Abstract
Vanillin is undoubtedly one of the most popular and widely used flavoring agents in the world. Taking into consideration the worldwide demand for natural vanillin and its limited supply, alternative routes for its production including biotransformation are being constantly explored. In this regard, a novel soil bacterium capable of converting isoeugenol to vanillin was isolated by conventional enrichment process from soils of Ocimum field. On the basis of morphological and physiochemical characteristics and 16S rRNA gene sequence analysis, the isolate was identified as Pseudomonas chlororaphis CDAE5 (EMBL # AM158279). Vanillin formation was analyzed by gas chromatography (GC), and its structure was confirmed by GC-mass spectrometry and nuclear magnetic resonance. After 24-h reaction, the vanillin concentration reached 1.2 g L−1 from 10 g L−1 isoeugenol in 20-mL reaction solution at 25°C and 180 rpm. The strain showed potential to be a good candidate for biotechnological production of vanillin from isoeugenol. Further studies for standardization and optimization for higher yield of vanillin production needs to be investigated.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Vanillin (4-hydroxy-3-methoxybenzaldehyde) is one of the most widely used flavor compounds in food, beverages, pharmaceuticals, perfumes, and medicinal industries [16]. More than 12,000 tons of vanillin is produced each year, but only 1% originates from its natural source, i.e., Vanilla planifolia. The main portion is produced by chemical synthesis from guaiacol or lignin [24]. The price of the chemically synthesized vanillin is very low (about US $12/kg) as compared to the price of natural vanillin (between US$ 1200 and 4000/kg) [10]. The difference between the prices combined with the increasing customer-led demand for natural flavors has stimulated the exploration of biotechnological routes for the production of natural vanillin. One such route is microbial or enzymatic transformation of natural precursors such as ferulic acid [2, 6, 12, 14, 15, 18], vanillic acid [22], eugenol [17, 25], or isoeugenol [8, 9, 11, 21, 26, 27]. With respect to isoeugenol, there have been few studies on its degradation pathway and efficient bioconversion system. Although Bacillus species capable of transforming isoeugenol to vanillin have been reported earlier [21, 26, 27], little has been investigated about other microorganisms [4, 16]. With the aim of exploring the microbial wealth of the Western Himalayan region for biotransformation processes, we herein report the isolation and identification of novel strain of Pseudomonas chlororaphis, labeled as CDAE5, capable of transforming isoeugenol to vanillin (Fig. 1).
Materials and Methods
Materials and Microorganisms
Standard isoeugenol (98%, cis-trans mixture) and vanillin (99%) for the biotransformation studies were obtained from Merck (Germany). Other chemicals were of analytical grade. P. chlororaphis CDAE5 and other strains were isolated from soils of the Western Himalayan region.
Enrichment Culture
Soil samples collected from Ocimum (Ocimum basilicum, O. sanctum, O. clocimum) fields at Chandpur farm (Institute of Himalayan Bioresource Technology, Palampur, Himachal Pradesh, India) were processed for isolation of bacteria by enrichment technique. Ten grams of each sample was suspended in 90 mL of sterile distilled water, and these suspensions were used as an inoculum for enrichment cultures. For enrichment procedures, nutrient broth (NB) medium (0.3% beef extract, 0.5% peptone, 0.5% NaCl, pH 7.0) supplemented with 0.1% isoeugenol was used. Cultures in 100-mL flask containing 20 mL of medium were inoculated with 3 mL of soil suspension and incubated shaking at 150 rpm and 28°C. Following four transfers (30 μL into 20 mL of fresh medium) after every 24 h, cultures were diluted (1 mL culture in 9 mL of distilled water) and plated on nutrient agar plates. After incubation for 24–48 h at 28°C, morphologically different colonies appearing on the plates were isolated and subjected to further purification by streaking on same medium.
Screening of Strains Transforming Isoeugenol to Vanillin
Isolates were grown in a 100-mL flask containing 20 mL of biotransformation (BT) medium (1.0% glycerol, 0.31% Na2HPO4, 0.25% KH2PO4, 0.25% (NH4)2SO4, 0.0025% FeSO4.7H2O, 0.2% 1M MgSO4, 0.3% 0.1 M CaCl2) [17] at 25°C and 180 rpm for 24 h and isoeugenol was added up to a concentration of 1.5%. After incubation for 72 h, the potential biotransformation products were extracted with dichloromethane and analyzed by thin layer chromatography using hexane:ethyl acetate (80:20) as solvent system. The products were detected by developing the plates in iodine vapors [26].
Biotransformation of Isoeugenol to Vanillin by a Growing Culture
Isolates, which were found positive for biotransformation of isoeugenol to vanillin in initial screening, were grown on BT medium at 25°C and 180 rpm for 24 h, and isoeugenol was added up to 2.0% concentration. The whole culture was extracted three times with dichloromethane. Organic fractions were collected, filtered, dried over anhydrous sodium sulphate, and concentrated at 40°C to remove solvent under vacuum on a rotary vacuum evaporator (Buchi, Switzerland).
Vanillin and Isoeugenol Analysis
The above obtained extract was then dissolved in analytical grade chloroform (Merck) and analyzed by FID gas chromatograph (Shimadzu 2010) equipped with BP-1 capillary column (30m × 0.25 mm id, 0.25 μm), carrier gas N2 at 0.5 mL/min, split ratio 40:1, injection temperature 250°C, detection temperature 280°C. The oven temperature was set at 220°C. The system was linked to a computerized integrator. Under these conditions, retention times recorded for vanillin and isoeugenol were 11.7 min and 13.6 min, respectively (Fig. 2). Gas chromatography (GC)-mass spectrometry (Shimadzu QP-2010) and nuclear magnetic resonance (Bruker Avance-300) were also used for confirmation of vanillin.
Morphological, Physiological, and Biochemical Characterization of Transforming Strains
Based on thin layer chromatography (TLC) and GC analysis, the strain giving highest vanillin yield was selected and characterized phenotypically and biochemically using standard techniques (Gram staining, motility, colony shape, size, and color on nutrient agar plate, growth on MacConkey agar, catalase, oxidase, O/F test, lipid and starch hydrolysis, etc.), according to the diagnostic table of Cowan and Steel (1974) [3] and Bergey’s Manual of Determinative Bacteriology (1994) [5].
Phylogenetic Analysis
Bacterial DNA was isolated from pure culture and polymerase chain reaction (PCR) amplification of almost the entire length 16S rRNA fragment was carried out [20]. The sequence of primers used for amplification were 5′-AGAGTTTGATCATGGCT CAGA-3′ and 5′-GTTACCTTGTTACGACTT-3′ corresponding to 8 to 28 and 1493 to 1510, respectively, which are parts of 16S rRNA gene of Escherichia coli and are useful for amplifying 16S rRNA gene from various kinds of bacteria. The amplified 16S rRNA gene was purified from agarose gel using Qiaquick Gel Extraction Kit (Qiagen, Germany), ligated into the cloning vector (pGEM®-T easy vector) with the cloning kit (Promega, USA) and used for biotransformation of E. coli DH5α [20].
The nucleotide sequencing of the gene was done by using Big DyeR Terminator Cycle Sequencing Kit (Applied Biosystems) and 3130xl Genetic Analyzer (Applied Biosystems). The BLASTN program (http://www.ncbi.nlm.nih.gov/BLAST/, NCBI, Bethesda, MD) was used for homology searches with the standard program default. Multiple alignments of the sequence were performed and a neighbor-joining phylogenetic tree [7, 19] was constructed using CLUSTAL W program [23]. The nucleotide sequence of the 16S rRNA gene of isolated bacterial strain is under EMBL Nucleotide Sequence Database Accession Number AM158279.
Results and Discussion
Isolation of Strains Transforming Isoeugenol to Vanillin
The present work was focused to isolate bacterial strains capable of transforming isoeugenol to vanillin. In earlier reported works, toxicity of the substrate and product to microorganisms and potential degradation of vanillin to vanillyl alcohol and vanillic acid has resulted in low yields of vanillin from isoeugenol [1, 15]. This strengthens the notion that strains having high tolerance to isoeugenol and lower vanillin degrading ability should be isolated [27]. Keeping this in view, microbial strains having high tolerance to isoeugenol were isolated by enrichment process from the soil samples taken from an Ocimum field, because Ocimum is a rich source of phenylpropanoids (about 75%), including isoeugenol. Based on their morphology, 21 different bacterial strains were selected and tested for biotransformation of isoeugenol into vanillin. Products were analyzed through TLC and GC (Fig. 2). Under the conditions (as mentioned under Materials and Methods), strain CDAE5 produced the highest amount of vanillin. Based on these results, strain CDAE5 was selected for further studies.
Identification and Phylogenetic Analysis of Strain CDAE5
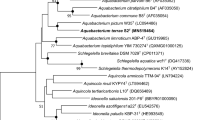
Morphological, physiological, and biochemical characters and 16S rRNA gene sequencing was used to identify strain CDAE5. Microbial characteristics of strain CDAE5 as studied are listed in Table 1. Cells are Gram-negative, motile rods. Growth occurred at temperature of 5°C to 37°C, pH of 5.0 to 10.0, and in a medium supplemented with 2.5-5% NaCl. These characteristics indicated that the strain belongs to the genus Pseudomonas. To confirm its phylogenetic relationship with Pseudomonas, genomic DNA was isolated from the bacterium and gene coding for 16S rRNA was amplified by polymerase chain reaction. The sequencing of almost full-length 16S rDNA showed that it was closely related to genus Pseudomonas showing 99.2% homology to P. chlororaphis (Fig. 3). In accordance with these data, the isolate was included in the genus Pseudomonas and named as Pseudomonas chlororaphis CDAE5. Strain CDAE5 is kept at Microbial Type Culture Collection and Gene Bank, Chandigarh, India as Pseudomonas chlororaphis CDAE5.
Phylogenetic tree indicating the estimated relationship between strain CDAE5 (Accession no. AM158279) and species of the genus Pseudomonas that shared highest 16S rRNA gene sequence similarities. Acinetobacter calcoaceticus (Z93434) was selected as the outgroup. Numbers indicate bootstrap values. Bar, 0.1 substitutions per site.
Biotransformation of Isoeugenol to Vanillin by a Growing Culture
Bacterial culture was grown for 24 h in 20 mL of BT medium. Different concentrations of isoeugenol ranging from 0.5% to 2.0% were then added to culture, and 1.0% isoeugenol was found to give the better yield (Fig. 4). This may be because of the toxicity of substrate to microorganisms at its higher concentrations, as reported earlier [15]. The maximum vanillin yield of 1.2 gL−1 from 10 gL−1 was achieved after 24 h in 20-mL reaction mixture, resulting in a molar efficiency of 12.64%. The first BT of isoeugenol to vanillin was achieved with Aspergillus niger having only 10% efficiency [1]. Other bacterial strains reported include those of genera Klebsiella, Enterobacter, Bacillus, and Serratia, with Serratia marcescens showing maximum conversion efficiency of 20.5% [17]. Recently, a strain of Bacillus fusiformis has been reported for conversion of isoeugenol to vanillin, whereby product inhibition was avoided with the addition of HD-8 resin, yielding a vanillin concentration of 8.10 gL−1 from 50 gL−1 isoeugenol [27]. Further studies for improving the yield of vanillin using Pseudomonas chlororaphis CDAE5 by addition of resins and process optimization are under progress.
Conclusion
A novel strain, P. chlororaphis CDAE5, was isolated from soils of Ocimum field, which was found to transform isoeugenol to vanillin. In earlier studies, Pseudomonas fluorescens was been reported to degrade ferulic acid to vanillin [13], and Pseudomonas putida converted isoeugenol to vanillic acid [4]. To the best of our knowledge, the present study gives the first evidence for conversion of isoeugenol to vanillin by P. chlororaphis CDAE5. Using 10 gL–1 isoeugenol as substrate, 1.2 gL–1 vanillin was produced in 20-mL reaction solution after 24 h, at 25°C and 180 rpm. Further studies in this direction for obtaining higher yields of vanillin are in progress.
Literature Cited
Abraham WR, Arfmann HA, Stumpf S, Washausen P, Kieslich K (1988) Microbial transformations of some terpenoids and natural compounds. In: Schreier P (ed) Bioflavour ‘87. Analysis, biochemistry, biotechnology. Proceedings of an International Conference. Berlin: de Gruyter, pp 399–414
Benedict CO, Victorio V (1999) Constructions of recombinants Pseudomonas putida BO14 and Escherichia coli QEFCA8 for Ferulic acid biotransformation to vanillin. J Biosci Bioeng 88:103–110
Cowan ST, Steel KJ (1974) Manual for the identification of medical bacteria, 2nd ed. London: Cambridge University Press
Furukawa H, Morita H, Yoshida T, et al. (2003) Conversion of isoeugenol into vanillic acid by Pseudomonas putida 158 cells exhibiting high isoeugenol-degrading activity. J Biosci Bioeng 96:401–403
Holt JG, Krieg NR, Sneath PHA, et al. (1994) In: Bergey’s manual of determinative bacteriology, 9th ed. Baltimore: Williams and Wilkins
Karmakar B, Vohra RM, Nandanwar H, et al. (2000) Rapid degradation of Ferulic acid via 4-vinylguiacol and vanillin by a newly isolated strain of Bacillus coagulans. J Biotechnol 80:195–202
Kimura M (1980) A simple method for estimating evolutionary rates base substitution through comparative studies nucleotide sequences. J Mol Evol 16:111–120
Li YH, Sun ZH, Zhao LQ, et al. (2005) Biotransformation of isoeugenol to vanillin by crude enzyme extracted from soybean. Appl Biochem Biotechnol 125:1–10
Li YH, Sun ZH, Zheng P (2004) Determination of vanillin, eugenol and isoeugenol by RP-HPLC. Chromatographia 60:709–713
Lomascolo A, Stentelaire C, Asther M, et al. (1999) Basidiomycetes as new biotechnological tools to generate natural aromatic flavours for the food industry. Trends Biotechnol 17:282–289
Markus PH, Peters ALJ, Roos R (1992) Process for the preparation of phenylaldehydes. European Patent EP 542348
Muheim A, Muller B. Munch T, et al. (2001) Microbiological process for producing vanillin. US Patent 6235507
Narbad A, Gasson MJ (1998) Metabolism of ferulic acid via vanillin using a novel CoA-dependent pathway in a newly isolated strain of Pseudomonas fluorescens. Microbiol 144:1397–1405
Narbad A, Rhodes MJC, Gasson MJ, et al. (2001) Production of vanillin. US Patent 2001014467
Overhage J, Priefert H, Rabenhorst J, et al. (1999) Biotransformation of eugenol to vanillin by a mutant of Pseudomonas sp. Strain HR199 constructed by disruption of the vanillin dehydrogenase (vdh) gene. Appl Microbiol Biotechnol 52:820–828
Priefert H, Rabenhorst J, Steinbüchel A (2001) Biotechnological production of vanillin. Appl Microbiol Biotechnol 56:296–314
Rabenhorst J, Hopp R (1991) Process for the preparation of vanillin. US Patent 5017388
Rabenhorst J, Hopp R (2000) Process for the preparation of vanillin and microorganisms suitable therefor. US Patent 6133003
Saitou N, Nei M (1987) The neighbor joining method: a new method for reconstructing the phylogenetic tree. Mol Biol Evol 4:406–425
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory
Shimoni E, Ravid U, Shoham Y (2000) Isolation of a Bacillus sp. capable of transforming isoeugenol to vanillin. J Biotechnol 78:1–9
Stentelaire C, Laurence L-M, Oddou J, et al. (2000) Design of a fungal bioprocess for vanillin production from vanillin acid at scalable level by Pycnoporus cinnabarinus. J Biosci Bioeng 89:223–230
Thompson JD, Higgins DG, Gibson DJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence weighing, position specific gap penalties and weight matrix choice. Nucleic Acid Res 22:4673–4680
Van den Heuvel RHH, Fraaije MW, Laane C, et al. (2001) Enzymatic synthesis of vanillin. J Agric Food Chem 49:2954–2958
Yukio W, Tetsushi A, Naomi H, et al. (1993) Production of vanillin and its related compound by fermentation. Japanese Patent 5227980
Zhao LQ, Sun ZH, Zheng P, et al. (2005) Biotransformation of isoeugenol to vanillin by a novel strain of Bacillus fusiformis. Biotechnol Lett 27:1505–1509
Zhao LQ, Sun ZH, Zheng P, et al. (2006) Biotransformation of isoeugenol to vanillin by Bacillus fusiformis CGMCC1347 with addition of resin H D-8. Process Biochem 41:1673–1676
Acknowledgments
This study was supported by a project (SMM-0002) sponsored by Council of Scientific & Industrial Research, New Delhi, India. The authors gratefully acknowledge the Director, IHBT, Palampur, for providing the necessary facilities during the course of the project.
Author information
Authors and Affiliations
Corresponding author
Additional information
†IHBT Communication No. 0676
Rights and permissions
About this article
Cite this article
Kasana, R.C., Sharma, U.K., Sharma, N. et al. Isolation and Identification of a Novel Strain of Pseudomonas chlororaphis Capable of Transforming Isoeugenol to Vanillin† . Curr Microbiol 54, 457–461 (2007). https://doi.org/10.1007/s00284-006-0627-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-006-0627-z