Abstract
The skin is the human body’s largest organ and is home to a diverse and complex variety of innate and adaptive immune functions that protect against pathogenic invasion. Recent studies have demonstrated that cutaneous commensal bacteria modulated the host immune system. For example, Staphylococcus epidermidis, a skin commensal bacterium, has been demonstrated to induce cutaneous interferon (IFN)-γ- and interleukin (IL)-17A-producing T cells. In addition, cutaneous microbiota changes occur in the chronic inflammatory skin disorders, such as atopic dermatitis, and may influence the activity of skin diseases. In this article, we will review the recent findings related to the interactions of the commensal bacteria with skin homeostasis and discuss the role of the dysbiosis of these bacteria in the pathogenesis of skin diseases.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Skin serves as a protective barrier that prevents the invasion of harmful organisms, such as viruses, bacteria, and fungi, as well as other antigenic particles. In addition to being a physical barrier, the skin is an immunological barrier [1]. For example, antigen-presenting cells make a tightly meshed network throughout the entire skin, capture foreign antigen, and induce T cell proliferation and differentiation. These tissue-tailored immunological networks are essential for the maintenance of tissue or exogenous tolerance and the development of protective and controlled immune responses.
Tissue-specific responses have been explored in depth in barrier tissues, such as the skin and the gastrointestinal tract sites that are constitutively colonized by highly diverse and site-specific flora. In the gastrointestinal tract, a part of the local immune responses is aimed at maintaining a peaceful coexistence with the resident microbiota. These microbes can control many aspects of both innate and adaptive responses. Skin commensal bacteria also modulate skin immune cell function and induce protective immunity to pathogens [2]. In this article, we will review the recent findings about the role of skin microbiota in host immunity and will also discuss how dysbiosis may contribute to the induction of various skin pathologies.
The profile of commensal bacteria in the skin
There exist about one trillion commensal bacteria in the human skin surface [3]. Until recently, our knowledge of the skin microbiota has largely depended on culture assays, although it is estimated that less than 1 % of bacterial species can be cultivated [3]. Recently, the use of a polymerase chain reaction-based genomic approach allowed to characterize skin bacteria, allowing to reveal a much greater diversity of organisms than previously revealed by culture-based methods [4]. Grice et al. characterized the topographical and temporal diversity of the human skin microbiome with the use of 16S ribosomal RNA gene phylotyping from samples of 20 diverse skin sites on each of 10 healthy humans [5]. The authors demonstrated that bacteria composition was dependent on the physiology of the skin site, with specific bacteria being associated with moist, dry, and sebaceous microenvironments (Table 1).
In general, bacterial diversity seems to be lowest in sebaceous sites. Sebaceous sites, including the forehead, the retroauricular crease, the back, and the alar crease, contain low phylotype richness [5, 6]. Propionibacterium spp. are the dominant organisms at these sites and other sebaceous areas, which confirms classical microbiological studies describing Propionibacterium spp. as lipophilic residents of the pilosebaceous unit [6].
Metagenomic analysis also revealed that Staphylococcus and Corynebacterium spp. were the most abundant organisms colonizing moist areas [5, 6], consistent with culture data suggesting that these organisms prefer areas of high humidity. These moist sites include the umbilicus, the axillary vault, the inguinal crease, the gluteal crease, the sole of the foot, the popliteal fossa, and the antecubital fossa.
The sites in which bacterial diversity is the highest are the dry areas, with mixed representation from the phyla Actinobacteria, Proteobacteria, Firmicutes, and Bacteriodetes [5, 6]. These sites include the forearm, buttock, and hand [5].
There is intra- and interpersonal variability in the composition of bacterial communities. Fierer et al. reported that hand microbiota from the same individual shared only 17 % of their phylotypes of intrapersonal variation, with different individuals sharing only 13 % [7]. Women had significantly higher diversity than men, and community composition was significantly affected by handedness, time since last hand washing, and an individual’s sex.
Cutaneous invaginations and appendages, including sweat glands, sebaceous glands, and hair follicles, are likely to be associated with their own unique microbiota [4]. Eccrine glands are found on virtually all skin surfaces and continuously bathe the skin surface with their secretions. The role of eccrine sweat is not only thermoregulation but also acidification of the skin, which prevents the colonization and growth of microorganisms. Sebaceous glands are relatively anoxic and support the growth of facultative anaerobes such as Propionibacterium acnes. This bacterium hydrolyzes the triglycerides in sebum, which releases free fatty acids onto the skin. These free fatty acids also contribute to the acidic pH (∼5) of the skin surface. The growth is inhibited by an acidic pH of many common pathogens, such as Staphylococcus aureus and Streptococcus pyogenes [4].
Skin microbiota and homeostasis
Recent studies demonstrated that symbiotic factors produced by intestinal commensal bacteria beneficially modulate the host immune systems. The gut microbiome influences the host immune response which controls autoimmune disorders and infectious inflammation. Mazmanian et al. reported that the prominent gut commensal bacteria Bacteroides fragilis protected animals from experimental colitis by producing polysaccharide A (PSA) [8]. Purified PSA administered to animals was required to suppress pro-inflammatory interleukin (IL)-17A production by intestinal immune cells. Furthermore, PSA protected from inflammatory bowel disease through a functional requirement for IL-10-producing CD4+ T cells. On the other hand, the gut microbiota can also promote protective immunity to pathogens or vaccines [9–11].
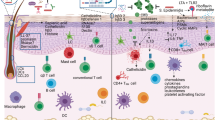
In the skin, Staphylococcus epidermidis, a commensal bacterium, has recently been demonstrated to modulate the host innate immune response [12]. Lai et al. demonstrated a mutually beneficial relationship between S. epidermidis and keratinocyte inflammatory responses [12] (Fig. 1a), which is mediated by staphylococcal lipoteichoic acid (LTA) that inhibits uncontrolled skin inflammation during skin injury. Staphylococcal LTA is recognized by keratinocyte Toll-like receptor (TLR) 2 and inhibits TLR3 signaling. Following skin injury, the host RNA from damaged cells activates TLR3 in the keratinocytes, which accounts for the release of inflammatory cytokines, resulting in inflammation. Staphylococcal LTA inhibits both inflammatory cytokine release from keratinocytes and inflammation triggered by injury. These findings reveal that defined microflora ligands can modulate cutaneous inflammatory responses.
Molecular interactions of microbial symbiosis in skin innate and adaptive immune systems. Factors produced by skin commensal bacteria modulate the skin immune system. a After skin injury, host RNA from damaged cells activates TLR3 in keratinocytes. Staphylococcal lipoteichoic acid (LTA) inhibits excess inflammatory cytokine release from keratinocytes and inflammation through a TLR2-dependent mechanism. The mechanism for LTA-TLR-mediated suppression of TLR3 signaling is by induction of the negative regulatory factor TNF receptor-associated factor-1 (TRAF1). b Staphylococcus epidermidis modulate IL-1 production from keratinocytes, which induce cutaneous T cell differentiation. Cutaneous T helper (Th)-1, Th17, and IL-17A-producing γδ T cells play an important role in the protection against foreign pathogens
Commensal bacteria and skin diseases
A shift in the skin microbiota composition has been shown in the context of skin inflammatory disorders, such as acne vulgaris, psoriasis, and atopic dermatitis [13]. In acne vulgaris, the primary process is sebaceous hyperplasia and lipid release into the follicular lumen, which leads to comedo formation and overgrowth of P. acnes. Such process results in follicular wall rupture, triggering neutrophil influx and pustule formation. Furthermore, P. acnes modulates the expression of antimicrobial peptides which induce overgrowth of P. acnes and S. epidermidis [13, 14]. P. acnes also induces keratinocyte TLR expression, which plays an essential role in acne-linked inflammation [15].
Recent observations indicate that psoriasis may be partly associated with alteration in the composition and representation of the cutaneous microflora. For example, in the psoriatic lesions, the representation of Propionibacterium and Actinobacteria species was lower than for normal control skin. In contrast, Firmicutes species were overrepresented in the psoriatic lesions [13]. Notably, overexpression of antimicrobial peptides is reported in psoriatic lesions [16]. These peptides are known for their integral role in killing pathogenic microorganisms; however, these peptides can also control host inflammatory responses via a variety of mechanisms [17].
One of these antimicrobial peptides, LL-37, has been highlighted as a modulator of psoriasis development (Fig. 2) [18]. Stressed cells stimulated by trauma or bacterial products release LL-37 and self-DNA. In the initiation phase, stressed keratinocytes release self-DNA that forms complexes with LL-37, which in turn activates plasmacytoid dendritic cells (pDCs) to produce interferon-α (IFN-α). Keratinocyte-derived IL-1β, IL-6, and tumor necrosis factor (TNF) and pDC-derived IFN-α activate dermal DCs. Activated dermal DCs then migrate to the skin-draining lymph nodes to present an as yet unknown antigen (either of self or of microbial origin) to naive T cells and promote their differentiation into T helper 1 (Th1) and/or Th17 cells that produce, respectively, IFN-γ/TNF-α and IL-17A/IL-17F. These cytokines can directly act on keratinocytes, leading to the activation, proliferation, and production of antimicrobial peptides or chemokines [19]. In this manner, antimicrobial peptides may contribute to the exacerbation of psoriasis.
LL-37 induces psoriasis dermatitis. Stressed cells stimulated by environmental triggers release LL-37 and self-DNA. Self-DNA form complexes with LL-37, which in turn activates pDCs to produce IFN-α. pDC-derived IFN-α activate dermal DCs. Activated dermal DCs then migrate to the skin-draining lymph nodes to present an antigen to naive T cells and promote their differentiation into Th1 and/or Th17 cells that produce, respectively, IFN-γ/TNF-α and IL-17A/IL-17F. These mediators act on keratinocytes, leading to the activation and proliferation
Perhaps, the example of dysbiosis is demonstrated in atopic dermatitis. The main symptom of atopic dermatitis is xerosis (dry skin). The direct evidence for xerosis is a recently discovered link between the incidence of atopic dermatitis and mutations in the gene encoding filaggrin (FLG) [20]. The FLG mutations seen in atopic dermatitis patients are a loss-of-function mutation of FLG, which determine major susceptibility to atopic dermatitis. FLG proteins are localized in the granular layers of the epidermis. These FLG proteins are degraded into natural moisturizing factors (NMFs). Filaggrin-derived NMFs are important to maintain skin hydration and low pH. Thus, in atopic dermatitis patients, the skin is dry and exhibits increased pH. As mentioned above, the change in skin moisture and pH induces are believed to lead to dysbiosis [21] (Fig. 3).
Atopic dermatitis induces Staphylococcus aureus colonization. Barrier dysfunction in line with filaggrin deficiency will lead to changes in the skin environment, which may play an important role in the development of S. aureus colonization. S. aureus produce δ-toxin which induces mast cell degranulation
Furthermore, dysregulation of antimicrobial peptides was proposed to also contribute to the induction of dysbiosis in atopic dermatitis. The expression of LL-37 mRNA is significantly lower in atopic dermatitis [22]. On the other hand, expression of other antimicrobial peptides such as RNase 7, psoriasin, and human beta defensin-2 is highly expressed in lesional skin of atopic dermatitis [23]. These observations support the idea that unusual antimicrobial peptide production during inflammatory states may induce a loss of skin bacterial diversity.
It has long been suspected that the development of atopic dermatitis was associated with S. aureus skin colonization and/or infection. S. aureus colonizes the skin lesions of more than 90 % of atopic dermatitis patients [24]. A recent study revealed that S. aureus exacerbates or contributes to persistent skin inflammation in atopic dermatitis by secreting toxins with superantigenic properties, resulting in marked activation of T cells and other immune cells [25]. Application of staphylococcal enterotoxin B to the skin induces eczematous changes accompanied by infiltration of T cells selectively expanded in response to the superantigen. In addition, atopic dermatitis patients produce specific IgE antibodies directed against the toxins found on their skin [26], with basophils from these patients releasing histamine following exposure to the relevant toxin [27].
Nakamura et al. showed that colonization of the skin with S. aureus triggers local allergic responses by releasing δ-toxin, which directly induces the degranulation of dermal mast cells, and in turn promotes both innate and adaptive responses [28]. Indeed, S. aureus isolates recovered from patients with atopic dermatitis produced large amounts of δ-toxin. Further investigation of the crosstalk that occurs between the microbiota, epithelial, and immune cells in the skin is an interesting issue to better clarify the mechanisms that regulate skin immune homeostasis and inflammation.
Decreased cutaneous IL-17A-producing cells in germ-free mice
Germ-free animals, which have no microorganisms living in or on them, are useful tools to analyze the cutaneous immune effect of skin commensal bacteria. Such animals are raised in the germ-free isolators in order to control their exposure to viral, bacterial, or parasitic agents. Naik et al. reported a significant reduction of IL-17A production by T cells in the skin of germ-free mice relative to that of specific pathogen-free mice [2] (Table 2). The most notable role of IL-17A is its involvement in inducing and mediating barrier immunity and pro-inflammatory responses. IL-17A induces the production of many cytokines, chemokines, and prostaglandins from many types of cells (fibroblasts, endothelial cells, epithelial cells, keratinocytes, and macrophages). IL-17A also significantly enhances protective inflammation during pathogen infection and chronic inflammation associated with autoimmune diseases.
In the murine skin, the main source of IL-17A is Th17 cells and dermal γδ T cells [29]. IL-17A-producing dermal γδ T cells express the IL-23 receptor and the transcription factor RORγt [30]. These cells produce IL-17A in response to IL-1β and IL-23, without T cell receptor engagement [31]. The number of IL-17A-producing γδ T cells was reduced in the skin of germ-free mice and was induced by skin commensal bacterial colonization. γδ T cells appear to be the dominant early source of IL-17A, following activation by external pathogens, such as S. aureus [32]. During an immune response, γδ T cells rapidly produce IL-17A in response to IL-23 and/or other dendritic cell (DC) products. Later, antigen-specific Th17 cells may also develop and contribute to the response [33].
Th17 induction in the skin by S. epidermidis
Ivanov et al. reported that colonization of a single commensal microbe, segmented filamentous bacterium (SFB), is sufficient to induce Th17 cells in the lamina propria of the small intestine in mice [10]. In contrast, the mice harboring SFB in their gastrointestinal tract did not restore effector cytokine production in the skin of germ-free mice. Colonization with the skin commensal S. epidermidis was sufficient to rescue IL-17A production in the skin but not in the gut. Furthermore, protective immunity to a cutaneous pathogen was found to be critically dependent on the skin microbiota but not on the gut microbiota [2]. These results suggest that skin commensals are important drivers and amplifiers of skin immunity.
It was reported that IL-6, TLR5, and TLR9 signals induce Th17 cells [9, 34, 35]. However, cutaneous IL-17A production in IL-6- and TLR 2/3/5/9-deficient mice was comparable with that in wild-type mice [2]. On the other hand, Th17 cells and IL-17A-producing γδ T cells were reduced in the IL-1R1- and its downstream signaling complex MyD88-deficient mice. Indeed, IL-1α production by cutaneous cells was significantly reduced in germ-free relative to specific pathogen-free (SPF) mice, and mono-association of germ-free mice with S. epidermidis restored the production of IL-17A (Fig. 1b). Additionally, keratinocytes from germ-free mice displayed increased levels of the IL-1 receptor antagonist (IL-1ra) mRNA relative to SPF mice, indicating that commensals control various aspects of functional IL-1 signaling. Complementing these observations, the addition of S. epidermidis to germ-free mice significantly reduced IL-1ra from cutaneous cells [2].
How commensal microbiota contributes to immune cell homeostasis at barrier surfaces is poorly understood. Ivanov et al. demonstrated that Th17 cell-inducing microbiota modifies lamina propria DC cytokine production [10]. On the other hand, how cutaneous commensals control Th17 induction is largely unknown.
Commensal bacteria modulate regulatory T cell differentiation
Regulatory T cells (Tregs) play a critical role in the maintenance of immune homeostasis. In the gut, Atarashi et al. reported that a significant decrease in the number of Tregs was observed in the colonic lamina propria of germ-free mice compared with SPF mice [36] (Table 2). However, the number of Tregs in other organs such as the small intestinal lamina propria, inguinal lymph nodes, Peyer’s patches, and mesenteric LNs was unchanged.
In the skin, Naik et al. reported that the skin of germ-free mice contains a high frequency of Foxp3+ Tregs compared with SPF mice [2]; however, the underlying mechanism remains unknown. Petermann et al. reported that IL-23 receptor-expressing γδ T cells suppress Treg differentiation [37], suggesting that the loss of IL-17A-producing-γδ T cells may enhance Treg differentiation in the skin of germ-free mice.
Volz et al. demonstrated that Vitreoscilla filiformis activate cutaneous Treg [38]. V. filiformis is a gram-negative bacterium originally found in thermal spa water. In vitro, exposure of DC to a lysate of V. filiformis promotes the production of the immunoregulatory cytokine IL-10 in a MyD88-dependent pathway, which is an essential mediator for signal transduction by TLRs and IL-1R. V. filiformis-induced IL-10+ DCs primed naive CD4+ T cells for differentiation to IFN-γlow IL-10high type 1 Treg cells with negative FoxP3 expression. Furthermore, epicutaneous application with V. filiformis lysate induced IL-10high T cells and inhibited antigen-specific T cell proliferation during atopic dermatitis-like inflammation in NC/Nga mice. These results suggest that transplantation of a certain type of bacteria may induce immune tolerance in the skin.
Commensal bacteria and systemic inflammatory diseases: Can skin microbiota modulate systemic immune responses?
It was reported that commensal bacteria in the gut could influence both local and systemic immunity. Lee et al. demonstrated that mice maintained under germ-free conditions developed significantly attenuated experimental autoimmune encephalomyelitis (EAE) compared with conventionally colonized mice [39]. Remarkably, germ-free animals harboring SFBs alone developed EAE, showing that gut bacteria can affect systemic neurologic inflammation. Thus, a single commensal microbe can drive an autoimmune disease.
The relationship between gut commensal bacteria and skin disorder is largely unknown. However, Oyoshi et al. reported that cutaneous exposure to food antigens can reprogram gut-homing effector T cells in lymph nodes to express skin-homing receptors, eliciting skin lesions upon food allergen contact in orally sensitized atopic dermatitis patients [40]. This observation raises the possibility that gut T cells may migrate to the skin and potentially modulate skin immunity. In addition, it has previously been demonstrated that skin-infiltrated T cells could return to the draining lymph nodes and modulate immune responses [41]. These findings suggest that cutaneous immune cells, and thereby the skin microbiota, may have the capacity to modulate systemic immune responses.
Probiotics for inflammatory diseases
The beneficial activities of commensal bacteria in immune homeostasis have led to several therapeutic strategies aimed at limiting inflammatory diseases. Some of the best evidence in support of probiotic health benefits is in the treatment of antibiotic-associated diarrhea (AAD). AAD results from an imbalance in the colonic microbiota caused by antibiotic therapy [42]. Probiotic treatment might reduce the incidence and severity of AAD as indicated in several meta-analyses. For example, Arvola et al. reported that treatment with Lactobacillus GG might reduce the risk of AAD and improved stool consistency during antibiotic therapy [43]. In addition, the transplantation of a mixture of commensal Clostridium species isolated from healthy human fecal samples successfully induced the accumulation and functional maturation of Treg cells in the colon, resulting in attenuation of disease in mouse models of colitis and allergic diarrhea [44]. These findings suggest that probiotics for skin flora may modulate inflammatory skin diseases, although there is no study on the effect of probiotics for skin flora thus far.
Recently, the protective role of probiotic therapy to prevent allergic disease has been tested in several clinical trials [45, 46]. In the skin, a significant improvement on the course of atopic dermatitis has been reported in infants given probiotic-supplemented elimination diets [47]. Rosenfeldt et al. examined the effect of two probiotic Lactobacillus strains given in combination for 6 weeks to 1- to 13-year-old children with atopic dermatitis. After the treatment, 56 % of the patients experienced improvement of the eczema, whereas only 15 % of symptoms had improved after placebo [48]. Kalliomäki et al. reported that prescribed Lactobacillus GG was given prenatally to mothers who had at least one first-degree relative with atopic dermatitis, allergic rhinitis, or asthma, and postnatally for 6 months to their infants. Atopic dermatitis was diagnosed in 46 of 132 (35 %) children aged 2 years. The frequency of atopic dermatitis in the probiotic group was half that of the placebo group [49]. These results suggest that Lactobacillus was effective in the prevention of early atopic disease in children at high risk. The mechanistic basis of skin effects induced by probiotic gut flora is thought to be represented by changes in systemic immune responses. In particular, modulation of specific T cell subsets, such as stimulation of Th1 cells in the gut mucosa which may subsequently influence immune responses in other tissues, may play a role [50].
Nonetheless, the preventive effect of probiotics remains controversial [51] and the effect of skin probiotic for the control of skin inflammation remains to be tested.
Concluding remarks
In contrast to studies on gut commensal bacteria, the studies on skin commensal bacteria are limited. In the gut, defined bacterial species induces Th17 while Clostridium species induce Tregs thereby modulating both the gut and systemic immune system [36]. In the skin, skin resident bacteria such as S. epidermidis can induce Th17. However, the link between defined skin bacteria and tissue inflammation and the systemic consequence of this control remain largely unknown. The next step in the field should allow to further explore the relationship between the skin microbiota and local immunity as well as the clinical association with defined microbial communities or bacterial products.
References
Egawa G, Kabashima K (2011) Skin as a peripheral lymphoid organ: revisiting the concept of skin-associated lymphoid tissues. J Invest Dermatol 131(11):2178–2185. doi:10.1038/jid.2011.198
Naik S, Bouladoux N, Wilhelm C, Molloy MJ, Salcedo R, Kastenmuller W, Deming C, Quinones M, Koo L, Conlan S, Spencer S, Hall JA, Dzutsev A, Kong H, Campbell DJ, Trinchieri G, Segre JA, Belkaid Y (2012) Compartmentalized control of skin immunity by resident commensals. Science 337(6098):1115–1119. doi:10.1126/science.1225152
Grice EA, Kong HH, Renaud G, Young AC, Program NCS, Bouffard GG, Blakesley RW, Wolfsberg TG, Turner ML, Segre JA (2008) A diversity profile of the human skin microbiota. Genome Res 18(7):1043–1050. doi:10.1101/gr.075549.107
Grice EA, Segre JA (2011) The skin microbiome. Nat Rev Microbiol 9(4):244–253. doi:10.1038/nrmicro2537
Grice EA, Kong HH, Conlan S, Deming CB, Davis J, Young AC, Program NCS, Bouffard GG, Blakesley RW, Murray PR, Green ED, Turner ML, Segre JA (2009) Topographical and temporal diversity of the human skin microbiome. Science 324(5931):1190–1192. doi:10.1126/science.1171700
Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R (2009) Bacterial community variation in human body habitats across space and time. Science 326(5960):1694–1697. doi:10.1126/science.1177486
Fierer N, Hamady M, Lauber CL, Knight R (2008) The influence of sex, handedness, and washing on the diversity of hand surface bacteria. Proc Natl Acad Sci U S A 105(46):17994–17999. doi:10.1073/pnas.0807920105
Mazmanian SK, Round JL, Kasper DL (2008) A microbial symbiosis factor prevents intestinal inflammatory disease. Nature 453(7195):620–625. doi:10.1038/nature07008
Hall JA, Bouladoux N, Sun CM, Wohlfert EA, Blank RB, Zhu Q, Grigg ME, Berzofsky JA, Belkaid Y (2008) Commensal DNA limits regulatory T cell conversion and is a natural adjuvant of intestinal immune responses. Immunity 29(4):637–649. doi:10.1016/j.immuni.2008.08.009
Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, Tanoue T, Imaoka A, Itoh K, Takeda K, Umesaki Y, Honda K, Littman DR (2009) Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 139(3):485–498. doi:10.1016/j.cell.2009.09.033
Biswas PS, Pedicord V, Ploss A, Menet E, Leiner I, Pamer EG (2007) Pathogen-specific CD8 T cell responses are directly inhibited by IL-10. J Immunol 179(7):4520–4528
Lai Y, Di Nardo A, Nakatsuji T, Leichtle A, Yang Y, Cogen AL, Wu ZR, Hooper LV, Schmidt RR, von Aulock S, Radek KA, Huang CM, Ryan AF, Gallo RL (2009) Commensal bacteria regulate Toll-like receptor 3-dependent inflammation after skin injury. Nat Med 15(12):1377–1382. doi:10.1038/nm.2062
Gallo RL, Nakatsuji T (2011) Microbial symbiosis with the innate immune defense system of the skin. J Invest Dermatol 131(10):1974–1980. doi:10.1038/jid.2011.182
Chronnell CM, Ghali LR, Ali RS, Quinn AG, Holland DB, Bull JJ, Cunliffe WJ, McKay IA, Philpott MP, Muller-Rover S (2001) Human beta defensin-1 and -2 expression in human pilosebaceous units: upregulation in acne vulgaris lesions. J Invest Dermatol 117(5):1120–1125. doi:10.1046/j.0022-202x.2001.01569.x
Jugeau S, Tenaud I, Knol AC, Jarrousse V, Quereux G, Khammari A, Dreno B (2005) Induction of toll-like receptors by Propionibacterium acnes. Br J Dermatol 153(6):1105–1113. doi:10.1111/j.1365-2133.2005.06933.x
de Jongh GJ, Zeeuwen PL, Kucharekova M, Pfundt R, van der Valk PG, Blokx W, Dogan A, Hiemstra PS, van de Kerkhof PC, Schalkwijk J (2005) High expression levels of keratinocyte antimicrobial proteins in psoriasis compared with atopic dermatitis. J Investig Dermatol 125(6):1163–1173. doi:10.1111/j.0022-202X.2005.23935.x
Gallo RL, Hooper LV (2012) Epithelial antimicrobial defence of the skin and intestine. Nat Rev Immunol 12(7):503–516. doi:10.1038/nri3228
Morizane S, Gallo RL (2012) Antimicrobial peptides in the pathogenesis of psoriasis. J Dermatol 39(3):225–230. doi:10.1111/j.1346-8138.2011.01483.x
Nestle FO, Di Meglio P, Qin JZ, Nickoloff BJ (2009) Skin immune sentinels in health and disease. Nat Rev Immunol 9(10):679–691. doi:10.1038/nri2622
Kabashima K (2013) New concept of the pathogenesis of atopic dermatitis: interplay among the barrier, allergy, and pruritus as a trinity. J Dermatol Sci 70(1):3–11. doi:10.1016/j.jdermsci.2013.02.001
Schommer NN, Gallo RL (2013) Structure and function of the human skin microbiome. Trends Microbiol 21(12):660–668. doi:10.1016/j.tim.2013.10.001
Mallbris L, Carlen L, Wei T, Heilborn J, Nilsson MF, Granath F, Stahle M (2010) Injury downregulates the expression of the human cathelicidin protein hCAP18/LL-37 in atopic dermatitis. Exp Dermatol 19(5):442–449. doi:10.1111/j.1600-0625.2009.00918.x
Harder J, Dressel S, Wittersheim M, Cordes J, Meyer-Hoffert U, Mrowietz U, Folster-Holst R, Proksch E, Schroder JM, Schwarz T, Glaser R (2010) Enhanced expression and secretion of antimicrobial peptides in atopic dermatitis and after superficial skin injury. J Investig Dermatol 130(5):1355–1364. doi:10.1038/jid.2009.432
Cho SH, Strickland I, Boguniewicz M, Leung DY (2001) Fibronectin and fibrinogen contribute to the enhanced binding of Staphylococcus aureus to atopic skin. J Allergy Clin Immunol 108(2):269–274. doi:10.1067/mai.2001.117455
Boguniewicz M, Leung DY (2011) Atopic dermatitis: a disease of altered skin barrier and immune dysregulation. Immunol Rev 242(1):233–246. doi:10.1111/j.1600-065X.2011.01027.x
Leung DY, Harbeck R, Bina P, Reiser RF, Yang E, Norris DA, Hanifin JM, Sampson HA (1993) Presence of IgE antibodies to staphylococcal exotoxins on the skin of patients with atopic dermatitis. Evidence for a new group of allergens. J Clin Invest 92(3):1374–1380. doi:10.1172/JCI116711
Bunikowski R, Mielke ME, Skarabis H, Worm M, Anagnostopoulos I, Kolde G, Wahn U, Renz H (2000) Evidence for a disease-promoting effect of Staphylococcus aureus-derived exotoxins in atopic dermatitis. J Allergy Clin Immunol 105(4):814–819. doi:10.1067/mai.2000.105528
Nakamura Y, Oscherwitz J, Cease KB, Chan SM, Munoz-Planillo R, Hasegawa M, Villaruz AE, Cheung GY, McGavin MJ, Travers JB, Otto M, Inohara N, Nunez G (2013) Staphylococcus delta-toxin induces allergic skin disease by activating mast cells. Nature 503(7476):397–401. doi:10.1038/nature12655
Pantelyushin S, Haak S, Ingold B, Kulig P, Heppner FL, Navarini AA, Becher B (2012) Rorgammat+ innate lymphocytes and gammadelta T cells initiate psoriasiform plaque formation in mice. J Clin Invest 122(6):2252–2256. doi:10.1172/JCI61862
Martin B, Hirota K, Cua DJ, Stockinger B, Veldhoen M (2009) Interleukin-17-producing gammadelta T cells selectively expand in response to pathogen products and environmental signals. Immunity 31(2):321–330. doi:10.1016/j.immuni.2009.06.020
Sutton CE, Lalor SJ, Sweeney CM, Brereton CF, Lavelle EC, Mills KH (2009) Interleukin-1 and IL-23 induce innate IL-17 production from gammadelta T cells, amplifying Th17 responses and autoimmunity. Immunity 31(2):331–341. doi:10.1016/j.immuni.2009.08.001
Cho JS, Pietras EM, Garcia NC, Ramos RI, Farzam DM, Monroe HR, Magorien JE, Blauvelt A, Kolls JK, Cheung AL, Cheng G, Modlin RL, Miller LS (2010) IL-17 is essential for host defense against cutaneous Staphylococcus aureus infection in mice. J Clin Invest 120(5):1762–1773. doi:10.1172/JCI40891
Roark CL, Simonian PL, Fontenot AP, Born WK, O’Brien RL (2008) Gammadelta T cells: an important source of IL-17. Curr Opin Immunol 20(3):353–357. doi:10.1016/j.coi.2008.03.006
Torchinsky MB, Garaude J, Martin AP, Blander JM (2009) Innate immune recognition of infected apoptotic cells directs T(H)17 cell differentiation. Nature 458(7234):78–82. doi:10.1038/nature07781
Uematsu S, Fujimoto K, Jang MH, Yang BG, Jung YJ, Nishiyama M, Sato S, Tsujimura T, Yamamoto M, Yokota Y, Kiyono H, Miyasaka M, Ishii KJ, Akira S (2008) Regulation of humoral and cellular gut immunity by lamina propria dendritic cells expressing Toll-like receptor 5. Nat Immunol 9(7):769–776. doi:10.1038/ni.1622
Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, Cheng G, Yamasaki S, Saito T, Ohba Y, Taniguchi T, Takeda K, Hori S, Ivanov II, Umesaki Y, Itoh K, Honda K (2011) Induction of colonic regulatory T cells by indigenous Clostridium species. Science 331(6015):337–341. doi:10.1126/science.1198469
Petermann F, Rothhammer V, Claussen MC, Haas JD, Blanco LR, Heink S, Prinz I, Hemmer B, Kuchroo VK, Oukka M, Korn T (2010) Gammadelta T cells enhance autoimmunity by restraining regulatory T cell responses via an interleukin-23-dependent mechanism. Immunity 33(3):351–363. doi:10.1016/j.immuni.2010.08.013
Volz T, Skabytska Y, Guenova E, Chen KM, Frick JS, Kirschning CJ, Kaesler S, Rocken M, Biedermann T (2014) Nonpathogenic bacteria alleviating atopic dermatitis inflammation induce IL-10-producing dendritic cells and regulatory Tr1 cells. J Invest Dermatol 134(1):96–104. doi:10.1038/jid.2013.291
Lee YK, Menezes JS, Umesaki Y, Mazmanian SK (2011) Proinflammatory T-cell responses to gut microbiota promote experimental autoimmune encephalomyelitis. Proc Natl Acad Sci U S A 108(Suppl 1):4615–4622. doi:10.1073/pnas.1000082107
Oyoshi MK, Elkhal A, Scott JE, Wurbel MA, Hornick JL, Campbell JJ, Geha RS (2011) Epicutaneous challenge of orally immunized mice redirects antigen-specific gut-homing T cells to the skin. J Clin Invest 121(6):2210–2220. doi:10.1172/JCI43586
Tomura M, Honda T, Tanizaki H, Otsuka A, Egawa G, Tokura Y, Waldmann H, Hori S, Cyster JG, Watanabe T, Miyachi Y, Kanagawa O, Kabashima K (2010) Activated regulatory T cells are the major T cell type emigrating from the skin during a cutaneous immune response in mice. J Clin Invest 120(3):883–893. doi:10.1172/JCI40926
Bartlett JG (2002) Clinical practice. Antibiotic-associated diarrhea. N Engl J Med 346(5):334–339. doi:10.1056/NEJMcp011603
Arvola T, Laiho K, Torkkeli S, Mykkanen H, Salminen S, Maunula L, Isolauri E (1999) Prophylactic Lactobacillus GG reduces antibiotic-associated diarrhea in children with respiratory infections: a randomized study. Pediatrics 104(5):e64
Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, Nishikawa H, Fukuda S, Saito T, Narushima S, Hase K, Kim S, Fritz JV, Wilmes P, Ueha S, Matsushima K, Ohno H, Olle B, Sakaguchi S, Taniguchi T, Morita H, Hattori M, Honda K (2013) Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature 500(7461):232–236. doi:10.1038/nature12331
Matsumoto M, Ebata T, Hirooka J, Hosoya R, Inoue N, Itami S, Tsuji K, Yaginuma T, Muramatsu K, Nakamura A, Fujita A, Nagakura T (2014) Antipruritic effects of the probiotic strain LKM512 in adults with atopic dermatitis. Ann Allergy Asthma Immunol Off Publ Am Coll Allergy Asthma Immunol. doi:10.1016/j.anai.2014.05.002
Yang HJ, Min TK, Lee HW, Pyun BY (2014) Efficacy of probiotic therapy on atopic dermatitis in children: a randomized, double-blind, placebo-controlled trial. Allergy Asthma Immunol Res 6(3):208–215. doi:10.4168/aair.2014.6.3.208
Krutmann J (2009) Pre- and probiotics for human skin. J Dermatol Sci 54(1):1–5. doi:10.1016/j.jdermsci.2009.01.002
Rosenfeldt V, Benfeldt E, Nielsen SD, Michaelsen KF, Jeppesen DL, Valerius NH, Paerregaard A (2003) Effect of probiotic Lactobacillus strains in children with atopic dermatitis. J Allergy Clin Immunol 111(2):389–395
Kalliomaki M, Salminen S, Arvilommi H, Kero P, Koskinen P, Isolauri E (2001) Probiotics in primary prevention of atopic disease: a randomised placebo-controlled trial. Lancet 357(9262):1076–1079. doi:10.1016/S0140-6736(00)04259-8
Pohjavuori E, Viljanen M, Korpela R, Kuitunen M, Tiittanen M, Vaarala O, Savilahti E (2004) Lactobacillus GG effect in increasing IFN-gamma production in infants with cow’s milk allergy. J Allergy Clin Immunol 114(1):131–136. doi:10.1016/j.jaci.2004.03.036
Abrahamsson TR, Jakobsson T, Bottcher MF, Fredrikson M, Jenmalm MC, Bjorksten B, Oldaeus G (2007) Probiotics in prevention of IgE-associated eczema: a double-blind, randomized, placebo-controlled trial. J Allergy Clin Immunol 119(5):1174–1180. doi:10.1016/j.jaci.2007.01.007
Author information
Authors and Affiliations
Corresponding author
Additional information
This article is a contribution to the Special Issue on Microbiome, Immunity and Inflammation - Guest Editor: Hiroshi Ohno
Rights and permissions
About this article
Cite this article
Nakamizo, S., Egawa, G., Honda, T. et al. Commensal bacteria and cutaneous immunity. Semin Immunopathol 37, 73–80 (2015). https://doi.org/10.1007/s00281-014-0452-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00281-014-0452-6