Abstract
Metarhizium anisopliae infects arthropods via a combination of specialized structures and cuticle degradation. Hydrolytic enzymes are accepted as key factors for the host penetration step and include chitinases. The characterization of the chi2 chitinase gene from M. anisopliae var. anisopliae is reported. The chi2 gene is interrupted by two short introns and is 1,542-bp long, coding a predicted protein of 419 amino acids with a stretch of 19 amino acid residues displaying characteristics of signal peptide. The predicted chitinase molecular mass is 44 kDa with a mature protein of 42 kDa and a theoretical pI of 4.8. The comparison of the CHI2 predicted protein to fungal orthologues revealed similarity to the glycohydrolase family 18 and a phylogenetic analysis was conducted. The chi2 gene is up-regulated by chitin as a carbon source and in conditions of fungus autolysis, and is down-regulated by glucose. This regulation is consistent with the presence of putative CreA/Crel/Crr1 carbon catabolic repressor binding domains on the regulatory sequence.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Metarhizium anisopliae is a well-known, broad-range arthropod pathogen, which is applicable in the biological control of several insect pests, including vectors for human diseases, and ticks [13, 14, 16]. During fungal penetration through the host cuticle, hydrolytic enzymes such as proteases, chitinases, and lipases are produced and secreted and are proposed to be important for the initiation of the infection process, leading to cuticle transposition [28]. This range of extracellular enzymes that degrade the components of the host cuticle is produced when M. anisopliae is grown in arthropod cuticle or chitin as the sole carbon and nitrogen source [3, 18]. Chitinases are among these extracellular enzymes and some have been purified and characterized [15, 23, 29]. In fungi, chitinases have a physiological role in hyphal growth and morphogenesis [30] and have also been shown to be produced during host infection by entomopathogenic fungi [8]. Analysis of secreted chitinases in M. anisopliae revealed at least six isoforms (30, 33, 43.5, 45, 60, and 110 kDa) and only one has both the protein and the gene isolated and characterized (chi3 gene and CHIT30 chitinase) [8]. Three genes coding for chitinases were described in Metarhizium: chit1 gene and the ortholog chi1, code a 42-kDa endochitinase [2, 5, 26]; chi2 (partial sequence, AJ293217); and, chi3, which codes for an endo/exo-acting 30-kDa chitinase (CAC07217.1) [8]. However, the role of chitinases in arthropod pathogenesis is still not completely understood.
One approach to understand their function is the isolation of chitinase genes and the evaluation of their overexpression in bioassays. Thus, the overexpression of the M. anisopliae chit1 gene did not show altered pathogenicity to Manduca sexta [26]. In contrast, the M. anisopliae CHIT30 chitinase (chi3 gene) was shown to be produced during tick infection [8] and the overexpression of a Beauveria bassiana chitinase, gene Bbchit1, enhanced the virulence for aphids [12]. These three chitinases, CHIT1, CHIT30, and Bbchit1, share very low levels of similarity and da Silva et al. [8], analyzing the sequences from chitinases whose function in cell morphogenesis/growth or in pathogenesis was assigned, showed that chitinases with similar cellular roles may diverge in sequence. In Metarhizium, only one of the chitinase genes, the chit1 gene, was fully characterized [5]. For genes chi2 (AJ293217) and chi3 (AJ293218), only ESTs sequences are deposited.
Aiming to contribute to the investigation of the role of Metarhizium chitinase genes in the host infection process, we isolated and characterized the genomic and cDNA copies of the chi2 ortholog from M. anisopliae var. anisopliae. We also studied its transcription regulation under different culture conditions, including the use of host cuticle as a carbon/nitrogen source.
Materials and Methods
Organisms and growth conditions
M. anisopliae var. anisopliae strain E6 from the Microbial Genetics Group (Escola Superior de Agronomia Luiz de Queiroz, USP, Brazil) was maintained in complete Cove’s medium (MCc) media as previously described [9]. For RNA extraction, the fungus was grown in liquid Cove’s medium [9] with NaNO3 0.6%, supplemented with glucose (1%), N-acetylglucosamine, GlcNAc, (0.1%), Boophilus microplus cuticle (1%) [9], or chitin (0.8%). E. coli XL1-Blue (Stratagene, La Jolla, CA, USA) was used for genomic library construction and propagation of pUC18 plasmid and clones. Bacterial cultivation was in LB agar or LB broth [24].
Southern hybridization and library construction
Genomic DNA from M. anisopliae was extracted from mycelium [4] and (10 μg) digested with BamHI, EcoRI, HindIII, KpnI, PstI, or XbaI restriction endonucleases and fractionated on 0.8% agarose gel electrophoresis. The DNA was transferred to nylon blotting membrane HybondTM-N+, probed with a 615-bp amplicon from chi2 gene and hybridized using the ECL kit. The probe was generated using primers (Chi2F-GTGTTGGCCTTGTTGGCCTG and Chi2R-TACTGGCCAATTTG CTCGGC) (Invitrogen, São Paulo, Brazil) based on the reported ortholog chi2 gene partial sequence from M. anisopliae var. acridum [AJ293217].
Nucleotide sequencing and computational analysis
Inserts and amplicons were sequenced at the ACTGene Laboratory (Centro de Biotecnologia, UFRGS, Porto Alegre, RS, Brazil in an ABI-PRISM3100 Genetic Analyzer and analyzed by Blast using the NCBI server at http://www.ncbi.nlm.nih.gov/BLAST/ [1]. Signalscan Program (at http://www.dna.affrc.go.jp/PLACE/signalscan.html) was used to find a putative signal peptidase cleavage site. Chitinase amino acid sequences from fungi (CAC07216.1; AAB81998; AAN41259.1; CAG86633.1; EAL03025.1; EAL00460; CAG62749.1; BAA36223.1; AAS55554; NP_013388; AAA92642.1) were aligned using ClustalX [31] and a phylogenetic tree was constructed using the Molecular Evolutionary Genetics Analysis (MEGA) software [19] by the neighbor-joining method. Phylogenetic tree architecture confidence was evaluated by 10,000 bootstrap replications.
RT-PCR analysis and characterization of transcription start site
Total RNA extraction was performed as described [9]. First-strand cDNA synthesis was performed with M-MLV Reverse Transcriptase (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol with oligo dT as primer, using 1 μg of RNA. Amplification of the chi2 gene transcripts was performed as described above and RNA quantity was normalized by the amplification of tef1-α gene [21]. Amplicons were resolved by electrophoresis in 1.0% agarose gel. For isolation and characterization of 5′ ends from the chi2 gene, a 5′ RACE System was used (Version 2.0, Invitrogen), with 1 μg of RNA extracted from mycelium grown for 48 h with chitin as carbon source. Primers were Chi2R (see above) and an antisense primer (Chi2IR-GAATTGGGTTGGCAGTAC). The amplified product was purified and sequenced.
Results and Discussion
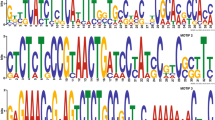
In order to clone the M. anisopliae var. anisopliae strain E6 complete chi2 gene sequence, PCR fragments were amplified (615 bp in size) using primers derived from the previously reported ortholog M. anisopliae var. acridum chi2 gene [AJ293217]. A recombinant clone with a 5.3-kb insert encompassing the chi2 gene was selected by colony hybridization from about 1,000 colonies from a plasmid library carrying M. anisopliae var. anisopliae strain E6 genomic DNA. The complete nucleotide sequence of the chi2 gene was determined (DQ011663) and the 419 amino acid residue ORF (CHI2) shows high similarity (97%) to the putative chitinase ortholog from M. anisopliae var. acridum [AJ293217]. The transcription initiation site was determined by sequencing a 311-bp cDNA amplicon generated by 5′ RACE reaction. The transcription initiation (G + 1) was identified and the ATG start codon was positioned at 95 bp from the transcription initiation site. The transcription initiation environment is ACATCAAG, which is similar to the consensus TCATCANC [10]. The chi2 gene is 1,542 bp long and is interrupted by two introns (210 and 72 bp long). In silico analysis of the 5′ flanking region revealed canonical CAAT and TATAA putative controlling elements at the appropriate distances. In addition, a consensus motif was found for the CreA/Crel/Crr1 carbon catabolic repressor, a negative regulator mediating carbon catabolism repression in A. nidulans and M. anisopliae (Fig. 1) [7, 25, 31].
M. anisopliae var. anisopliae strain E6 chi2 gene 5′ flanking sequence. Capitalized letters mark the first ATG. Putative CAAT and TATAA sequences and a putative CREA element (CCCCAC) are underlined. The transcription start site is shown in bold and the conserved transcription initiation sequence is shaded.
Chitinase CHI2 has a predicted molecular mass of 44 kDa and a putative signal peptidase cleavage site at V19, rendering a mature protein of 42 kDa with theoretical pI of 4.8. A chitinase with a similar molecular mass is coded by the chit1 gene from M. anisopliae var. anisopliae (CHIT42 endochitinase, AF027498) [2]. However, there is very little amino acid identity between the two chitinases (CHIT42 and CHI2) and their theoretical pIs differ (Fig. 2) [5, 26].
Phylogenetic neighbor-joining tree of M. anisopliae var. anisopliae strain E6 chi2 deduced amino acid sequences. Tree confidence was confirmed by 10,000 bootstraps and the numbers on the branches represent values for the bootstrap. The scale bar indicates the number of amino acid substitutions. For the GenBank accession number for the chitinase sequences analyzed, see Materials and Methods.
The comparison of the predicted CHIT2 chitinase to fungal orthologs revealed a similarity to the glycohydrolase family 18 [Pfam database; 11]. The consensus motif SXGG corresponding to a substrate-binding site was identified; however, the catalytic domain consensus motif (D1XXD2XD3XE), highly conserved among fungal chitinases [22, 25], has one amino acid substitution (D1→N) in the CHI2 sequence. A characteristic fungal-type cellulose-binding domain (CBD) present in the chi2 C-terminal sequence is similar to that of the 33-kDa chitinase gene in Trichoderma virens, predicted to encode a protein C-terminus with homology to the conserved family I cellulose-binding domain [17]. Apparently, the CDB in endochitinases increases hydrolytic activity towards insoluble substrates such as chitin-rich fungal cell walls in Trichoderma harzianum strains [20].
In order to evaluate the evolutionary relationships and to classify the predicted chitinase CHIT2 into bacterial-like or plant-like classes, a neighbor-joining phylogenetic tree was constructed. As shown in Figure 2, the tree collapsed in two clusters, one encompassing Metarhizium CHIT2 orthologues and CHIT30, as a plant-like class, and the other cluster with CHIT42 (coded by chit1 gene), a bacterial-like chitinase [8].
Previously, we reported the effect of different carbon sources on both total chitinase synthesis and secretion in M. anisopliae and the dual regulation depending on the GlcNAc concentration in the culture medium [3, 5, 18, 23]. To investigate the regulation of the chi2 gene, RT-PCR was conducted using RNA extracted from cultures amended with different carbon sources: 1% glucose, 0.1% GlcNAc, 1% tick cuticle, or 0.8% chitin. In glucose-added cultures, the sugar was supplemented every 24 hours to ensure its availability throughout fungal growth. The primers were targeted to a region spanning the first intron of the chi2 gene, generating an amplicon of 402 bp when cDNA was used as template for amplification, and a 615-bp amplicon for genomic DNA. To normalize RNA quantities, a 1,031-bp amplicon generated by primers directed to the tef 1-α gene (AY445082) was used. As shown in Figure 3, after 48 h M. anisopliae culture, chi2 gene transcripts were only detected when chitin was the carbon source. After 72 h, chi2 gene transcripts were also detected in cultures in the presence of GlcNAc or tick cuticle whilst transcripts were still not detected in the presence of glucose. Similar results were reported for the chit36 gene from T. harzianum [32] and for the Bchit1gene from B. bassiana [12]. In early cultures (18 or 30 h), chi2 gene transcripts were not detected (data not shown). In cultures with 0.1% GlcNAc, the chi2 gene transcripts were only detected after 72 hours of fungal growth, when the amino sugar was exhausted. This suggests that the expression of chi2 gene may be triggered by autolysis. Similar results were described for the ech42 gene that encodes chitinase ECH42 in T. harzianum, in which significant ech42 expression was detected only after prolonged carbon starvation [6]. In tick cuticle and chitin, the M. anisopliae chi2 gene transcription was induced, indicating that synthesis is subject to regulation by the substrate.
RT-PCR of chi2 gene transcripts in different culture conditions. M. anisopliae var. anisopliae strain E6 was cultured for 48 h (A) or 72 h (B). Upper gel in each panel represents chi2 gene transcripts. Lower gel in each panel represents tef1α gene transcripts, used to normalize RNA quantity. Numbers at the left represent size in bp. Numbers at the right represent the amplicon size in bp. Lanes: (1) control with no template added; (2) genomic DNA from gene chi2 template, resulting in a 615-bp product; RNA from cultures in minimal medium added of: (3) 1% glucose; (4) 0.1% GlcNAc; (5) 1% tick cuticle; (6) 0.8% chitin. cDNA amplicon from chi2 gene is 402 bp long.
In fungi, chitinases have a physiological role in hyphal growth and morphogenesis. The relevance of chitinase production and secretion during the penetration of host cuticle by fungal pathogens is not fully understood. To date the exo/endochitinase CHIT30 of M. anisopliae strain E6 was shown to be present during B. microplus infection [8] and only the chit1 and Bbchit1 chitinase genes, from M. anisopliae and B. bassiana, respectively, have been investigated in the insect fungus pathogenic context. The CHIT42 (chit1 gene) chitinase from M. anisopliae was shown to have no effect on virulence to insects [26], while overproduction of Bbchit1 did increase the virulence of B. bassiana for aphids [12].
Seidl et al. [27] showed that both chi2 and chi3 genes from Metarhizium are related to chitinase genes from mycoparasites (Trichoderma) and to no other chitinases described in all other ascomycetous genomes. The authors suggest that these chitinases probably have special functions in host chitin degradation during parasitism. Indeed, the related Hypocrea jecorina (anamorph: Trichoderma reesei) chitinase gene chi18-13 is up-regulated in the presence of host cell wall [27] as is the Metarhizium chi2 gene in the presence of host cuticle (Fig. 3B).
The cloning and characterization of the chitinase genes is important to elucidate the relationships between chitinases and virulence in insects/ticks or in the fungus morphogenesis. In situ immunodetection of the protein and overexpression and gene silencing experiments are necessary to elucidate its biological role in Metarhizium.
Literature Cited
Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Baratto CM, da Silva MV, Santi L, Passaglia L, Schrank IS, Vainstein MH, Schrank A (2003) Expression and characterization of the 42 kDa chitinase of the biocontrol fungus Metarhizium anisopliae in Escherichia coli. Can J Microbiol 49:723–726
Barreto CC, Staats CC, Schrank A, Vainstein MH (2004) Distribution of chitinases in the entomopathogen Metarhizium anisopliae and effect of N-acetylglucosamine in protein secretion. Curr Microbiol 48:102–107
Bogo MR, Queiroz MV, Silva DM, Gimenez Pecci MP, Azevedo JL, Schrank A (1996) Double-stranded RNA and isometric virus-like particles in the entomopathogenic fungus Metarhizium anisopliae. Mycol Res 100:1468–1472
Bogo MR, Rota CA, Pinto H Jr., Ocampos M, Correa CT, Vainstein MH, Schrank A (1998) A chitinase encoding gene (chit1 gene) from the entomopathogen Metarhizium anisopliae: isolation and characterization of genomic and full-length cDNA. Curr Microbiol 37:221–225
Carsolio C, Gutierrez A, Jimenez B, Van MM, Herrera-Estrella A (1994) Characterization of ech-42, a Trichoderma harzianum endochitinase gene expressed during mycoparasitism. Proc Natl Acad Sci USA 91:10903–10907
Cubero B, Scazzocchio C (1994) Two different, adjacent and divergent zinc finger binding sites are necessary for CREA-mediated carbon catabolite repression in the proline gene cluster of Aspergillus nidulans. EMBO J 13:407–415
da Silva MV, Santi L, Staats CC, da Costa AM, Colodel EM, Driemeier D, Vainstein MH, Schrank A (2005) Cuticle-induced endo/exoacting chitinase CHIT30 from Metarhizium anisopliae is encoded by an ortholog of the chi3 gene. Res Microbiol 156:382–392
Dutra V, Nakazato L, Broetto L, Silveira SI, Henning VM, Schrank A (2004) Application of representational difference analysis to identify sequence tags expressed by Metarhizium anisopliae during the infection process of the tick Boophilus microplus cuticle. Res Microbiol 155:245–251
Eberle J, Russo VE (1994) Neurospora crassa blue light-inducible gene bli-3. Biochem Mol Biol Int 34:737–744
Falquet L, Pagni M, Bucher P, Hulo N, Sigrist CJ, Hofmann K, Bairoch A (2002) The PROSITE database, its status in 2002. Nucleic Acids Res 30:235–238
Fang W, Leng B, Xiao Y, Jin K, Ma J, Fan Y, Feng J, Yang X, Zhang Y, Pei Y (2005) Cloning of Beauveria bassiana chitinase gene Bbchit1 and its application to improve fungal strain virulence. Appl Environ Microbiol 71(1):363–370
Frazzon AP, da Silva VJI, Masuda A, Schrank A, Vainstein MH (2000) In vitro assessment of Metarhizium anisopliae isolates to control the cattle tick Boophilus microplus. Vet Parasitol 94:117–125
Gillespie JP, Bateman R, Charnley AK (1998) Role of cuticle-degrading proteases in the virulence of Metarhizium spp. for the desert locust, Schistocerca gregaria. J Invertebr Pathol 71:128–137
Kang SC, Park S, Lee DG (1999) Purification and characterization of a novel chitinase from the entomopathogenic fungus, Metarhizium anisopliae. J Invertebr Pathol 73:276–281
Kanzok SM, Jacobs-Lorena M (2006) Entomopathogenic fungi as biological insecticides to control malaria. Trends Parasitol 22:49–51
Kim DJ, Baek JM, Uribe P, Kenerley CM, Cook DR (2002) Cloning and characterization of multiple glycosyl hydrolase genes from Trichoderma virens. Curr Genet 40:374–384
Krieger de MC, Schrank A, Vainstein MH (2003) Regulation of extracellular chitinases and proteases in the entomopathogen and acaricide Metarhizium anisopliae. Curr Microbiol 46:205–210
Kumar S, Tamura K, Jakobsen IB, Nei M (2001) MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17:1244–1245
Limon MC, Chacon MR, Mejias R, gado-Jarana J, Rincon AM, Codon AC, Benitez T (2004) Increased antifungal and chitinase specific activities of Trichoderma harzianum CECT 2413 by addition of a cellulose binding domain. Appl Microbiol Biotechnol 64:675–685
Nakazato L, Dutra V, Broetto L, Staats CC, Vainstein MH, Schrank A (2006) Development of an expression vector for Metarhizium anisopliae based on the tef-1alpha homologous promoter. Appl Microbiol Biotechnol 10:1–8
Orikoshi H, Baba N, Nakayama S, Kashu H, Miyamoto K, Yasuda M, Inamori Y, Tsujibo H (2003) Molecular analysis of the gene encoding a novel cold-adapted chitinase (ChiB) from a marine bacterium, Alteromonas sp. strain O-7. J Bacteriol 185:1153–1160
Pinto AS, Barreto CC, Schrank A, Ulhoa CJ, Vainstein MH (1997) Purification and characterization of an extracellular chitinase from the entomopathogenic Metarhizium anisopliae. Can J Microbiol 43:322–327
Sambrook J, Russel DW (2001) Molecular cloning: A laboratory manual, 3rd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory
Screen S, Bailey A, Charnley K, Cooper R, Clarkson J (1997) Carbon regulation of the cuticle-degrading enzyme PR1 from Metarhizium anisopliae may involve a trans-acting DNA-binding protein CRR1, a functional equivalent of the Aspergillus nidulans CREA protein. Curr Genet 31:511–518
Screen SE, Hu G, St. Leger RJ (2001) Transformants of Metarhizium anisopliae sf. anisopliae overexpressing chitinase from Metarhizium anisopliae sf. acridum show early induction of native chitinase but are not altered in pathogenicity to Manduca sexta. J Invert Pathol 78:260–266
Seidl V, Huemer B, Seiboth B, Kubicek CP (2005) A complete survey of Trichoderma chitinases reveals three distinct subgroups of family 18 chitinases. FEBS J 272:5923–5939
St. Leger RJ, Charnley AK, Cooper RM (1986) Cuticle-degrading enzymes of entomopathogenic fungi: Mechanisms of interaction between pathogen enzymes and insect cuticle. J Invert Pathol 47:295–302
St. Leger RJ, Cooper RM, Charnley AK (1991) Characterization of chitinase and chitobiase produced by the entomopathogenic fungus Metarhizium anisopliae. J Invert Pathol 58:415–426
Takaya N, Yamazaki D, Horiuchi H, Ohta A, Takagi M (1998) Cloning and characterization of a chitinase-encoding gene (chiA) from Aspergillus nidulans, disruption of which decreases germination frequency and hyphal growth. Biosci Biotechnol Biochem 62:60–65
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Viterbo A, Haran S, Friesem D, Ramot O, Chet I (2001) Antifungal activity of a novel endochitinase gene (chit36) from Trichoderma harzianum Rifai TM. FEMS Microbiol Lett 200:169–174
Acknowledgments
This work was supported by FAPERGS (Fundação de Amparo a Pesquisa do Estado do Rio Grande do Sul), CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico), PADCT (Programa de Apoio ao Desenvolvimento Científico e Tecnológico), and CAPES (Coordenação de Aperfeiçoamento de Pessoal de Ensino Superior). We thank Irene Schrank for a critical reading of the manuscript and Giancarlo Pasqualli for the use of sequencing facilities.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Baratto, C.M., Dutra, V., Boldo, J.T. et al. Isolation, Characterization, and Transcriptional Analysis of the Chitinase chi2 Gene (DQ011663) from the Biocontrol Fungus Metarhizium anisopliae var. anisopliae . Curr Microbiol 53, 217–221 (2006). https://doi.org/10.1007/s00284-006-0078-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-006-0078-6