Abstract
LysGH15, a phage endolysin, exhibits a particularly broad lytic spectrum against Staphylococcus aureus, especially methicillin-resistant S. aureus (MRSA). Sequence analysis reveals that this endolysin contains a C-terminal cell wall binding domain (SH3b), which causes the endolysin to bind to host strains. In this study, the substrate binding affinity of the SH3b domain (LysGH15B) was evaluated. A fusion protein of LysGH15B and green fluorescent protein (LysGH15B–GFP) were cloned and expressed in Escherichia coli. Laser scanning confocal microscopy was used to detect the fluorescence of the treated cells irradiated at different excitation wavelengths and to determine the binding activity of LysGH15B–GFP and GFP. We found that LysGH15B–GFP not only generated green fluorescence, but, more importantly, also displayed specific affinity to staphylococcal isolates, especially MRSA. In contrast, the single GFP did not display any binding activity. The high affinity was attributed to the portion of LysGH15B and the binding activity of the fusion protein was specific to staphylococci. This study provides an insight into the SH3b domain of LysGH15. The specific binding activity may cause LysGH15B to serve as an anchoring device, and offer an alternative approach for cell surface attachment onto staphylococci.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Endolysins (also known as lysins) from bacteriophages are cell wall hydrolases synthesized at the end of the phage lytic life cycle that are involved in cell lysis and the release of the progeny particles from host cells [16, 22]. One common feature of lysin that infects gram-positive bacterial species is a modular structure, consisting of at least two separate functional domains: an N-terminal catalytic domain and a C-terminal cell wall binding domain [12].
We recently reported a novel staphylococcal phage lysin, LysGH15, which displayed a particularly broad host range and strong lytic activity against Staphylococcus aureus, especially methicillin-resistant S. aureus (MRSA) [6, 7]. Analysis of its amino acid sequence revealed that LysGH15 has a modular structure similar to those of LysK, phi11, MV-L, and LysH5, with two catalytic domains, specifically a CHAP (amidohydrolase/peptidase) domain and a central amidase-2 (N-acetylmuramoyl-l-alanine amidase) domain, as well as a C-terminal SH3b cell-binding domain (CBD) [14–16, 18].
Detailed studies of the CHAP domain of LysK have been conducted previously, and found that this domain sustains lytic activity against staphylococci [10]. However, to the best of our knowledge, there was only one cell wall-targeting (CWT) domain of staphylococcal phage lysin (ClyS) which has been used for binding to staphylococci via fluorescence tag [3, 17]. In this study, to evaluate the substrate binding affinity of the LysGH15B (the SH3b domain of LysGH15), we generated the fusion protein LysGH15B–GFP and determined its specific affinity to staphylococci.
Materials and Methods
Construction of Expression Vectors
LysGH15B (Fig. 1a) corresponds to the SH3b portion of LysGH15 (GenBank accession no. HM015284). To amplify the lysGH15B and GFP genes for cloning and plasmid constructions, the full genome of the phage GH15 and the pEGFP vector were, respectively, used as templates. The lysGH15B gene was PCR-amplified using SH3b-SalI (ACGCGTCGACAGGTGGAACTCCCTGACAAG) and SH3b-EcoRI (CGGAATTCTGGAAAAAGAACCAATACG) primers (enzyme sites are underlined), whereas the GFP gene was amplified using GFP-SalI (ACGCGTCGAC GGGGG TTCTGTGAGCAAGGGCGAGGAG) and GFP-HindIII (CCCAAGCTTGTACAGCTCGTCCA) primers (the bold type indicate the linkers). The PCR products were digested with the corresponding restriction enzymes and cloned into the EcoRI/SalI (lysGH15B) and SalI/HindIII (GFP) sites of a pET-28a (+) vector, generating the plasmid pET28a-LysGH15B-GFP. The LysGH15B–GFP gene was amplified using SH3b-XhoI (CCGCTCGAGTGGAAAAAGAACCAATACG) and GFP-BamHI (CGGGATCCCTACTTGTACAGCTCGTCCA) primers. This fragment was cloned into the XhoI/BamHI sites of the vector pET15b, generating the expression vector pET15b-LysGH15B-GFP. The GFP gene was also inserted into the same sites of pET15b using GFP-XhoI (CCGCTCGAG GGGGGTTCTGTGAGCAAGGGCGAGGAG) and GFP-BamHI primers.
Expression and Purification of LysGH15B–GFP and GFP
Exponentially growing cultures of Escherichia coli BL21 containing pET15b-LysGH15B-GFP or pET15b-GFP were induced by incubation with 1 mmol l−1 isopropyl-β-d-thiogalacto-pyranoside (IPTG) for 6 h at 25°C with shaking. Subsequently, bacterial cells were washed with 20 mmol l−1 sterile PBS and disrupted with an ultrasonic disintegrator. The resultant clear lysates were centrifuged at 10,000 rpm for 15 min at 4°C. For protein purification, the supernatant was dialyzed against PBS, added to (Ni–NTA, nickel matrix) His·Bind slurry, and eluted according to the manufacturer’s instructions (Merck-Novagen). The protein file was analyzed by SDS-polyacrylamide gel electrophoresis (SDS-PAGE).
Binding of LysGH15B–GFP to S. aureus
1-ml aliquot of stationary phase S. aureus culture (YB57, MRSA strain) was collected and washed twice using PBS. The cells were dyed with 20 μmol l−1 Hoechst No. 33342 fluorescent dyes for 10 min at 37°C, washed five times with PBS, and then resuspended in 100 μl of PBS. The cells were then incubated with either GFP or the LysGH15B–GFP protein for 15 min at 37°C. After incubation, the cells were collected by centrifugation at 8000 rpm for 5 min, washed five times with PBS, and resuspended in 100 μl of PBS. Laser scanning confocal microscopy (LSCM) was used to detect the fluorescence of the treated cells that were irradiated at different excitation wavelengths and to determine the binding activity of LysGH15B–GFP and GFP.
The Effectiveness of LysGH15B–GFP to Inhibit the Lytic Activity of LysGH15
Mid-log-phase cells of the YB57 strain were washed and resuspended in sterile PBS, and the cell concentration was modified to an OD600 of 1.2. The YB57 substrate was pre-incubated with LysGH15B–GFP (final concentration, 40 μg ml−1) for 15 min at 37°C, and the excess fusion was washed out with sterile PBS. The phage lysin LysGH15 (40 μg ml−1) was added to the bacterial suspension and incubated for 30 min at 37°C. The OD600 was again determined. As controls, the washed YB57 was treated with 40 μg ml−1 LysGH15, 40 μg ml−1 LysGH15B–GFP, or elution buffer alone.
Binding Specificity of LysGH15B–GFP
To determine the binding range of LysGH15B–GFP, 53 S. aureus strains [6] and 6 Staphylococcus epidermidis were treated with Hoechst No. 33342 fluorescent dye and LysGH15B–GFP, as described above. LSCM was used to observe the fluorescence of these strains after treatment with the fusion protein. Streptococcus, Bacillus subtilis, Salmonella enteritidis, Klebsiella pneumoniae, and E. coli were also included in experiments to determine the specificity of the binding activity of LysGH15B–GFP.
Results and Discussion
The Expression and Purification of LysGH15B–GFP
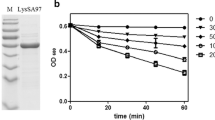
The fusion protein contained the His-tag, LysGH15B, linker, and GFP components in order from the N-terminal to the C-terminal (Fig. 1b). A His-tag was also located in the N-terminal region of single GFP. Full expression vectors, pET15b-GFP and pET15b-LysGH15B-GFP, were identified by PCR and sequence detection, and were introduced into competent E. coli BL21 cells. SDS-PAGE analysis revealed protein bands at about 28 and 38 kDa in the E. coli BL21 cells that contained pET15b-GFP and pET15b-LysGH15B-GFP (Fig. 2), respectively, after being induced by IPTG. The homogeneous band also emerged in the sample containing the purified His-tagged protein. The bands accorded with the predicted sizes of GFP and LysGH15B–GFP.
Protein profiles of N-terminal His·6-tagged LysGH15B–GFP and GFP extracts from expression bacterial strains, respectively. Every lane was added with 15 μl total volume. Lane 1 E. coli BL21 (Coden Plus) cells containing a pET15b-GFP construct induced with IPTG. Lane 2 the purified GFP fraction (about 28 kDa). Lane 3 pET15b-LysGH15B-GFP-containing E. coli BL21 (Coden Plus) cells induced with IPTG. Lane 4 the purified LysGH15B–GFP fraction (about 38 kDa). Lane M the molecular mass marker (kDa)
LysGH15B–GFP Binds to S. aureus Isolate
Blue fluorescence emitted by Hoechst No. 33342 fluorescent dyes was found near the center of the cells of the YB57 strain when examined at 405 nm wavelength, and green fluorescence emitted by GFP was observed around the LysGH15B–GFP-treated YB57 cells at 488 nm wavelength (Fig. 3). However, no green fluorescence was present around YB57 cells treated with GFP under the same conditions. These results demonstrate that LysGH15B–GFP binds to YB57 cells, and that the binding activity and green fluorescence of LysGH15B–GFP were due to the LysGH15B and GFP components, respectively.
Localization of LysGH15B–GFP on YB57. YB57 was dyed with a 20 μmol l−1 Hoechst No. 33342 fluorescent dye for 10 min at 37°C and incubated with LysGH15B–GFP for 15 min at 37°C. a Localization at 405 nm wavelength (blue fluorescence, emitted by Hoechst No. 33342 fluorescent dye). b Localization at 488 nm wavelength (green fluorescence, emitted by GFP). c Image of ordinary ray (normal light). d Overlay of the pictures shown in a, b, and c
Effectiveness of GH15B–GFP in Inhibiting the Lytic Activity of LysGH15
LysGH15 displayed a strong lytic activity toward the MRSA strain YB57; the OD600 of YB57 cultures decreased about 65% after exposure to LysGH15 (Fig. 4). However, the YB57 cultures treated with LysGH15B–GFP were insensitive to LysGH15; the decrease of OD600 only reached about 30%. As a control, we determined that LysGH15B–GFP alone had no effect on the OD600 of YB57 cultures, similar to buffer treatment. Hence, LysGH15B–GFP displayed competitive inhibition to the lytic activity of LysGH15, and indirectly indicated the binding activity of LysGH15B–GFP to YB57 cells.
Competitive inhibition of LysGH15B–GFP on the lytic activity of LysGH15. YB57, which had been treated with 40 μg ml−1 LysGH15B–GFP for 15 min, was washed by PBS and then exposed to 40 μg ml−1 LysGH15 for 30 min. As controls, YB57 was treated with 40 μg ml−1 LysGH15, 40 μg ml−1 LysGH15B–GFP, or elution buffer alone for 30 min. Values represent mean ± SD (n = 3)
Binding Specificity
The LysGH15B–GFP protein sufficiently bound to 43 S. aureus strains and 4 S. epidermidis strains. More importantly, the fusion protein bound to all 24 MRSA strains examined. In contrast, no green fluorescence was detected around Streptococcus, B. subtilis, S. enteritidis, K. pneumoniae, or E. coli strains after incubation with LysGH15B–GFP (data not shown). Therefore, the affinity of LysGH15B–GFP was apparently specific to staphylococci, at least based on these limited studies.
This study revealed that LysGH15B–GFP, which contains the SH3b domain of LysGH15, displayed a broad binding host range that is essentially consistent with the lytic spectra of LysGH15. Since, we have determined that the specific affinity of LysGH15B–GFP to staphylococci is attributed to its LysGH15B portion, it might be concluded that the lytic range of lysin is also decided by the SH3b portion [4, 5]. The ability of binding is closely related to the lytic activity of lysin [19]. Hence, the host range and lytic activity of lysin may be changed by modifying the structure of the SH3b domain.
Phages have previously been used to detect corresponding bacterial strains [1, 8, 9]. However, there are some limitations to the development of fluorescence-tagged bacteriophages, such as the environmental contamination of phages. Phage lysins are not subjected to this limitation and, more importantly, they manifest a broader host range than the whole virus [2, 6]. Thus, compared with phages, the SH3b domain of lysin might be a better detection tool. LysGH15B–GFP not only generates green fluorescence but also maintains specific binding activity toward staphylococci. In addition, it is relatively inexpensive and convenient for mass production by industrial fermentation. Therefore, LysGH15B–GFP possesses great potential as a detection tool of staphylococci.
Because, LysGH15B has been demonstrated to possess the ability to target the heterologous protein GFP on the surface of staphylococci, and because there is no mutual effect between LysGH15B and GFP, it might be possible to use LysGH15B as an anchoring motif for the display of other proteins, such as antigens or functional enzymes, on the surface of staphylococci [13, 20, 21]. This strategy has practical applications for the development of oral vaccines, biocatalysts, whole-cell absorbents, and other purposes [11]. In conclusion, LysGH15B represents a new type of anchoring device and offers an alternative approach for cell surface attachment onto staphylococci.
References
Balasubramanian S, Sorokulova IB, Vodyanoy VJ, Simonian AL (2007) Lytic phage as a specific and selective probe for detection of Staphylococcus aureus—a surface plasmon resonance spectroscopic study. Biosens Bioelectron 22:948–955
Borysowski J, Weber-Dabrowska B, Gorski A (2006) Bacteriophage endolysins as a novel class of antibacterial agents. Exp Biol Med (Maywood) 231:366–377
Daniel A, Euler C, Collin M, Chahales P, Gorelick KJ, Fischetti VA (2010) Synergism between a novel chimeric lysin and oxacillin protects against infection by methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 54:1603–1612
Fischetti VA (2005) Bacteriophage lytic enzymes: novel anti-infectives. Trends Microbiol 13:491–496
Garcia E, Garcia JL, Garcia P, Arraras A, Sanchez-Puelles JM, Lopez R (1988) Molecular evolution of lytic enzymes of Streptococcus pneumoniae and its bacteriophages. Proc Natl Acad Sci USA 85:914–918
Gu J, Xu W, Lei L, Huang J, Feng X, Sun C, Du C, Zuo J, Li Y, Du T, Li L, Han W (2011a) LysGH15, a novel bacteriophage lysin, protects a murine bacteremia model efficiently against lethal methicillin-resistant Staphylococcus aureus infection. J Clin Microbiol 49:111–117
Gu J, Zuo J, Lei L, Zhao H, Sun C, Feng X, Du C, Li X, Yang Y, Han W (2011b) LysGH15 reduces the inflammation caused by lethal methicillin-resistant Staphylococcus aureus infection in mice. Bioeng Bugs 2:96–99
Guntupalli R, Sorokulova I, Krumnow A, Pustovyy O, Olsen E, Vodyanoy V (2008) Real-time optical detection of methicillin-resistant Staphylococcus aureus using lytic phage probes. Biosens Bioelectron 24:151–154
Hennes KP, Suttle CA, Chan AM (1995) Fluorescently labeled virus probes show that natural virus populations can control the structure of marine microbial communities. Appl Environ Microbiol 61:3623–3627
Horgan M, O’Flynn G, Garry J, Cooney J, Coffey A, Fitzgerald GF, Ross RP, McAuliffe O (2009) Phage lysin LysK can be truncated to its CHAP domain and retain lytic activity against live antibiotic-resistant staphylococci. Appl Environ Microbiol 75:872–874
Hu S, Kong J, Kong W, Guo T, Ji M (2010) Characterization of a novel LysM domain from Lactobacillus fermentum bacteriophage endolysin and its use as an anchor to display heterologous proteins on the surfaces of lactic acid bacteria. Appl Environ Microbiol 76:2410–2418
Loessner MJ (2005) Bacteriophage endolysins—current state of research and applications. Curr Opin Microbiol 8:480–487
Marraffini LA, Dedent AC, Schneewind O (2006) Sortases and the art of anchoring proteins to the envelopes of gram-positive bacteria. Microbiol Mol Biol Rev 70:192–221
Navarre WW, Ton-That H, Faull KF, Schneewind O (1999) Multiple enzymatic activities of the murein hydrolase from staphylococcal phage phi11. Identification of a d-alanyl-glycine endopeptidase activity. J Biol Chem 274:15847–15856
O’Flaherty S, Coffey A, Meaney W, Fitzgerald GF, Ross RP (2005) The recombinant phage lysin LysK has a broad spectrum of lytic activity against clinically relevant staphylococci, including methicillin-resistant Staphylococcus aureus. J Bacteriol 187:7161–7164
Obeso JM, Martinez B, Rodriguez A, Garcia P (2008) Lytic activity of the recombinant staphylococcal bacteriophage PhiH5 endolysin active against Staphylococcus aureus in milk. Int J Food Microbiol 128:212–218
Pastagia M, Euler C, Chahales P, Fuentes-Duculan J, Krueger JG, Fischetti VA (2011) A novel chimeric lysin shows superiority to mupirocin for skin decolonization of methicillin-resistant and -sensitive Staphylococcus aureus strains. Antimicrob Agents Chemother 55:738–744
Rashel M, Uchiyama J, Ujihara T, Uehara Y, Kuramoto S, Sugihara S, Yagyu K, Muraoka A, Sugai M, Hiramatsu K, Honke K, Matsuzaki S (2007) Efficient elimination of multidrug-resistant Staphylococcus aureus by cloned lysin derived from bacteriophage phi MR11. J Infect Dis 196:1237–1247
Sass P, Bierbaum G (2007) Lytic activity of recombinant bacteriophage phi11 and phi12 endolysins on whole cells and biofilms of Staphylococcus aureus. Appl Environ Microbiol 73:347–352
Steen A, Buist G, Leenhouts KJ, El Khattabi M, Grijpstra F, Zomer AL, Venema G, Kuipers OP, Kok J (2003) Cell wall attachment of a widely distributed peptidoglycan binding domain is hindered by cell wall constituents. J Biol Chem 278:23874–23881
Steidler L, Viaene J, Fiers W, Remaut E (1998) Functional display of a heterologous protein on the surface of Lactococcus lactis by means of the cell wall anchor of Staphylococcus aureus protein A. Appl Environ Microbiol 64:342–345
Wang S, Kong J, Zhang X (2008) Identification and characterization of the two-component cell lysis cassette encoded by temperate bacteriophage phiPYB5 of Lactobacillus fermentum. J Appl Microbiol 105:1939–1944
Acknowledgment
The authors thank the National Natural Science Foundation of China (Key Program, No. 31130072) for the financial support of this investigation.
Author information
Authors and Affiliations
Corresponding author
Additional information
Jingmin Gu and Rong Lu have contributed equally to this study.
Rights and permissions
About this article
Cite this article
Gu, J., Lu, R., Liu, X. et al. LysGH15B, the SH3b Domain of Staphylococcal Phage Endolysin LysGH15, Retains High Affinity to Staphylococci. Curr Microbiol 63, 538 (2011). https://doi.org/10.1007/s00284-011-0018-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00284-011-0018-y