C and rainfall ranged from 0 to 6.5 mm (2008–2009). All tested cultivars of crucifers were challenged by using leaf clipping of young plants with concentrated suspension of Xcc, Xac, and Xoo to establish the extent and types of resistance in all tested species of Brassica and R. sativus, which clearly favors the pathogen and the rapid development of symptoms. The black rot symptoms appeared within six days in susceptible cultivars, while symptoms development was delayed >10 days in resistant cultivars. Based on the pathogenic reaction of the crucifer hosts, the strains of Xcc were grouped into 6 groups viz, Group A, B, C, D, E, and F. Group A having 13 strains of Xcc were collected from different agro-climatic conditions from cole crops and radish; while strains Xcc-C12 and Xcc-C13 from B. juncea host at Laxminagar, (Delhi) collected in two consecutive years 2006 and 2007 formed two different groups i.e., Group B and C, respectively. However, in this case, we could not identify the races of Xcc, but the pathogenic variability was recorded.
Several specific genome regions, including hrpF [2, 8, 10, 17] have been used as targets for diagnostics of Xanthomonas species. Among them hrp gene is too highly conserved to enable differentiation of the pathovars, primers targeting the 3′ end of hrpF successfully amplified a 619 bp product only from X. campestris. The product was neither amplified from extracts of other bacterial genera nor from other species of Xanthomonas like Xoo and Xac. Non pathogenic species of Xanthomonas from brassicas showed lack of hrpF gene, as indicated by the failure to amplify any portion of the gene using range of primer combinations [17]. The strains of Xcc are morphologically indistinguishable causing black rot and amplified at 619 bp, which further confirmed causal agent of black rot disease before going for pathogenic and genomic diversity analysis.
Genetic diversity of plant pathogenic bacteria has been studied by DNA based approaches to generate evidence of genome plasticity, ecological distribution, dispersal, and evolution. Knowledge of the existence of genetic variability in the pathogen population is important for plant breeding and consequently in crop improvement program. Crucifers especially cole crops, tubers, and oliferous crops belonging to Brassica and Raphanus species are affected by black rot incited by Xcc. This genus comprises 20 DNA homology groups, which were considered as genomic species. Sixteen of them consist of former X. campestris pathovars. The data obtained from pathogenicity and genetic variability analysis in this study confirms previous findings describing the heterogeneity within Xcc strains. Genetic diversity within other pathovars of Xanthomonads infecting rice [24], citrus [25] are also reported. No extensive analysis of genome of the strains belonging to Xcc from different geographical area of India has been performed earlier. For genomic variability, rep-PCR fingerprinting using primer sets (REP, ERIC and BOX) are highly conserved repetitive sequences [26] showed different banding pattern among Xcc isolates of different cruciferous hosts in India. The analysis clearly reveals potential of the Rep-PCR to cluster the strains of Xcc to their pathogenic behavior as indicated in Fig. 2. Specific fingerprinting patterns were linked to geographical distribution and also to their hosts. Genomic diversity of Xcc from Oklahoma, 45 local strains of Xcc belonged to a single BOX genotype similar to known crop strain, PHW117, representative for the haplotype 1 [27]. Group B was similar to the strain NCPPB 528T of the haplotype 3 [27] and five strains belonged to exotic groups C, D, and E from the United States and East Asia. Fingerprinting of Xcc was done since it is faster, cheaper and usually more discriminative than other techniques like PFGE and AFLP [12]. Among three Rep-PCR techniques, it was found that the ERIC primers are the most important for discriminating among Xcc strains. This primer also revealed a higher level of genetic diversity than that of REP and BOX primers in previous studies on Xcc [1] and X. axonopodis pvs. phaseoli and phaseoli var. fuscans [28]. This further confirms the utility of rep-PCR for differentiation of closely related strains of bacteria and the potential usefulness for studying bacteria evolution in specific ecological area. The unique fingerprinting profiles generated by using rep-PCR could a be useful tool in diagnosis and differentiation of strains, without a data base the utility of rep-PCR for routine identification of Xcc strains is limited. These limitations can be overcome by continuous improvement of techniques and creation of such databases as well as inclusion of pathogenicity testing, using universal cultivars, as an integrated part of strain identification.
The present findings establish the existence of strains of Xcc in India with genetic and pathogenic variability. Pathogenic variants in Xcc have also been reported in other countries by various workers [1, 6, 29]. However, we could not distinguish the Xcc variants in races because of non availability of standard differentials. The information generated on genetic and pathogenic variability of Indian strains of Xcc will provide a sound basic input for development of management strategies for black rot of crucifers vis-a-vis host resistance.
References
Jensen BD, Vicente JG, Manandhar HK, Roberts SJ (2010) Occurrence and diversity of Xanthomonas campestris pv. campestris in vegetable Brassica fields in Nepal. Plant Dis 94:298–305
Singh D, Dhar S (2011) Bio-PCR based diagnosis of Xanthomonas campestris pathovars in black rot infected leaves of crucifers. Indian Phytopathol 64(1):7–11
William PH (1980) Black rot: a continuing threat to world crucifers. Plant Dis 64:736–737
Ignatov A, Sechler A, Schuenzel EL, Agarkova I, Oliver B, Vidavar AK, Schaad NW (2007) Genetic diversity in populations of Xanthomonas campestris pv. campestris in cruciferous weeds in central coastal California. Phytopathology 97:803–812
Kamoun S, Kamda HV, Tola E, Kado CI (1992) Incompatible reactions between crucifers and Xanthomonas campestris involve a vascular hypersensitive response: role of hrpX locus. Mol Plant-Micro Inter 5:22–23
Taylor JD, Conway J, Roberts SJ, Astley D, Vicente JG (2002) Sources and origin of resistance to Xanthomonas campestris pv. campestris in Brassica genomes. Phytopathology 92:105–111
Vicente JG, Taylor JD, Sharpe AG, Parkin IAP, Lydiate DJ, King GJ (2002) Inheritance of race-specific resistance to Xanthomonas campestris pv. campestris in Brassica genomes. Phytopathology 92:1134–1141
Zaccardelli M, Francesco C, Annalisa S, Massimo M (2007) Detection and identification of the crucifer pathogen, Xanthomonas campestris pv. campestris, by PCR amplification of the conserved Hrp/type III secretion system gene hrc C. Euro J Plant Pathol 118:299–306
Goncalves ER, Rosato YB (2002) Phylogenetic analysis of Xanthomonas species based upon16S-23S rDNA intergenic spacer sequences. Int J Syst Evol Microbiol 52:355–361
Leite RP Jr, Minsavage GV, Bonas U, Stall RE (1994) Detection and identification of phytopathogenic Xanthomonas strains by amplification of DNA sequences related to the hrp genes of Xanthomonas campestris pv. vesicatoria. Appl Environ Microbiol 60:1068–1077
Louws FJ, Fulbright DW, Stephens CT, de Bruijn FJ (1994) Specific genomic fingerprinting of phytopathogenic Xanthomonas and Pseudomonas pathovars and strains generated with repetitive sequences and PCR. Appl Environ Microbiol 60:2286–2295
Valverde A, Hubert T, Stolov A, Dagar A, Kopelowitz J, Burdman S (2007) Assessment of genetic diversity of Xanthomonas campestris pv. campestris isolates from Israel by various DNA fingerprinting techniques. Plant Pathol 56:17–25
Higgins CF, Ames GFL, Barnes WM, Clement JM, Hofnung M (1992) A novel intercistronic regulatory element of prokaryotic operans. Nature (London) 298:760–762
Martin C, Briese T, Hakenbeck R (1992) Nucleotide sequences of genes encoding penicillin binding proteins from Streptococcus pneumonia and Streptococcus oralis with high homology to Escherichia coli penicillin binding proteins 1A and 1B. J Bacteriol 174:4517–4523
Pitcher DG, Saunelers NA, Queen RJ (1989) Rapid extraction of bacterial genomic DNA with guanidium thiocyanate. Lett App Microbiol 8:151–156
Schaad NW, Jones JB, Lacy GH (2001) Xanthomonas. In: Schaad NW, Jones JB, Chun W (eds) Laboratory guide for the identification of plant pathogenic bacteria. American Phytopathological Society, St Paul, pp 175–200
Berg T, Tesoriero L, Hailstones DL (2005) PCR-based detection of Xanthomonas campestris pathovars in Brassica seeds. Plant Pathol 54:416–427
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Tamura K, Nei M, Kumar S (2004) Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Nati Acad Sci USA 11030–11035
Ignatov A, Kuginuki Y, Hida K (1998) Race specific reaction of resistance to black rot in Brassica oleracea. Euro J Plant Pathol 104:821–827
Massomo SMS, Mabagala RB, Swai IS, Hockenhull J, Mortensen CN (2004) Evaluation of varietals resistance in cabbage against the black rot pathogen Xanthomonas campestris pv. campestris in Tanzania. Crop Prot 23:315–325
Ignatov A, Kuginuki Y, Hida K (2000) Distribution and inheritance of race-specific resistance to Xanthomonas campestris pv. campestris in Brassica rapa and B. napus. J Russian Phytopathol Soc 1:89–94
Leach JE, Rhoads ML, Vera Cruz CM, White FF, Mew TW, Leung H (1992) Assessment of genetic diversity and population structure of Xanthomonas oryzae pv. oryzae with repetitive DNA element. Appl Environ Microbiol 58:21–25
Jan LW, Rademaker JLW, Hoste B, Louws FJ, Karel K, Swings J, Vauterin L, Vauterin P, de Bruijn FJ (2000) Comparison of AFLP and rep-PCR genomic fingerprinting with DNA–DNA homology studies: Xanthomonas as a model system. Int J Syst Evol Microbiol 50:665–677
Versalovic J, Koeuth T, Lupski JR (1991) Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res 19:6823–6831
Tsygankova SV, Ignatov AN, Boulygina ES, Kuznetsov BB, Korotkov EV (2004) Genetic relationships among strains of Xanthomonas campestris pv. campestris revealed novel rep-PCR primers. Euro J Plant Pathol 110:845–853
Lopez R, Asensio C, Gilbertson RL (2006) Phenotypic and genetic diversity in strains of common blight bacteria (Xanthomonas campestris pv. phaseoli and X. campestris pv. phaseoli var. fuscans) in a secondary center of diversity of the common bean host suggests multiple introduction events. Phytopathology 96:1204–1213
Vicente JG, Conway J, Roberts SJ, Taylor JD (2001) Identification and origin of Xanthomonas campestris pv. campestris races and related pathovars. Phytopathology 91:492–499
Acknowledgments
Authors are thankful to Dr. R. K. Jain, Head, Division of Plant Pathology, IARI, New Delhi for financial and administrative support to conduct the experiments and also grateful to the Director, NBPGR, New Delhi and the Head, Division of Genetics, IARI, New Delhi for providing seed materials for this study.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Singh, D., Dhar, S. & Yadava, D.K. Genetic and Pathogenic Variability of Indian Strains of Xanthomonas campestris pv. campestris Causing Black Rot Disease in Crucifers. Curr Microbiol 63, 551 (2011). https://doi.org/10.1007/s00284-011-0024-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00284-011-0024-0