Abstract
Mealybugs have an association with prokaryotic endosymbionts that are located in specialized cells called bacteriocytes. In order to compare the phylogeny of the host with that of the previously published phylogeny of the endosymbionts, 3.1 to 3.2 kilobase DNA fragments containing mitochondrial cytB (part), nd1,16S ribosomal DNA(rDNA), and 12S rDNA (part) were amplified and sequenced. A phylogenetic analysis of the data and a comparison with the trees obtained from endosymbiont genes and host 18S and 28S rDNA indicated that all the trees were similar. This result is consistent with an infection of a mealybug ancestor with a precursor of the endosymbiont followed by the vertical transmission of the endosymbiont to progeny. Comparison of the guanine + cytosine (G + C) contents of the mealybug mitochondrial genes with the same genes from other members of Sternorrhyncha and Arthropoda indicated that the mealybug genes had unusually low G + C contents in their DNAs (10.2 to 11.1 mol%).
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Mealybugs (Hemiptera, Coccoidea, Pseudococcidae) are plant sap-sucking insects within the suborder Sternorrhyncha that also contains psyllids, whiteflies, and aphids [2, 3]. All of these insects are composite organisms in that they consist of an obligatory association of the host with different prokaryotic endosymbionts [2, 14]. The function of the endosymbionts appears to be primarily the synthesis of essential amino acids that are lacking in plant sap, the diet of these insects [4, 7]. There is evidence in psyllids, whiteflies, and aphids that the association is the result of an infection of the insect host with a different prokaryote followed by cospeciation, that is, vertical transmission of the endosymbiont to the progeny. This conclusion is derived from the observation that phylogenetic trees based on endosymbiont 16S-23S ribosomal DNA (rDNA) are congruent with trees based on host genes [2, 6, 9, 11]. In a recent study of whitefly endosymbionts, we used mitochondrial DNA fragments of about 3 kb containing cytB (part)-nd1-16S rDNA-12S (part) and obtained a phylogenetic tree that was similar to that obtained with 16S-23S rDNA of Candidatus Portiera aleyrodidarum, the primary (P-) endosymbiont of whiteflies [11]. More limited studies with psyllids and aphids indicated that the phylogenetic trees obtained using these genes were also congruent with the trees obtained using P-endosymbiont rDNA [12].
Mealybugs have within their body cavity aggregates of specialized cells called bacteriocytes that contain within them the P-endosymbiont Candidatus Tremblaya princeps (here referred to as Tremblaya) that is a member of the β-subdivision of the Proteobacteria [10]. This P-endosymbiont has the unusual property in that it may contain within its cytoplasm secondary (S-) endosymbionts that are members of the γ-subdivision of the Proteobacteria [13]. The acquisition of these different organisms has occurred multiple times and once the association has been established, there was cospeciation between Tremblaya and the S-endosymbionts [10]. In this study, we compare the phylogenetic trees obtained from Tremblaya 16S-23S rDNA to those obtained from mealybug mitochondrial cytB-nd1-16S rDNA. In addition, we compare these trees to those obtained from mealybug 18S-23S rDNA from a recent extensive study on the phylogeny of mealybugs and related organisms [5]. Comparisons of the mealybug mitochondrial DNA fragment with those from other members of the Sternorryncha and other arthropods indicates that mealybug mitochondrial DNA has some unusual properties.
Materials and Methods
The source of the DNA samples and their preparation have been described [10]. The methods used for the purification of polymerase chain reaction (PCR)-amplified fragments, their cloning into pBluescript (Stratagene Inc., La Jolla, CA) or direct nucleotide sequence determination, as well as phylogenetic analyses have been previously described [9, 12]. For each mealybug species, two overlapping DNA fragments were obtained by PCR consisting of cytB-nd1-16S rDNA and 16S-12S. For the cytB-nd1-16S rDNA fragments, the oligonucleotide primers used were 5′-GAG GAT CCG GAA AAA TAT TAT TAA ATT GAA TTT GAG G-3′ (BamHI) and 5′-GAG GTA CCA TTA CTT TAG GGA TAA CAG G-3′ (KpnI). For the 16S-12S DNA fragments, the following oligonucleotide primers were used: 5′-GAG GTA CCG ATA GAA ACC AAC CTG GCT TAC ACC GG-3′ (KpnI) in conjunction with either 5′-GTG GAT CCG TGC CAG CAG TTG CGG TTA AAC-3′ (BamHI) or 5′-CAG TAA TAA ATT TTA AGG GGA-3′. PCR amplification was carried out in 10 μL reaction mixture containing 10 ng insect DNA, 3.5 mM MgCl2, 0.15 mM deoxynucleoside triphosphate mix, 5 pmol of each primer, 0.4 U Bio-X-Act DNA polymerase in Opti Buffer (Bioline, London, UK). The following conditions were used: initial denaturation 95°C, 5 min; 30 cycles of 94°C, 20 s (denaturation); 45°C, 30 s (annealing); 68°C, 2.5 min (extension), followed by 68°C, 10 min.
The GenBank accession numbers of the sequences determined in this study as well as those used in the phylogenetic analyses are given in Table 1.
Results and Discussion
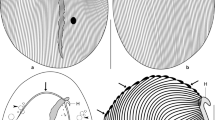
Figure 1 presents the results of the phylogenetic analysis using (a) Tremblaya 16S-23S rDNA (4102 characters), (b) mealybug mitochondrial cytB-nd1-16S (1710 characters), and (c) mealybug 18D-23S rDNA (2050 characters). More groupings are resolved with the endosymbiont genes (Fig. 1a), followed by mitochondrial (Fig. 1b) and host genes (Fig. 1c). All of the three trees are similar and there are no major contradictions in the order of branching. The one difference between Fig. 1a and b is based on the relatively low bootstrap value of 70%. A phylogenetic analysis of the combined data from Fig. 1a–cresults in a tree identical with Fig. 1a. These results are similar to those previously observed with the P-endosymbionts and their hosts in whiteflies, psyllids, and aphids, and are consistent with an infection of a mealybug ancestor with a free-living Tremblaya precursor followed by cospeciation between the P-endosymbiont and the host [6, 9, 10, 11].
Comparisons of phylogenetic trees based on maximum-likelihood analyses of mealybug-associated genes from (a) Tremblaya 16S-23S rDNA, (b) mitochondria cytB-nd1-16S rDNA, and (c) insect 18S-23S rDNA. The numbers at the nodes are bootstrap percentages based on 1000 replicates; only bootstrap values of 70% or over are shown. The outgroups are (a) Neisseria meningitidis, (b) Schizaphis graminum, (c) Puto albicans. (d) Approximate position of the runs of “Ts” that disrupt the open reading frame corresponding to nd1; +, addition of a “T” results in a “correct” ORF; −, removal of a “T,” results in a correct ORF; arrow, indicates similar position of required “correction” in two or more species.
The guanine + cytosine (G + C) contents of the DNAs of the 3.1 to 3.2 kb mealybug cytB (part)-nd1-16S rDNA-12S rDNA (part) fragments were 9.6 to 10.5 mol%. We have combined the DNA corresponding to mitochondrial cytB (part), nd1, and 16S rDNA from mealybugs, whiteflies, psyllids, and aphids as well as 48 arthropods and determined their G + C contents. A plot of the distribution of the % G + C content for the different members of the Sternorrhyncha is presented in Fig. 2. The most extensive data is for whiteflies (19 species) that span the range of 13.6–33.0 mol%. The combination of aphids and phyloxera (5 species) and psyllids (4 species) spanned the range of 15.7–17.0 and 24.1–27.9 mol%, respectively. Mealybugs (nine species) spanned the range of 10.2–11.1 mol%, below that of the other members of the Sternorrhyncha (Fig. 2). The G + C span of 48 representative arthropods was 14.6–38.0 mol%. These results indicate that, in their low mol% G + C contents, the mitochondria of mealybugs are rather unusual.
In cytB of Planococcus ficus, there is a stop codon that disrupts the open reading frame (ORF). The manual insertion of an “A” in a row of 9 “As” “corrects” this frameshift and makes a complete peptide. The nd1 from the mitochondria of three mealybug species has an ORF corresponding to the expected ND1 protein. In the case of the remaining species, there are multiple frameshifts in nd1 that are summarized in Fig. 1d. In all of these cases, the defect is in a region corresponding to rows of “Ts” and can be corrected by the manual insertion or removal of a “T.” Some of these defects are found in the same region and are grouped in the same manner as the mealybug species based on relationship of Tremblaya, mitochondrial or host genes (Fig. 1a–c). For example, the run of “Ts” at about nucleotide (nt) position 525 is found in the mitochondria of five mealybug species. Five runs of “Ts” at about nucleotide positions 310, 350, 525, 670, and 725 are found in the closely related species Planococcus citri and P. ficus. Some of the “corrections” in approximately the same position in the gene involve the removal of a “T” in one species and the addition of a “T” in another species (Fig. 1d). Of the total of 17 runs of “Ts” that require a “correction” to obtain an intact ORF, two runs have 10 nt, five have 9 nt, eight have 7 nt, and one run each of 7, 6, and 4 nt.
The significance of these observations is not understood. They do not appear to be simple PCR errors since five separate PCR amplifications of Planaococcus citri cytB-nd1-16S rDNA-12S rDNA resulted in the detection of the same defects. The fact that some defects are grouped according to relationships suggests a single occurrence in an ancestor and subsequent inheritance by the progeny. nd1 is an essential component of the mitochondria of cells and it would seem unlikely that it is replaced by another protein in some mealybug species but not in others. A very similar situation was found in the case of pseudogenes for cell wall and cysteine synthesis of Buchnera (the P-endosymbiont of aphids) [8]. On the basis of an analysis of the ratios of non-synonymous and synonymous substitutions in defective and non-defective DNAs, it was concluded that the frameshift is overcome and that in Buchnera an intact protein is made. The rationale for this interpretation is that if the encoding regions are defective and do not make a functional protein, the frequency of non-synonymous substitutions would rise since the functional constrains that restrict changes in the sequence of proteins would be eliminated. This and other possible interpretations are considered in [1, 8]. A resolution of this issue would require the isolation of a functional protein from the organisms that have the frameshift.
Literature Cited
JO Andersson (2000) ArticleTitleEvolutionary genomics: is Buchnera a bacterium or an organelle? Curr Biol 10 R866–R868
Baumann P, Moran NA, Baumann L. (2000) Bacteriocyte-associated endosymbionts of insects. In: Dworkin M (ed) The prokaryotes. New York: Springer. http://link.springer.de/link/service/books/10125
P Buchner (1965) Endosymbiosis of animals with plant microorganisms Interscience New York 207–432
AE Douglas (1998) ArticleTitleNutritional interactions in insect-microbial symbioses: aphids and their symbiotic bacteria Buchnera Annu Rev Entomol 43 17–37
DA Downie PJ Gullan (2004) ArticleTitlePhylogenetic analysis of mealybugs (Hemiptera: Coccoidea: Pseudococcidae) based on DNA sequences from three nuclear genes, and a review of the higher classification Syst Entomol 29 238–259
D Martinez-Torres C Bauades A Latorre A Moya (2001) ArticleTitleMolecular systematics of aphids and their primary endosymbionts Mol Phylogen Evol 20 437–449
NA Moran GR Plague JP Sandström JL Wilcox (2003) ArticleTitleA genomic perspective on nutrient provisioning by bacterial symbionts of insects Proc Natl Acad Sci USA 100 14543–14548
I Tamas JP Sandström SGE Andersson (2002) Natural frameshift mutants in Buchnera: are they functional? I Tamas (Eds) Comparative genomics of endosymbiotic bacteria. Comprehensive summaries of Uppsala dissertations from the Faculty of Science and Technology Acta Universitatis Upsaliensis Uppsala, Sweden
ML Thao NA Moran P Abbot EB Brennan DH Burckhardt P Baumann (2000) ArticleTitleCospeciation of psyllids and their prokaryotic endosymbionts Appl Environ Microbiol 66 2898–2905
ML Thao PJ Gullan P Baumann (2002) ArticleTitleSecondary (γ) endosymbionts infect the primary (β) endosymbionts of mealybugs multiple times and coevolve with their hosts Appl Env Microbiol 68 3190–3197
ML Thao P Baumann (2004) ArticleTitleEvolutionary relationships of primary prokaryotic endosymbionts of whiteflies and their hosts Appl Environ Microbiol 70 3401–3406
ML Thao L Baumann P Baumann (2004) ArticleTitleOrganization of the mitochondrial genomes of whiteflies, aphids, and psyllids (Hemiptera, Sternorrhyncha) BMC Evol Biol 4 25
CD Dohlen Particlevon S Kohler ST Alsop WR McManus (2001) ArticleTitleMealybug β-proteobacterial endosymbionts contain γ-proteobacterial symbionts Nature (London) 412 433–436
JJ Wernegreen (2002) ArticleTitleGenome evolution in bacterial endosymbionts of insects Nat Rev Gen 3 850–861 Occurrence Handle10.1038/nrg931 Occurrence Handle1:CAS:528:DC%2BD38Xotlersbg%3D Occurrence Handle12415315
Acknowledgments
This material is based on work supported by National Science Foundation Awards DEB-9978518 (N. A. Moran, P. Baumann) and MCB-9807145 (P. Baumann) and the University of California Experiment Station (P. Baumann).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Baumann, L., Baumann, P. Cospeciation Between the Primary Endosymbionts of Mealybugs and Their Hosts. Curr Microbiol 50, 84–87 (2005). https://doi.org/10.1007/s00284-004-4437-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-004-4437-x