Abstract
A new Bacillus thuringiensis strain, Bt185, was isolated from HeBei soil samples in China. Observations after transmission electron microscopy found that the strain produced spherical parasporal inclusions similar to that of the B. thuringiensis subsp. japonensis Buibui strain, which showed toxicity to both Anomala corpulenta and Popillia japonica. The plasmid profile seen on an agarose gel revealed that Bt185 contained six large bands of 191 kb, 161 kb, 104 kb, 84 kb, 56 kb, and 37 kb. Sodium dodecyl sulfate–polyacrylamide gel electrophoresis analysis revealed one major band with an estimated molecular mass of 130 kDa. Polymerase chain reaction–restriction fragment length polymorphism results showed that a novel cry8-type gene sequence was found in the Bt185 strain. When we screened for this novel gene sequence, an additional novel cry8-type gene was isolated, having a partial sequence of 2340 bp and encoding a protein of 780 amino acids. Bioassay results showed that Bt185 had no toxicity against several Coleopteran and Lepidopteran pests. However, Bt185 exhibited toxicity against larvae of the Asian cockchafer, Holotrichia parallela. This is the first report of the occurrence of a Bacillus strain that has insecticidal activity against Holotrichia parallela larvae.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Bacillus thuringiensis is a Gram-positive bacterium that produces one or more insecticidal proteins, deposited in an intracellular parasporal crystal form, during sporulation [16]. These crystalline inclusions often exhibit strong and specific toxicity against several orders of insects. This characterization makes B. thuringiensis one of the most promising and environmentally sound microbial agents for control of insect pests in both agriculture and medicine [6].
The cry gene family is a large family of homologous genes, encoding proteins active against a large number of insect larvae [16]. Proteins of the Cry3, Cry6, Cry7, Cry8 [7, 11, 12, 15], Cry18 [24], and Cry43 [23] classes, as well as the binary toxins, Cry34A-Cry35A, are active against insects of the order Coleoptera (beetles and weevils) [3]. Among these classes, cry8-type, cry18-type, and cry43-type genes are toxic to Scarabaeidae larvae [12, 18, 23, 24]. Ohba et al. [12] first isolated a B. thuringiensis subsp. japonensis strain, Buibui, which was specific to only Scarabaeid larvae. In 1994, a new cry gene, cry8Ca, was cloned from this strain and its encoded protein exhibited larvicidal activity against Anomala cuprea Hope, Anomala rufocuprea Motschulsky, and Popillia japonica Newman [15]. Currently, approximately 10 cry8-type genes have been found, including cry8Aa and cry8Ba that were cloned by the American Mycogen company from the B. thuringiensis subsp. kumamotoensis, and have activity against Cotinis sp (such as Cotinis nitida, June beetle) (US Patent 5554534). The cloned cry8Bb1 and cry8Bc1 proteins were found to possess activity against Western corn rootworm and have been used in transgenic corn studies [1].
The larvae of cockchafers are important insect pests in agriculture, horticulture, and forestry in both Europe and Asia (China) [17, 22]. In China, Holotrichia parallela is one of the severe pests of the Scarab family and is currently endangering large areas of peanut, soybean, and sweet potato crops, especially in the HeBei, AnHui, and JiangSu provinces [22]. The larvae destroy the underground parts of the plants and cause a significant reduction of output and great economic loss. Several approaches for both biological and chemical pest control are under investigation; however, all methods examined so far are only minimally effective [8, 13, 17]. Scarab beetles have evolved efficient mechanisms of defense [21]. Thus, they were difficult to control [8, 13, 17]. Until recently, no B. thuringiensis strain was found to control Holotrichia parallela effectively.
In this report, we isolated and characterized the new B. thuringiensis strain Bt185 from Chinese soil samples that exhibited toxicity against the larvae of Holotrichia parallela (Scarabaeidae). Furthermore, two novel cry8-type genes were found in this strain. Analyses performed include transmission electron microscopy, sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE), plasmid profiling, polymerase chain reaction (PCR) identification of novel gene, and insect bioassays.
Materials and Methods
Bacteria and growth conditions
The B. thuringiensis strain used in this study was isolated from soil samples in Shunping county, HeBei province, China. The B. thuringiensis subsp. japonensis strain Buibui was used as a reference strain. B. thuringiensis strains were incubated for 3 days in cross-baffled flasks at 30°C with shaking at 250 rpm in LB medium (1% tryptone, 0.5% yeast extract, and 1% NaCl; pH 7.0).
Transmission electron microscopy
Bt185 strain spores and crystals were harvested by centrifugation at 12,000 rpm for 10 min, washed three times in distilled water, and then fixed in 3% glutaraldehyde in phosphate buffer. The sample was subsequently dehydrated in an ethanol–propylene oxide dilution series and embedded in an Epon resin mixture (Sangon Ltd., China). Ultrathin sectioning was done on a Reichert Ultracut ultramicrotome (Leika Aktiengesellschaft, Wien, Austria), and the sections were stained with uranyl acetate and lead citrate, examined, and photographed using a New Bio-TEM H-7500 electron microscope (Hitachi Ltd., Japan), operating at an accelerating voltage of 80 kV.
Plasmid isolation and plasmid DNA pattern
Plasmid DNA was isolated according to the method of Song et al. [19]. The plasmid DNA was fractionated in a 1% (wt/vol) SeaPlaque GTG agarose pulsed-field gel (FMC BioProducts) using the CHEF Mapper XA (BioRad, USA) in 0.5 × TBE with parameters of 6 V/cm, 12.5°C, and 120° angle, using a 90-s pulse for 18 h.
SDS-PAGE
At appropriate intervals (approximately every 2–4 h), Bt185 strain culture was collected by centrifugation and SDS-PAGE was performed on an 8% gel, according to the method of Laemmli [10]. The molecular weights were estimated by comparison with the protein ladder.
Identification of cry genes by PCR–restriction fragment length polymorphism
The Bt185 strain was incubated overnight at 30°C at 230 rpm in LB medium. After centrifugation, the 1-mL pellet was resuspended in 50 μL sterile water, boiled for 10 min, and centrifuged at 12,000 rpm for 5 min. The supernatant was collected as a template for PCR amplification. The primer pairs used for identification were from the sequences of cry1 genes [9], cry11 genes [19], cry2-4 genes, cry10 genes [20], and cry5-9 genes (Table 1), and the primer pairs were general primers. PCR was carried out in a 50-μL volume, containing 1-μL template DNA, 0.4 mM deoxynucleotide triphosphates, 0.2 μM primer, 1.5 U Taq DNA polymerase (Promega), and reaction buffer. Amplification was carried out for 30 cycles of denaturation at 94°C for 1 min, annealing at 54°C for 1 min, and extension at 72°C for 2 min, with an additional step of extension at 72°C for 10 min. A restriction endonuclease reaction was performed in a 20-μL volume containing the PCR products (0.5–1.0 μg) and 0.5 U restriction endonucleases DraI and EcoO109I, using the protocol described by Sambrook et al. [14].
Partial cloning of the novel cry8-type gene
Bt185 plasmid DNA was digested by endonuclease KpnI. DNA fragments were ligated into the pBlueScript II SK (+) cloning vector. The recombinant plasmids were transformed into Escherichia coli JM110 cells. Transformed E. coli cells were grown on LB plates containing 100 μg ampicillin per milliliter to construct a DNA library. The cry8-specific primers S5un8/S3un8 were used to screen the library. A 2.0-kb KpnI fragment was subsequently cloned into the pBlueScript II SK (+) vector to produce a recombinant plasmid pSS162 and was sequenced using an automated DNA sequencer (ABI-3730XL). The sequences were analyzed using a BLAST database search program [2].
Insect bioassays
Spores and crystals of Bt185 were harvested and resuspended in sterilized water. The amount of spores in suspensions was calculated, and then 40-mL suspensions with twofold serial dilutions were added to 200 g soil containing potato pieces sterilized under ultraviolet light in a plastic tube and containing 20 5-day-old Holotrichia parallela larvae and 20 15-day-old larvae of Anomala corpulenta. Bioassays were conducted at 25°C with a soil humidity of 18–20%. As a negative control, distilled water was mixed with the soil containing potato pieces sterilized under ultraviolet light. Larval mortality was scored after incubation for 7 days and again after 14 days. Insecticidal activity against larvae of Helicoverpa armigera (first-instar), Tribolium castaneum (first-instar), and Tenebrio molitor (first-instar) was measured after incorporation of a suspension of twofold serial dilutions of the spore-crystal mixture into the larval artificial diet. Toxicity studies on third-instar larvae of the diamondback moth (Plutella xylostella) were conducted on fresh cabbage disk leaves by leaf dip bioassay [19]. Ten larvae were each given the artificial diet or placed on a leaf disk. Bioassays were repeated at least twice, and LC50 values were calculated using probit analysis [5].
Results and Discussion
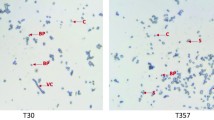
The parasporal crystals of Bt185 were observed to be spherically shaped (Fig. 1), similar to B. thuringiensis strain Buibui that has previously been shown to be toxic to both Anomala corpulenta and Popillia japonica [12]. The crystals were approximately 500 nm in length when observed with transmission electron microscopy. The plasmid profile of Bt185 revealed six large bands of 191 kb, 161 kb, 104 kb, 84 kb, 56 kb, and 37 kb. The plasmid profile of Bt185 was found to be clearly different from B. thuringiensis strain Buibui (Fig. 2), especially in regard to the small plasmid bands.
For detection of the crystal genes in the Bt185 strain, PCR analysis was performed with 11 specific primer pairs. The PCR results showed a DNA fragment similar to the cry8-type genes, whereas the cry1, cry1I, cry2, cry3, cry4, cry5, cry6, cry7, cry9, and cry10 genes were not detected. The amplified PCR fragment of the cry8 gene was about 1.212 kb in both the Bt185 and Buibui strains, and restriction fragment length polymorphism patterns showed that the Bt185 cry8 gene was different from that from the reference strain (Fig. 3). When we screened for novel cry8-type genes, we identified two novel cry8-type genes in strain Bt185. One was cloned and named cry8Ea1, which was confirmed by the B. thuringiensis Pesticidal Crystal Protein Nomenclature Committee [4]. The other 2.0-kb KpnI fragment (2.340 kb), from recombinant plasmid pSS162, was sequenced (GenBank accession number AY897354) and found to be the partial gene. Further study should include cloning of the full-length gene. The obtained sequences encoded the C-terminus of a protein with 780 amino acids. Protein Blast analysis showed that the C-terminus had maximal sequence identities with Cry8Ea (80%) (unpublished data), Cry8Aa (72%), and Cry8Ba (70%). In the C-termini of this protein and Cry8Ea1 (unpublished data), 519 amino acids were identical, but the 211 amino acids near the N-termini showed only 30% sequence identity. Thus, another potential novel cry8-type gene appears to exist in strain Bt185. Previously reported cry8Aa and cry8Ba proteins, isolated from B. thuringiensis subsp. kumamotoensis in 1994, were found to possess high activity to Cotinis sp (June beetle) (US Patent 5554534), and two novel cry8-type genes were also found in strain Bt185. These results suggest that the cry8-type genes often co-exist in B. thuringiensis strains toxic to the Scarabs.
Bacillus thuringiensis strains Bt185 and Buibui after polymerase chain reaction (PCR)–restriction fragment length polymorphism analysis. Lanes 1 and 2: Bt185 and Buibui PCR products, respectively (using cry8-specific primers); Lanes 3 and 4: Bt185 and Buibui PCR products, respectively, digested with endonucleases DraI and EcoO109; Lane 5: DNA ladder.
Cry8 proteins are composed of 1160–1210 amino acids and have a molecular weight in the range of 128–137 kDa [12, 18]. The protein profile of Bt185 was similar to the reference strain Buibui, as observed by Sato et al. [15]. Both strains have one major band with an estimated molecular mass of 130 kDa (Fig. 4). The crystal proteins of Bt185 could be detected after 20 h of growth, as shown in Fig. 4 (Lane 2). With an increase in the time of growth, the concentration of toxin protein was seen to increase gradually, and after 28 h (Lane 4), no additional significant changes in protein concentration were observed. The growth curve of strain Bt185 showed that during the initial 14 h, the optical density of the culture, measured at 600 nm, increased exponentially with time before reaching stationary phase. Combining the protein profile with the growth curve of strain Bt185, we came to the conclusion that the expression of the cry genes of Bt185 was sporulation dependent.
Sodium dodecyl sulfate–polyacrylamide gel electrophoresis profile of parasporal inclusion proteins from Bacillus thuringiensis strains Bt185 and Buibui. Lanes 1–5: Bt185 parasporal inclusion protein at 18 h, 20 h, 24 h, 28 h, and 32 h growth; Lanes 6 and 7: Buibui parasporal inclusion protein at 28 h and 32 h growth. M: molecular weight marker.
Bioassay results showed that the Bt185 strain possessed activity against Holotrichia parallela larvae with an LC50 of 0.9464 (108 cfu/mL), but no toxicity against Anomala corpulenta, Tribolium castaneum, Tenebrio molitor, cotton bollworm (Helicoverpa armigera), or diamondback moth (Plutella xylostella) (data not shown). In previous reports, B. thuringiensis strain Buibui was found to express the cry8Ca gene that exhibits high larvicidal activity against Anomala corpulenta and Popillia japonica [12]. B. thuringiensis strain SDS-502 was shown to possess the cry8Da2 gene that was highly toxic to Anomala cuprea [18], and B. thuringiensis subsp. kumamotoensis was shown to express cry8Aa and cry8Ba, which both were shown to have activity against Cotinis sp. (June beetle) (US Patent 5554534). The cry8Bb1 and cry8Ca1 proteins were found to be toxic to Leptinotarsa decemlineata, Diabrotica virgifera virgifera, and Diabrotica undecimpunctata howardi (WO Patent 02/347742 A2). Until recently, no B. thuringiensis strain was reported to control Holotrichia parallela. Holotrichia parallela and Anomala corpulenta have been shown to often damage crops simultaneously [22]. Scarab beetles have evolved efficient mechanisms of defense [21]. Thus, they were difficult to control [8, 13, 17]. Strain Bt185 could provide a new tool to control Scarabaeidae by the development of a new B. thuringiensis formulation.
In the present study, one of the most striking aspects that we found was a new B. thuringiensis strain, Bt185, which had high insecticidal activity against the Asian cockchafer Holotrichia parallela larvae. To our knowledge, no bacterium but this strain has been found to be toxic to the cockchafer until now. We also found two novel cry8 genes in strain Bt185. We predict the use of the novel cry8 gene in transgenic plants to control Holotrichia parallela. Future studies will include the cloning and expression of the entire cry8-type gene in Bt185 strain, and the analysis of the expression of the two cry8-type genes at both the transcriptional and translational levels in order to understand the role of these two genes in the insecticidal activity against Holotrichia parallela larvae.
Literature Cited
Abad AR, Duck NB, Feng X, Flannagan RD, Kahn TW, Sims LE (2002) Genes encoding novel proteins with pesticidal activity against coleopterans. WO 02/34774 A2. 02 May 2002. E.I. DuPont de Nemours and company (US)
Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Baum JA, Chu CR, Rupar M, Brown GR, Donovan WP, Huesing JE, Ilagan O, Malvar TM, Pleau M, Walters M, Vaughn T (2004) Binary Toxins from Bacillus thuringiensis Active against the Western Corn Rootworm, Diabrotica virgifera virgifera LeConte. Appl Environ Microbiol 70:4889–4898
Crickmore N, Zeigler DR, Feitelson JE, Schnepf JVR, Lereclus D, Baum J, Dean DH (1998) Revision of the nomenclature for the Bacillus thuringiensis pesticidal crystal proteins. Microbiol Mol Biol Rev 62:807–813
Finney DJ (1971) Probit analysis. Cambridge, United Kingdom: Cambridge University Press
Glare TR, O’Callaghan M (2000) Bacillus thuringiensis: biology, ecology and safety. London, United Kingdom: John Wiley & Sons, p 27
Herrnstadt C, Soares GG, Wilcox ER, Edward DL (1986) A new strain of Bacillus thuringiensis with activity against coleopteran insects. BioTechnology 4:305–308
Keller S, David-Henriet AI, Schweizer C (2000) Insect pathogenic soil fungi from Melolontha melolontha control sites in the canton Thurgau. In: Keller S (ed), Integrate control of soil pest subgroup “Melolontha”. Conference Proceeding IOBC/WPRS, vol. 23, pp 73–78
Kuo WS, Chak KF (1996) Identification of novel cry-type genes from Bacillus thuringiensis strains on the basis of restriction fragment length polymorphism of the PCR-amplified DNA. Appl Environ Microbiol 62:1369–1377
Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685
Lambert B, Hofte H, Annys K, Jansens S, Soetaert P, Peferoen M (1992) Novel Bacillus thuringiensis insecticidal crystal protein with a silent activity against coleopteran larvae. Appl Environ Microbiol 58:2536–2542
Ohba M, Iwahana H, Asano S, Suzuki N, Sato R, Hori H (1992) A unique isolate of Bacillus thuringiensis serovar japonensis with a high larvicidal activity specific for Scarabaeid beetles. Lett Appl Microbiol 14:54–57
Peters A (2000) Susceptibility of Melolontha mololontha to Heterorhabditis bacteriophora, H. megidis and Steinernema glaseri. In: Keller S (ed) Integrated control of soil pest subgroup “Melolontha”. Conference Proceeding IOBC/WPRS, vol. 23, pp 39–45
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
Sato R, Takeuchi K, Ogiwara K, Minami Masayosi, Kaji Y, Suzuki N, Hori H, Asano S, Ohba M, Iwahana H (1994) Cloning, heterologous expression, and localization of a novel crystal protein gene from Bacillus thuringiensis serovar japonensis strain Buibui toxic to Scarabaeid insects. Curr Microbiol 28:15–19
Schnepf E, Crickmore N, Rie JV, Lereclus D, Baum J, Feitelson J, Zeigler DR, Dean DH (1998) Bacillus thuringiensis and its pesticidal crystal proteins. Microbiol Mol Biol Rev 62:775–806
Schnetter W, Mitterüller R, Fröschle M (1996) Control of the cockchafer Melolontha in the Kraichgau with NeemAzal-T/S. In: Keller S (ed), Integrated control of soil pests. Conference Proceeding IOBC/WPRS, vol. 19, pp 95–99
Shin-ichiro Asano, Yamashita C, Iizuka T, Takeuchi K, Yamanaka S, Cerf D, Yamamoto T (2003) A strain of Bacillus thuringiensis subsp. Galleriae containing a novel cry8 gene highly toxic to Anomala cuprea (Coleoptera: Scarabaeidae). Biol Control 28:191–196
Song FP, Zhang J, Gu A, Wu Y, Han LL, He KL, Chen ZY, Yao J, Hu YQ, Li GX, Huang DF (2003) Identification of cry1I-type genes from Bacillus thuringiensis strains and characterization of a novel cry1I-type gene. Appl Environ Microbiol 69:5207–5211
Song FP, Zhang J, Huang DF, Xie TJ, Yang ZW, Dai LY, Li GX (1998) Establishment of PCR-RFLP identification system of cry genes from Bacillus thuringiensis. Sci Agric Sin 31:19–24
Wagner W, Möhrlen F, Schnetter W (2002) Characterization of the proteolytic enzymes in the midgut of the European Cockchafter, Melolontha melolontha (Coleoptera: Scarabaeidae). Insect Biochem Molec Biol 32:803–814
Xintian W, XuDan X, Deloach CJ (1995) Biological control of white grubs (Coleopera: Scarabaeidae) by larvae of Promachus yesonicus (Diptera: Asilidae) in China. Bio Control 5:290–296
Yokoyama T, Tanaka M, Hasegawa M (2004) Novel cry gene from Paenibacillus lentimorbus strain Semadara inhibits ingestion and promotes insecticidal activity in Anomala cuprea larvae. J Invertebr Pathol 85:25–32
Zhang J, Hodgman TC, Krieger L, Schnetter W, Schairer HU (1997) Cloning and analysis of the first cry gene from Bacillus popilliae. J Bacteriol 179:4336–4341
Acknowledgments
We are grateful to professor Shuliang Feng and Rongyan Wang (the Institute of Plant Protection, HeBei Academy of Agricultural and forestry Sciences) for bioassay. This study was supported by grants from the National Basic Research Program (973 Program) 2003CB114201, State Plant Transformation and Industrialization Program JY03A2201, and National Natural Science Foundation (30200184) in P. R. China.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Yu, H., Zhang, J., Huang, D. et al. Characterization of Bacillus thuringiensis Strain Bt185 Toxic to the Asian Cockchafer: Holotrichia parallela. Curr Microbiol 53, 13–17 (2006). https://doi.org/10.1007/s00284-005-0097-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-005-0097-8