Abstract
Vinegar production is based on the acetification process by indigenous acetic acid bacteria (AAB). Among vinegar technologies, solid-state fermentation (SSF) processes are widespread in Asian countries to produce vinegar at small-scale. In this study, 21 AAB strains isolated from Chinese cereal vinegars produced by SSF collected in different regions of China were characterized by enterobacterial repetitive intergenic consensus (ERIC)–PCR fingerprinting. Isolates exhibited high degree of phenotypic variability as well as suitable traits for their uses as selected strains in SSF vinegar production (growth modality by superficial biofilm, no production of cellulose, ability to growth on ethanol media). 16S rRNA gene sequencing analysis of representative strains showed that strains of Acetobacter pasteurianus have a close association to cereal vinegars, whereas Gluconacetobacter europaeus population is not favoured. Selection of single or multiple strains culture within A. pasteurianus species was predicted in view of their application in SSF technology. This seems to be the first report showing phenotypic and genetic variability of AAB strains involved in SSF processes. Results can be exploited for the implementation of large-scale SSF processes by selected strains for vinegar production and other innovative biotechnological applications.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Recently, the interest on acetic acid bacteria (AAB) related to vinegar production has been increased and several works on the ecology and functionality of culturable AAB have been published. Gluconacetobacter europaeus, Acetobacter pasteurianus and Acetobacter aceti species were mainly detected in wine and other vinegars produced by static and submerged cultures [7]. In China, Japan and other Asian countries, an age-old and traditional process, known as solid-state fermentation (SSF), is largely used to produce vinegars from cereals at small-scale. The basic process steps of these kinds of vinegar are: (1) crushing and steaming of cereals; (2) addition of water and Qu or Koji (respectively in Chinese and Japanese) that is a specific cereal preparation containing moulds, yeasts and bacteria; (3) alcoholic fermentation; (4) acetic acid oxidation during which wheat bran and rice (or other cereals) hull are mixed with old Pei (acetic acid fermented product from last batch as seed vinegar) [2]. Recently, SSF processes have attracted interest due to its potential not only in vinegar field, but also for the production of food and pharmaceuticals [11]. However, to date, no large-scale application of SSF is achieved mainly due to limited microbiologic knowledge about the ecology, physiology and function of the involved AAB as well as about the genetic variability among strains. Among DNA typing methods, enterobacterial repetitive intergenic consensus–PCR (ERIC/PCR) is based on the occurrence of repetitive element containing a highly conserved central inverted repeat located in non-coding extragenic regions of chromosome. The robustness of ERIC/PCR to discriminate bacterial strains under species level generating highly specific genomic DNA fingerprinting has been extensively stated for individual Eubacteria strains [19]. This technique has been also used to type AAB strains from rice and wine vinegar [6, 9, 14].
The present study was designed to collect representative AAB strains of cereal vinegars produced in SSF and to investigate suitable traits, which can be used as basic information to select strains for starter culture implementation.
Materials and Methods
Samples and Bacterial Strains
Three kinds of vinegar Pei (semi-finished acetic acid fermented product in SSF) samples, labelled as DL, SX and ZJ, were collected from Tianjin Duliu Old Aged Vinegar, Shanxi Old Aged Vinegar and Zhenjiang Aromatic Vinegar factories, respectively, in different regions in China. To avoid stress to cells, samples were diluted in GYE containing (per litre) glucose 10 g, yeast extract 10 g, ethanol 30 ml with the ratio of 1:1 (sample/broth). Cultures were incubated at 28°C for 24 h with shaking at 120 rpm. Enriched cultures were serially diluted in GYE ranging from 1 × 101 to 1 × 107 cell/ml and 500 μl of each dilution was spread onto GYEC plates (glucose 10 g, yeast extract 10 g, ethanol 30 ml, calcium carbonate 15 g, agar 8 g, 1 l distilled H2O). After incubating at 28°C for 3 days, colonies showing clear zone on GYEC plates from 105 to 107 dilutions were picked up and purified. Type strain A. pasteurianus DSM 3509T and reference strains Gluconacetobacter xylinus DSM 2004 and A. pasteurianus CICC 7009 were cultivated and preserved according to Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH (DSMZ) and China Centre of Industrial Culture Collection (CICC) procedures, respectively.
Detection of Isolates Variability
Bacterial strains diversity was determined by ERIC/PCR. To perform ERIC/PCR, genomic DNA was extracted by enzymatic lyses according to method reported before [8] and quantified by Nanodrop Spectrophotometer (ND-1000). ERIC reactions were carried out with DNA polymerase from Takara Bio, Inc., (Japan) as described previously [9, 19]. PCR amplifications were performed using Hybaid Thermal Cycler (Celbio, Italy) with initial denaturation at 94°C for 5 min, followed by 30 cycles of the denaturation at 94°C for 30 s, annealing at 57°C for 30 s, and polymerization at 65°C for 4 min. The polymerization was completed by a final cycle of 8 min at 65°C for extension. PCR products were resolved on 2% agarose gel, stained with ethidium bromide and visualized by UV illumination. Reproducibility of ERIC/PCR was assessed by the use of DNAs from different extractions and by repetition in at least three independent assays. Amplicons size was estimated by BioDocAnalyze (BDA) analysis software (Germany).
Strain Identification
Selected strains were identified by 16S rRNA gene sequencing. Genomic DNA was extracted by fresh culture as reported above and PCR reaction was performed on template DNAs using Ex Taq DNA polymerase (Takara Bio, Inc., Japan) according to the manufacturer’s conditions and procedures. Primers 16S 616 5′-TACGGGAGGCAGCAG-3′ (position 342–356 on 16S rRNA gene, Escherichia coli numbering) and 16S 1492 5′-GGCTACCTTGTTACGACTT-3′ (position 1490–1510 on rRNA gene, Escherichia coli numbering) were used and PCR reactions performed [7]. The amplified PCR products were purified with Montage® PCR Cleanup Kit, according to the manufacturer’s instructions and quantified by Nanodrop. Direct sequencing of 16S rRNA gene was performed by automated sequencing service of Eurofins MWG Operon (Ebersberg, Germany). Sequences contigs were assembled using CHROMASPro (Version 1.41), and similarities searched using the software BLAST (http://blast.ncbi.nlm.nih.gov/Blast).
Nucleotide Sequence Accession Number
The nucleotide sequence data have been deposited into EMBL databases under the accession numbers listed in Table 2.
Phenotypic and Technological Features of Strains
Cell shape was observed after incubating cultures in GYE broth at 28°C for 3 days. Gram-staining and catalase production were tested as described previously [15]. KOH test was performed emulsifying cell cultures on slide containing 3.0% KOH solution. Oxidation of ethanol to acetic acid was tested EYC medium containing (per litre) ethanol 30 ml, yeast extract 10 g, calcium carbonate 20 g, agar 8 g; plates were observed after 3 days incubation at 28°C. Cellulose production was examined boiling pellicles in 4 ml of 5.0% NaOH solution for 2 h and Ga. xylinus (DSM 2004) was used as positive control. Growth on 30% d-glucose and tolerance to ethanol (5, 10, 15 and 20%, v/v) were tested according to previously study [16]. Growth on single carbon sources was carried out by a modification of the previous method [18]. The carbon sources were sterilized by filtration (0.2 μm) and added to the sterile basal medium (yeast extract, 0.5 g; vitamin-free casamino acid, 3 g) to a final concentration of 0.3%.
Results
Strains Isolation and Characterization
Strains were obtained from colonies of 1 × 105–1 × 107 cell/ml dilutions of each vinegar sample showing clear zone in an area up to 10 mm from the edge of GYEC plates. All the selected colonies appeared small, circle and pale on GYEC medium after incubation at 28°C for 5 days. To characterize different strains, the isolates were screened by ERIC/PCR. Among all the isolates, 21 of them provided unique ERIC/PCR patterns. The number of ERIC amplicons varied from 3 (strain SX461) to 14 (strain ZJ361B) and the size from 93 to 4317 bp (Table 1). To verify that the resolution power was restricted to the strain level, A. pasteurianus strain CICC 7009 was used as species-specific control (Fig. 1). Amplification of genomic DNA from some strains (SX463, SX861, ZJ172, ZJ25B and ZJ555) provided background smearing also after several attempts were performed on genomic DNA from different extractions.
ERIC/PCR of representative acetic acid bacteria strains. M1. 100 bp DNA ladder (Takara, Japan), 1. ZJ153A, 2. ZJ171, 3. ZJ271, 4. ZJ273, 5. ZJ361B, 6. ZJ362, 7. 7009 (A. pasteurianus, reference strain), 8. DL13, 9. DL15, 10, DL21A, 11. SX363, 12. SX461, 13. SX561, 14. SX563, 15. SX862, M2 500 bp DNA ladder (Takara, Japan)
Strains Identification
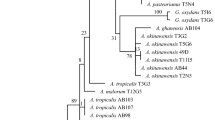
At least three isolates from each sample were identified to the species level on the basis of the nearly complete 16S rRNA gene sequencing (1374–1404 nucleotides). We identified strains able to grow starting from 5% (v/v) of ethanol because the ability to grow on it is one of main phenotypic traits to select AAB for vinegar production [7]. Moreover, strains DL13 and ZJ362, not growing on ethanol medium, were identified. A total of 12 representative strains sequences of 16S rRNA gene were obtained and compared with those of the recognized Acetobacter species and type strains of the other genera in the family Acetobacteraceae retrieved from EMBL. On this basis strains DL13, DL15, DL21A, SX363, SX461, SX862, ZJ153A, ZJ25B, ZJ273, ZJ361B and ZJ362 were assigned to A. pasteurianus species and strain ZJ555 to Ga. europaeus (Table 2).
Phenotypic and Technological Traits
All the strains showed the basic characteristic of AAB: Gram-negative, KOH positive and catalase positive reactions, mainly occurring in pairs or in short chains and all oxidized ethanol to acetic acid. Technological tests showed that among A. pasteurianus strains there is a high degree of variability for almost all the considered traits. In particular, the ability to grow at different ethanol concentrations showed that increased concentrations from (5–20% (v/v)) inhibited bacterial growth and 15% was tolerated only by strains SX861, SX862, ZJ25B, ZJ273 and ZJ555. Six strains grew on 30% d-glucose. Concerning the growth on single carbon sources, except from strains DL13, ZJ153A and SX561 that showed weak or no growth on all carbon sources tested, a heterogeneous behaviour was observed. No strains produced cellulose (Table 3).
Discussion
In this study, AAB from SSF Chinese vinegars were typed by ERIC/PCR. Among highly conserved repetitive DNA elements that are widely distributed in the genome of bacteria, ERIC/PCR has been demonstrated to be effective in detecting intraspecific variability of bacterial strains including AAB [4, 6, 13]. In our study, apart from some variations in band intensity, no differences were observed between the ERIC profiles obtained from each strain’s DNA analyzed in different assays. These differences were tested in triplicate assays and all the band patterns proved highly reproducible allowing the direct generation of strain specific genomic fingerprinting. From our data, the most culturable species recovered was A. pasteurianus. This evidence was consistent with previously studies showing the dominance of A. pasteurianus in cereal vinegars [10, 14]. We also detected one strain belonging to Ga. europaeus species, which strains are responsible for high acid (10–14% acetic acid) production in industrial wine vinegar. Ga. europaeus has distinctive phenotypic traits such as absolute requirement of acetic acid for growth, low biomass production and short viability out side vinegar environment. The recovery of only one Ga. europaeus strain suggests that discontinuous acetic acid availability affects the colonization by Ga. europaeus.
Within the species A. pasteurianus, we recovered different strains also from the same vinegar sample. This was in agreement with previously studies reporting that high complex strain microflora occurs in vinegars produced by biofilm on mechanical supports and on spoiled wines, whereas single dominant strains in submerged vinegar fermentations [1, 12, 17]. In our study, one of the most probable cause of strains diversity could be due to the effect of raw material composition and/or technological steps as source of variability. According to the producing practice, the addition of wheat bran or hull of cereals to induce the acetification can promote the establishment of less stringent environmental conditions that favourite the occurrence and functionality of different indigenous AAB. Furthermore, the AAB belonging to A. pasteurianus species showed a high degree of phenotypic variability such as tolerance to ethanol, growth on 30% of d-glucose and growth on single carbon sources. Some of these features were considered previously as discriminative tools for taxonomic purposes, for instances, growth on d-mannitol to differentiate Acetobacter from Gluconobacter and partly Gluconacetobacter genera and 30% of d-Glucose to discriminate Acetobacter malorum from A. pasteurianus. However, it is well known that phenotypic features are too controversial for the identification at species level and comparative phenotypic analysis of AAB is not considered a suitable tool [3]. Our data confirmed the evidence that the resolution power of phenotypic information is not reliable to taxonomically differentiated AAB. In contrast, phenotypic characterization still remains of basic relevance to screen strains in selection strategies providing data to cluster strains according to suitable metabolic profiles and allowing the exploitation of selected strains on specific fermentative substrates [5].
The application of strains of this study in selected starter cultures implementation can be predicted since they possess both a number of basic and specific phenotypic suitable traits. In this respect, growth modality by superficial biofilm, joint to no production of cellulose, contribute to avoid oxidation stuck due to oxygen limitation in static conditions, undesired sensorial profile and low acidity of the final product. The ability to grow on ethanol media, as preferred substrate, favourite efficient oxidation followed by a fast production of acetic acid that prevents the restarting of alcoholic fermentation and avoids spoilage by other organisms.
In this study, for the first time, phenotypic and genetic variability of AAB strains involved in SSF processes was stated. Results can be exploited for the implementation of large-scale SSF processes by selected strains for vinegar production and for innovative biotechnological applications.
References
Bartowsky EJ, Henschke PA (2008) Acetic acid bacteria spoilage of bottled red wine—a review. Int J Food Microbiol 125:60–70
Chen FS, Li L, Qu J, Chen CX (2009) Cereal vinegars made by solid-state fermentation in China. In: Giudici P, Solieri L (eds) Vinegar of the world. Springer, Milan, pp 243–259
Cleenwerck I, De Vos P (2008) Polyphasic taxonomy of acetic acid bacteria: an overview of the currently applied methodology. Int J Food Microbiol 125:2–14
De Vuyst L, Camu N, De Winter T, Vandemeulebroeckeb K, Van de Perre V, Vancanneyt M, De Vos P, Cleenwerck I (2008) Validation of the (GTG)5-rep-PCR fingerprinting technique for rapid classification and identification of acetic acid bacteria, with a focus on isolates from Ghanaian fermented cocoa beans. Int J Food Microbiol 125:79–90
Giudici P, Solieri L, Pulvirenti AM, Cassanelli S (2005) Strategies and perspectives for genetic improvement of wine yeasts. Appl Microbiol Biotechnol 66:622–628
Gonzalez A, Hierro N, Poblet M, Rozes N, Mas A, Guillamon JM (2004) Application of molecular methods for the differentiation of acetic acid bacteria in a red wine fermentation. J Appl Microbiol 96:853–860
Gullo M, Giudici P (2008) Acetic acid bacteria in traditional balsamic vinegar: phenotypic traits relevant for starter cultures selection. Int J Food Microbiol 125:46–53
Gullo M, Caggia C, De Vero L, Giudici P (2006) Characterization of acetic acid bacteria in “traditional balsamic vinegar”. Int J Food Microbiol 106:209–212
Gullo M, De Vero L, Giudici P (2009) Succession of selected strains of Acetobacter pasteurianus and other acetic acid bacteria in traditional balsamic vinegar. Appl Environ Microbiol 75:2585–2589
Haruta S, Ueno S, Egawa I, Hashiguchi K, Fujii A, Nagano M, Ishii M, Igarashi Y (2006) Succession of bacterial and fungal communities during a traditional pot fermentation of rice vinegar assessed by PCR-mediated denaturing gradient gel electrophoresis. Int J Food Microbiol 109:79–87
Liu DR, Zhu Y, Beeftink R, Ooijkaas L, Rinzema A, Chen J (2004) Chinese vinegar and its solid-state fermentation process. Food Rev Int 20:407–424
Mariette I, Schwarz E, Vogel RF, Hammes WP (1991) Characterization by plasmid profile analysis of acetic acid bacteria from wine, spirit and cider acetators for industrial vinegar production. J Appl Bacteriol 71:134–138
Mondal KK, Mani C (2009) ERIC/PCR-generated genomic fingerprints and their relationship with pathogenic variability of Xanthomonas campestris pv. punicae, the incitant of bacterial blight of pomegranate. Curr Microbiol. Published online: 25 August, 2009. doi:10.1007/s00284-009-9482-z
Nanda K, Taniguchi M, Ujike S, Ishihara N, Mori H, Ono H, Murooka Y (2001) Characterization of acetic acid bacteria in traditional acetic acid fermentation of rice vinegar (Komesu) and unpolished rice vinegar (Kurosu) produced in Japan. Appl Biochem Biotechnol 67:986–990
Navarro RR, Komagata K (1999) Differentiation of Gluconacetobacter liquefaciens and Gluconacetobacter xylinus on the basis of DNA base composition. J Gen Appl Microbiol 45:7–15
Sievers M, Swings J (2005) Family Acetobacteraceae, Ed. In: Garrity GM (ed) 2nd Bergey’s manual of systematic bacteriology, vol 2. Springer, New York, pp 41–95 (Part C)
Sievers M, Teuber M (1995) The microbiology and taxonomy of Acetobacter europaeus in commercial vinegar production. J Appl Bacteriol 79:84S–95S
Swings J, Gillis M, Kersters K (1992) Phenotypic identification of acetic acid bacteria. In: Board RG, Jones D, Skinner FA (eds) Identification methods in applied and environmental microbiology. Blackwell Scientific, Oxford, pp 103–110
Versalovic J, Koeuth T, Lupski RJ (1991) Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res 19:6823–6831
Acknowledgement
This research was supported by China Scholarship Council joint PhD Project and a grant of Manodori Foundation, Italy. The authors thank the Chinese vinegar factories for providing samples and factory technical assistance.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Wu, J., Gullo, M., Chen, F. et al. Diversity of Acetobacter pasteurianus Strains Isolated From Solid-State Fermentation of Cereal Vinegars. Curr Microbiol 60, 280–286 (2010). https://doi.org/10.1007/s00284-009-9538-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-009-9538-0