Abstract
A dual culture-based and non–culture-based approach was applied to characterize predator bacterial groups in surface water samples collected from Apalachicola Bay, Florida. Chemotaxis drop assays were performed on concentrated samples in an effort to isolate predator bacteria by their chemotactic ability. Yeast extract (YE) and casamino acids (CA) proved to be strong chemoattractants and resulted in three visibly distinct bands; however, dextrose, succinate, pyruvate, and concentrated cells of Vibrio parahaemolyticus P5 as prey did not elicit any response. The three distinct bands from YE and CA were separately collected to identify the chemotactic microbial assemblages. Plaque-forming unit assays from different chemotaxis bands with P5 as prey indicated 5- (CA) to 10-fold (YE) higher numbers of predator bacteria in the outermost chemotactic bands. Polymerase chain reaction–restriction fragment length polymorphism and 16S rDNA sequencing of clones from different chemotaxis bands resulted in identification of Pseudoalteromonas spp., Marinomonas spp., and Vibrio spp., with their numbers inversely proportional to the numbers of predators—i.e., Bdellovibrio spp. and Bacteriovorax spp—in the chemotaxis bands. This study indicates that predatorial bacteria potentially respond to high densities of microbial biomass in aquatic ecosystems and that chemotaxis drop assay may be an alternate culture-independent method to characterize predatorial bacterial guilds from the environment.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
The fairly less-studied predator group of Bdellovibrio and like organisms (BALOs) “make their living” by attacking and devouring other bacteria for their own propagation and are found in diverse environments, including estuaries, oceans, rivers, sewage facilities, runoff water, and manmade water supplies [7]. Our objective in this study was to investigate the response of aquatic bacteria and BALOs to chemotactic agents. The study site was the surface waters of Apalachicola Bay ecosystem, which is a fairly shallow subtropical estuary in the northeastern Gulf of Mexico (Fig. 1). It is considered to be relatively pristine and constitutes a part of the National Estuarine Research Reserve (NERR) system. However, this pristine ecosystem is highly vulnerable to eutrophication because the surrounding areas have witnessed dramatic urbanization during recent years. Little is known of the microbial processes, such as bacterial mortality, in Apalachicola Bay. Predators such as BALOs are highly motile and can attack and prey on other Gram-negative bacteria [6, 7, 12] and are likely one of the factors contributing to bacterial mortality in aquatic ecosystems [14]. However, few studies are available on the physiology and biodiversity of BALOs. One of the main reasons for the difficulties in the characterization of BALOs is that for them to grow, they require being cultured in a dual system with prey bacteria [12, 18].
The lack of a “universal” gene probe targeting halophilic BALOs prompted us to investigate whether chemotaxis can be used to study the predatorial response from environmental samples. Chemotaxis is the movement of a microorganism either toward or away from a chemical. B. bacteriovorus has been reported to be chemotactic toward prey cells [20]; amino acids [9]; pure compounds such as acetate, propionate, etc. [21]; and yeast extract [19]. Lambert et al. have shown that B. bacteriovorus possess a chemotaxis system to trigger predatorial response [10], and with the genome sequencing of B. bacteriovorus HD100, almost 20 methyl-accepting chemotaxis proteins and 2 component-regulatory chemotaxis genes have now been identified and fully characterized [13]. These findings clearly indicate a role of chemotaxis in predation by BALOs. However, all of these studies focused on axenic pure strains of BALOs. Here we report on the response of BALO assemblages, directly from concentrated environmental samples, toward yeast extract (YE) and casamino acids (CA) as well as the motile environmental bacteria that are attracted to these chemotactic agents. The characterization of BALOs by way of this novel approach of applying chemotaxis could serve as a culture-independent method for characterization of the biodiversity of predator bacteria. This could be applied to a variety of aquatic sources to rapidly screen the activity of BALOs predating on indigenous prey bacteria, but potential limitations may include false-negative results if incubations are run for longer durations, which will lead to growth of bacteria but not chemotaxis. To our knowledge, ours is the first such report.
Materials and Methods
Sampling site details
Surface-water samples were collected from three sites in February 2006 from a pier located in the mid-section of Apalachicola Bay as shown in Fig. 1 (latitude/longitude 29° 42.128′ north, 84° 52.811′ west).
Three water samples were obtained within an area of approximately 15 m2 (Fig. 1). Samples were collected by submerging a sterile container to an approximate depth of 1 m. After collection, the samples were placed in a cooler and transported to the laboratory at Florida A & M University. A single composite sample representing the sampling site was obtained by mixing the three replicate samples and filtered through a 0.8-μm filter to exclude debris and protozoan grazers [8, 12], and stored them at 4°C. Further experiments were set up within 2 days of sampling.
Sampling-site biogeochemical parameters
Apalachicola Bay water was assayed for selected parameters by a Portable Water Checker U-10 probe (Horiba, Kyoto, Japan). The probe was held under the water before sample collection to measure the parameters at the time of sampling.
Chemotaxis drop assay
The samples were filtered through a 0.45-μm filter to further concentrate the small-sized predator bacteria [8, 12], and a chemotaxis drop assay was performed as previously described [15] with minor modifications. Initial chemotaxis drop assays were set up using the environmental samples, which resulted in a weak chemotaxis response from the microbial community (indistinct chemotaxis rings were observed). The experiments were repeated using samples concentrated by centrifugation at 10,000 rpm at room temperature for 15 minutes. This resulted in a concentrated biomass from 4 L sample down to 40 mL. Melted agarose was mixed with each of these bay samples to yield a final concentration of 0.3% to achieve semiviscosity [15]. The concentrated samples were poured into large Petri dishes to accommodate the larger sample size (40 ml/plate). Chemotactic response was determined by placing crystals of dextrose, yeast extract, succinate, pyruvate, concentrated cells of P5, and CA at the center of each plate as previously described [2, 15]. The negative control did not contain any chemoattractant. Turbid, distinct rings were observed around the periphery of the chemoattractants within 5 to 8 hours of incubation at room temperature (25°C); however, it took almost 12 hours to show good contrast, at which point samples from the outer, middle, and inner chemotaxis bands were collected using 1-ml sterile tips that had been cut in half for collection of viscous bands. The plates were scanned to capture the images.
Plaque-forming unit assay
A 1-ml portion of each chemotaxis band sample was added to tubes containing 3.5 ml Pp20 top agar and 0.5 ml Vibrio parahaemolyticus P5 prey suspension as previously reported [8, 12]. The contents of the tubes were inverted once to mix and overlaid onto Pp20 bottom agar plates. The plates were left undisturbed to solidify and then incubated at 25°C. The plates were examined daily for 1 week for the presence of Bdellovibrio plaques, and plaque-forming units (PFUs) were counted after 7 days of incubation [8, 17].
Nucleic acid extraction and polymerase chain reaction amplification
DNA from chemotaxis band samples was extracted using the UltraClean GelSpin Kit per the manufacturer’s instructions (MoBio, Solana Beach, CA), except the DNA was eluted in sterile polymerase chain reaction (PCR)-grade water for downstream processing. Quality of the DNA was evaluated by electrophoresis through a 0.7% agarose gel with Tris-acetate-ethylenediaminetetraacetic acid (TAE) buffer, and concentrations of total DNA were estimated by ultraviolet absorbance at 260 nm [16]. Primers used for PCR amplification of bacterial 16S rDNA gene sequences were 27F (5′-AGAGTTTGATCMTGGCTCAG-3′) and 1492R (5′-TACGGYTACCTTGTTACGACTT-3′) [11]. All amplifications were performed in an iCycler thermocycler (Bio-Rad, Hercules, CA) using HotStarTaq Master Mix (Qiagen, Valencia, CA). Different dilutions of the DNA samples were desaturated at 95°C for 15 minutes, followed by 35 cycles of 94°C for 30 seconds, 55°C for 30 seconds, and 72°C for 30 seconds, followed by an extension step of 72°C for 7 minutes [4]. PCR products (5 μl) were analyzed by electrophoresis through a 1% agarose gel in TAE buffer.
Cloning bacterial 16S rDNA and RFLP analyses
Cloning 16S ribosomal gene was carried out with fresh PCR amplicons, which were ligated into pCRII-TOPO cloning vector and transformed into Escherichia coli TOP10F’ cells according to the manufacturer’s instructions (Invitrogen, Carlsbad, CA). Clones were screened with 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside (X-Gal)– and isopropyl-β-D-thiogalactopyranoside (IPTG) indicator Luria-Bertani agar plates supplemented with 50 μg/ml kanamycin. This was followed by direct-colony PCR on white colonies with the promoter-specific SP6 and T7 primers (Invitrogen, Carlsbad, CA) using the above-mentioned PCR program as previously reported [3]. Restriction fragment length polymorphism (RFLP) analyses were conducted using restriction enzyme HhaI and analyzed in 2% agarose gel [3, 4]. Clone libraries were subjected to rarefaction using aRarefactWin (version 1.3; S. Holland, Stratigraphy Laboratory, University of Georgia, Athens, GA [http://www.uga.edu/approximatelystrata/software/]) to confirm that sufficient numbers of RFLP groups were selected to represent the microbial diversity in the clone libraries from different chemotaxis bands.
DNA sequencing and phylogenetic analysis
Restriction profiles of the clone libraries were compared by RFLP and grouped in operational taxonomic units (OTUs). Selected clones were then sequenced at the DNA Sequencing Laboratory of Florida State University with 27F primer. Chimera detection was carried out by Chimera_Check, version 2.7, from the RDP-II Web site [5]. Sequences were also compared with previously identified sequences in the National Center of Biotechnology Information (NCBI) database using BLAST [1] and were aligned by ClustalX v. 1.8 [22]. Phylogenetic tree was generated with TREECON for Windows, version 1.3b (http://bioinformatics.psb.ugent.be/software.php) using neighbor joining with default settings [23]. Bootstrap resampling analysis for 100 replicates was performed to estimate the confidence of tree topologies.
Nucleotide sequence accession numbers
The partial 16S rDNA gene sequences from this study have been deposited in GenBank with the accession numbers DQ659738 to DQ659774.
Results and Discussion
Biogeochemical parameters
Selected biogeochemical parameters were noted at the time of sampling in February 2006. The three samples collected from the site had mid-salinity of 8.4 ppt, and pH was 7.5 to 8.0. The temperature of the sample was 15.5°C to 16°C; conductivity was between 14.8 and 15.0; and dissolved oxygen ranged between 50 and 54 mg/L.
Chemotaxis drop assay
Initial chemotaxis decrease assays were set up directly on the filtered bay samples, which elicited a weak chemotaxis response (the chemotactic ring was not distinct) against YE and CA (data not shown). The same samples were then concentrated 100 times to increase the density of microbial biomass, and chemotaxis drop assays were set up against the chemoattractants. Distinct chemotaxis bands could be visualized after incubation for 5 to 8 hours, but these were left undisturbed until 12 hours, and the results are shown in Figs. 2A through 2C. Three distinct bands were observed against YE and CA; however, dextrose, succinate, pyruvate, and concentrated cells of V. parahaemolyticus P5 (a prey usually employed for halophilic BALOs) [12], failed to elicit any response (i.e., no distinct chemotactic ring was observed). V. parahaemolyticus may have been unable to form a concentration gradient of dense biomass, which did not trigger a chemotactic response from the predatorial guilds of bacteria. YE and CA as chemoattractants would have the ability to attract all of the motile biomass in the samples, creating a dense concentration gradient to which BALOs could respond.
The negative control (no chemoattractant) did not show any chemotaxis, indicating that YE and CA served as chemoattractants for the microbial community. Portions representing the three distinct bands from YE and CA were collected separately for identification of chemotactic microbial community by culture-dependent and independent methods. Incubations for chemotaxis were conducted for 12 hours so that chemotaxis bands were clearly observed before their collection. This length of time may be sufficient for the microbial biomass to grow at the expense of substrate gradients formed on the plate. However, proof that microorganisms identified from different bands were likely chemotactic came later after 16S rDNA phylogenetic identification of the assemblages clearly showed that the chemotactic community consisted of motile guilds of marine bacteria (see section on bacterial phylogenetic analysis).
PFU estimation from different chemotaxis bands
PFU assay was set up to screen the presence of predatorial bacteria and further quantitate the differences, if any, in the three different chemotaxis bands (Table 1). Chemotaxis bands 1, which were the first bands to appear and were closest to the chemoattractants, yielded far less PFUs than did bands 2 and 3, which appeared after incubating for a longer duration (approximately 12 hours), and were farthest from the chemoattractants (Figs. 2A and 2B); this difference varied from 5- (CA) to 10-fold (YE) higher numbers of predator bacteria in the outermost chemotactic bands (Table 1). Although this assay targeted only those predatorial bacteria able to attack and consume V. parahaemolyticus P5 as prey, it indicated the presence and quantitative differences of predatorial bacteria in the chemotactic bands. This was followed by molecular analyses of motile community in the chemotaxis bands obtained from YE and CA.
Bacterial phylogenetic analysis on chemotaxis band samples
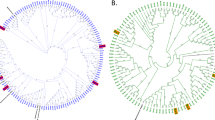
Motile microbial community from the three different bands from YE and CA were characterized by 16S rDNA gene-sequence analysis. For all libraries, rarefaction curves reached to a complete plateau, indicating that clone libraries represented the entire biodiversity of the samples (data not shown). Of 48 total clones screened from the YE library obtained from different chemotaxis bands, 4 OTUs were observed from YE-bands 1 and 2 and 5 OTUs from YE-band 3. Phylotypes were assigned the same OTUs based on similar RFLP pattern. Distribution of sequences within the individual clone libraries from YE are presented in Figs. 3A through 3C. Of the 48 total clones screened from the CA library, only 3 OTUs were observed from each of the CA bands, indicating less diversity in this library. Distribution of sequences within the individual clone libraries from CA are presented in Figs. 4A through 4C.
Phylogenetic analyses from the clone libraries are represented in Fig. 5. In YE-band 1, 35% of the clones were closely related with Pseudoalteromonas spp., 25% to Marinomonas spp., 24% to Vibrio spp., and 16% to Bdellovibrio spp., respectively. This representation changed slightly to Pseudoalteromonas spp. 30%, Marinomonas spp. 20%, and Vibrio spp. 10% in chemotaxis band 2; however Bdellovibrio spp. were higher in this band, which accounted for approximately 40% of phylotypes in the library (Figs. 3A through 3C). In band 3 (Figs. 2A and 2B), this situation was different, with Bdellovibrio spp. being represented by almost 50%, and a new OTU could be identified that was represented by Bacteriovorax spp., accounting for 19% of the clone library (Fig. 3C). Also of interest in this band library was the low representation of Pseudoalteromonas spp. 15%, Marinomonas spp. 10%, and Vibrio spp. 6%.
Phylogenetic tree represents partial 16S rDNA gene sequences from domain “Bacteria” constructed with TREECON for Windows version 1.3b using neighbor joining. A total of 48 clones were compared from each chemotaxis band library, two representatives from each phylotype were sequenced, and 16S rDNA gene sequences were compared with their closest cultured phylogenetic relative from the NCBI database. Clones suffixed with YE and CA originated from yeast extract or Casamino acids. Numbers at nodes represent bootstrap values (100 times resampling analysis); only values > 50% are presented. Arthrobacter globiformis was used as outgroup.
In CA libraries (Figs. 4A through 4C), band 1 consisted of 43% of Pseudoalteromonas spp., 39% of Marinomonas spp., and 18% of Bdellovibrio spp., respectively. Similarly, band 2 consisted of Pseudoalteromonas spp. 35%, Marinomonas spp. 30%, and Bdellovibrio spp., which were higher at 35% of the phylotypes in the library. In band 3, however, the same situation was observed as that in YE, with Pseudoalteromonas spp. 19%, Marinomonas spp. 6%, and Bdellovibrio spp., which were higher at 75%. Of particular interest in the CA libraries was the complete absence of sequences associated with Vibrio spp., indicating that CA may not be a good chemoattractant or that Vibrio spp. were consumed to extinction by the predators (Figs. 4A through 4C).
Although this study is based on a single composite sample from three samples that were collected within the same site, it has important ecologic conclusions. This is one of the first clues as to how BALOs may potentially respond to shifts in natural populations of bacteria in the environment. As the bacterial population concentrates in an area driven by migration toward a possible source of nutrition, BALOs are potentially attracted to the area of bacterial concentration. This situation may be analogous to the chemotaxis assays from our study, indicating that the motile bacterial assemblages first converge around YE and CA, leading to higher population densities around the chemoattractant, triggering predator bacterial communities to sense and respond to the presence of high numbers of prey bacterial communities, resulting in predation. This may be one of the reasons that BALOs were found in high numbers from aquatic biofilms or submerged surfaces but not in surrounding waters [8, 24]. BALOs potentially sensed and responded to high populations of bacterial prey associated within biofilms compared with low numbers found in the water column. In fact, B. bacteriovorus has been shown to be chemotactic to high concentrations (108 cells/mL) of prey cells [20], and, more recently, methyl-accepting chemotaxis proteins have been identified from this microorganism, which indicate that they sense prey bacteria [10].
As motile bacteria migrated toward the attractants, they formed three concentric bands. The inner band closest to the attractant was found to have greater numbers of the motile heterotrophic bacteria and fewer numbers of BALOs. This is reversed in the outermost bands, in which the numbers of predator bacteria drastically increased and, correspondingly, the other motile bacteria decreased (Figs. 3 and 4 and Table 1). It is highly likely that the bacterial assemblages closest to the chemotactic agents were exposed to a greater concentration of the nutrient, which may have promoted growth of the bacterial assemblages during the 12-hour incubation period. To the contrary, the bacteria in the outer band farthest away from the chemotactic agent were exposed to a lower concentration of the nutrient and did not exhibit an equivalent level of growth. Previous observations have shown that BALOs show greater growth, as measured by larger plaque size on plates or more rapid clearing of prey in broth-enrichment cultures when grown on a minimal rather than an enriched medium. Based on this, it appears that the more nutrient-enriched inner band of bacteria yielded lower production of BALOs than the less nutrient-rich outer bands. Although the experiment was designed using a time frame to avoid bacterial growth, it is possible that one or two rounds of cell division(s) could have occurred in the inner bands during the 12-hour incubation period.
Pseudoalteromonas spp., Marinomonas spp., and Vibrio spp. identified from YE and CA bands are Gram-negative and motile [6], and therefore, all could serve as potential prey for Bdellovibrio and Bacteriovorax spp. However, the data also indicate a prey preference. After comparing outer chemotaxis band 3 from YE and CA (where predatorial groups are more prevalent), the clone libraries are represented by Pseudoalteromonas spp. > Marinomonas spp. > Vibrio spp. The most likely explanation for this is that Vibrio spp. are preferentially predated, as previously reported by other investigators [7, 12, 14, 24].
This study indicates that predator bacteria sense and respond to the changes in the shifts of microbial prey population in the environment and that chemotaxis can be used as a culture-independent tool to characterize motile, predatorial bacteria without the need to culture them in dual predator–prey systems from aquatic ecosystems.
Literature Cited
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Bhushan B, Samanta SK, Chauhan A, Chakraborti AK, Jain RK (2000) Chemotaxis and biodegradation of 3-methyl-4-nitrophenol by Ralstonia sp. SJ98. Biochem Biophys Res Commun 275:129–133
Chauhan A, Ogram A (2006) Fatty acid-oxidizing consortia along a nutrient gradient in the Florida Everglades. Appl Environ Microbiol 72:2400–2406
Chauhan A, Ogram A, Reddy KR (2004) Syntrophic-methanogenic associations along a nutrient gradient in the Florida Everglades. Appl Environ Microbiol 70:3475–3484
Cole JR, Chai B, Marsh TL, Farris RJ, Wang Q, Kulam SA, et al. (2003) The Ribosomal Database Project (RDP-II): Previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy. Nucleic Acids Res 31:442–443
Dworkin M (2005) Prokaryotic life cycles. In: Dworkin M, et al. (eds) The prokaryotes: An evolving electronic resource for the microbiological community, 3rd ed. Online BETA–release 3.20 (12/31/2005)
Jerkovitch E (2000, November 2) The Genus Bdellovibrio. In: Dworkin M, et al. (eds) The prokaryotes: An evolving electronic resource for the microbiological community, 3rd ed. Release 3.7. New York, NY, Springer-Verlag. Available at: http://link.springer-ny.com/link/service/books/10125/
Kelley JI, Williams HN (1992) Bdellovibrios in Callinectus sapidus, the blue crab. Appl Environ Microbiol 58:1408–1410
LaMarre AG, Straley SC, Conti SF (1997) Chemotaxis toward amino acids by Bdellovibrio bacteriovorus. J Bacteriol 131:201–207
Lambert C, Smith MC, Sockett RE (2003) A novel assay to monitor predator-prey interactions for Bdellovibrio bacteriovorus 109 J reveals a role for methyl-accepting chemotaxis proteins in predation. Environ Microbiol 5:127–132
Lane DJ (1991) 16S/23S rRNA sequencing. In: E. Stackebrandt E, Goodfellow M (eds) Nucleic acid techniques in bacterial systematics. New York: NY, Wiley, pp 115–175
Pineiro SA, Sahaniuk GE, Romberg E, Williams HN (2004) Predation pattern and phylogenetic analysis of Bdellovibrionaceae from the Great Salt Lake, Utah. Curr Microbiol 48:113–117
Rendulic S, Jagtap P, Rosinus A, Eppinger M, Baar C, Lanz C, et al. (2004) A predator unmasked: Life cycle of Bdellovibrio bacteriovorus from a genomic perspective. Science 303:689–692
Rice TD, Williams HN, Turng BF (1998) Susceptibility of bacteria in estuarine environments to autochthonous Bdellovibrios. Microbiol Ecol 35:256–264
Samanta SK, Bhushan B, Chauhan A, Jain RK (2000) Chemotaxis of a Ralstonia sp. SJ98 toward different nitroaromatic compounds and their degradation. Biochem Biophys Res Commun 269:117–123
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: A laboratory manual, 2nd ed. Cold Spring Harbor, NY, Cold Spring Harbor Laboratory Press
Schoeffield AJ, Williams HN (1990) Efficiencies of recovery of Bdellovibrios from brackish-water environments by using various bacterial species as prey. Appl Environ Microbiol 56:230–236
Starr MP (1975) Bdellovibrio as symbiont: The associations of Bdellovibrios with other bacteria interpreted in terms of a generalized scheme for classifying organismic associations. Symp Soc Exp Biol 93–124
Straley SC, Conti SF (1974) Chemotaxis in Bdellovibrio bacteriovorus. J Bacteriol 120:549–551
Straley SC, Conti SF (1977) Chemotaxis by Bdellovibrio bacteriovorus toward prey. J Bacteriol 132:628–640
Straley SC, LaMarre AG, Lawrence LJ, Conti SF (1979) Chemotaxis of Bdellovibrio bacteriovorus toward pure compounds. J Bacteriol 140:634–642
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The ClustalX windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 24:4876–4882
Van de Peer Y, De Wachter R (1994) TREECON for Windows: A software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput Applic Biosci 10:569–570
Williams HN, Sheffield AJ, Guether D, Kelley J, Shah D, Falker WA (1995) Recovery of bdellovibrios from submerged surfaces and other aquatic habitats. Microb Ecol 29:39–48
Acknowledgments
Funding for this study was provided by National Science Foundation Grant No. OCE0455276 and National Oceanic and Atmospheric Administration Grant No. NA17AE1624. Dr. S. A. Piñeiro, University of Maryland, is acknowledged for help during the course of this study. We acknowledge the anonymous reviewers who contributed immensely in strengthening this manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Chauhan, A., Williams, H.N. Response of Bdellovibrio and Like Organisms (BALOs) to the Migration of Naturally Occurring Bacteria to Chemoattractants. Curr Microbiol 53, 516–522 (2006). https://doi.org/10.1007/s00284-006-0292-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-006-0292-2