Abstract
Purpose
Gemcitabine resistance is the main problem in pancreatic adenocarcinoma patients. Hence, we aimed to identify the correlation between expression of RRM1 and CDA as the resistance genes and their predicted targeting miR-608 in the resistant pancreatic cancer cell lines to gemcitabine.
Methods
Dual luciferase assay was performed to determine whether both RRM1 and CDA are targeted by miR-608 in 293T and pancreatic cancer cell lines. AsPC-1 and MIA PaCa-2 cell lines became gradually resistant to gemcitabine by exposing to the increasing doses of gemcitabine. After RNA and miRNAs extraction and cDNA conversion, the expressions of RRM1, CDA and miR-608 in all cell lines were studied by quantitative PCR. Pre-miR-608 transfection to the cell lines was done by calcium phosphate method. MTT assay was performed for analyzing the chemo sensitivity of different cell lines to gemcitabine.
Results
Luciferase assays showed that miR-608 targeted RRM1 and CDA genes in 293T, AsPC-1 and MIA PaCa-2 cell lines. Compared to parental cell line, resistant MIA PaCa-2 and AsPC-1 cells demonstrated increased expression of RRM1 and CDA. On the other hand the expression of miR-608 in resistant MIA PaCa-2 and AsPC-1 cells was lower than parental cells. Furthermore, transfection of MIA PaCa-2 and AsPC-1 cells by miR-608 lead to decreased expression of RRM1 and CDA and lowered viability of the cells in comparison with scrambled microRNA transfected cells.
Conclusion
During resistance induction in pancreatic cancer cells, miR-608 which is targeting RRM1 and CDA is downregulated which leads to upregulation of these genes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Pancreatic adenocarcinoma (PDAC) is one of the most devastating malignancy in human [1]. Only in about 20% of the patients surgical resection could be effective because the disease is diagnosed in advanced stages [2]. Even though gemcitabine is the most popular approach for PDAC treatment, there are still therapeutic challenges because of high degree of inherent and acquired chemoresistance [3, 4]. It is believed that variable responses of patients to a medication may be due to the genetic and epigenetic factors which are known as determinants of drug efficacy and toxicity [5]. Alterations in the gene sequence (including SNPs or single nucleotide polymorphisms) or expression may be the most important underlying mechanism for different reactions to a drug [6].
Several studies have reported many genes underlying chemoresistance in pancreatic cancer cells [7, 8]. One of these genes playing role in the metabolism of gemcitabine is ribonucleotide reductase (RNR) which reduces nucleotides into deoxynucleotides. Increased activity or expression of RNR which is essential for DNA synthesis, increases the dNTP pool in the cells and by competition with gemcitabine-diphosphate for incorporation into DNA counteracts the activity of gemcitabine [9]. The active site of this multimeric enzyme is located in the ribonucleotide reductase large subunit (RRM1) which is encoded by rrm1 [10]. Upregulation of rrm1 mRNA as a target of gemcitabine has been implicated in gemcitabine resistance [11]. Hence, studying genetic variants and mRNA expression level of this gene has been discussed as a prognostic marker in different types of malignancies, including breast, non-small-cell lung, biliary tract and pancreatic cancer [12].
Cytidine deaminase (CDA) is an enzyme metabolizing the majority of administered gemcitabine to the inactive form of 2′2′-difluorodeoxyuridine (dFdU) [13]. Several SNPs have been reported in this metabolizing enzyme. Polymorphisms in this gene results in various functional enzymatic activities leading to significantly differential sensitivity to gemcitabine [14]. For instance, reduced enzyme activity was shown in pancreatic cancer patients carrying cda 79A > C (Lys27Gln) SNPs [15]. Also, high levels of CDA mRNA in advanced pancreatic cancer patients after treatment with gemcitabine were associated with delayed response to gemcitabine [16].
MicroRNAs (miRNAs) belong to small noncoding RNA molecule family (21–23 nucleotides) which play important roles in post transcriptional control of protein-coding genes. MiRNA pharmacogenomics states that overexpression of specific miRNAs down-regulate the genes encoding proteins that promote drug efficacy while overexpression of some other miRNAs inhibits drug efficiency [17]. Both processes may impact drug function making miRNAs as the regulators of drug action [6]. It is assumed that each miRNA can regulate several messenger RNAs while each mRNA can be regulated by different miRNAs. Despite the fact that many computational analyses have demonstrated the interaction between miRNAs and their targets, few studies have experimentally examined this relevance in vivo/vitro [18].
As we reviewed recently [5], some other studies have proven the role of miRNAs in gemcitabine resistance, including miR-15a [19], miR-21 [20], miR-34 [21], miR-200b, miR-200c [22], miR-214 [19], miR-221 [23] as well as some members of the let7 family [22]. However, it seems that studying miRNAs with common target genes is still untouched area. Several studies have illustrated the role of different genes in cellular uptake and metabolism of gemcitabine as the resistance biomarkers. Herein RRM1 and CDA were chosen as they showed increased activity/expression in gemcitabine-resistant cells. Thereafter, we hypothesized that any microRNA regulating both of these genes would also affect the response of pancreatic cancer cells to gemcitabine. In the current study, we developed gemcitabine-resistant cells from human pancreatic cancer cell lines and performed comparative studies on the expression level of RRM1 and CDA as well as their predicted commonly targeting miRNA (miR-608).
Materials and methods
Cell culture
MIA PaCa-2 and 293-T acquired from National Cell Bank of Iran (Pasteur Institute of Iran, Tehran, Iran) were cultured in Dulbecco’s modified Eagle’s medium (DMEM) while AsPC cell line was Roswell Park Memorial Institute (RPMI) 1640 medium. All media were supplemented with 10% heat-inactivated fetal bovine serum, l-glutamine (2 mM) and antibiotics (100 units/ml of penicillin and 100 μg/ml of streptomycin) (all from Gibco, Invitrogen). The cells were maintained at 37 °C in 5% CO2. Gemcitabine was obtained from Eli Lilly (Italy).
Establishment of gemcitabine-resistant cells
For establishment of gemcitabine-resistant cells, the cells were gradually adapted to gemcitabine through exposing to increasing concentrations of the drug. Based on the previous articles we chose the least and the most reported IC50 for gemcitabine for these two cell lines and we carried out three independent triplicate MTT assay experiments to find a proper inhibitory concentration of gemcitabine in MIA PaCa-2 and AsPC-1 cells [4, 24]. To make resistant cell lines, the cell lines were incubated with 80% of inhibitory concentration 50% (IC50) for at least 14 days. In this process, the flasks refreshed by gemcitabine-contained-medium every 3 days for 2 weeks. After 2 weeks the medium was replaced with medium lacking gemcitabine supplemented with l-glutamine and non-essential amino acids. The alive cells after 3–4 weeks were considered as resistant cell lines and this was proved by proliferation assay. After each period of exposition MTT assay was done to detect any probable change in IC50 values of two cell lines. The next period of exposition was done on the alive cells after the previous exposition process by 80% of the new IC50. These processes continued four times for MIA PaCa-2 cells and two times for AsPC-1 cell line until no cells can endure the drug (80% of new IC50) and no viable cells were seen 3 weeks after discontinuing the gemcitabine incubation. Following schedules were applied for incubation of the cells with gemcitabine consecutively: (a) MIA PaCa-2: 10, 15, 25 and 40 μM, (b) AsPC-1: 25 and 35 μM. They were named MIA PaCa2-S, MIA PaCa2-RG1, MIA PaCa2-RG2, MIA PaCa2-RG3, and MIA-PaCa2-RG4. AsPC-1 clones contained AsPC-S and its two resistant clones AsPC-RG1 and AsPC-RG2 cells according to the consecutive resistance to gemcitabine.
RNA isolation and cDNA synthesis
TRIZOL reagent (Life Technologies) was applied for isolation of small RNAs containing total RNA from MIA PaCa-2 and AsPC-1 after each period of incubation, followed by cDNA synthesis according to the previously described methods [25, 26].
Real-time quantitative RT-PCR for analysis of mRNA and miRNA expression level
Duplicate qRT-PCR assays were fulfilled in a StepOnePlus ™ Real-Time PCR System (Applied Biosystems) with SYBR Green PCR Master Mix and TaqMan universal PCR Master Mix (both from Takara, Japan) for mRNA and miRNA, respectively. Primer sequences are shown in Table 1.
Relative expressions of mRNAs/miRNA were calculated by mean CT value for replicates. Then the test gene mean CT was taken away from mean CT of the housekeeping gene (GAPDH for mRNAs and U47 for miRNA). After obtaining ΔCT, ΔΔCT was calculated as ΔCT of exposed samples from ΔCT of non-exposed samples. As the efficiency of all real-time PCR conditions were 100% ±3, the relative expression was calculated by 2−ΔΔCT. Relative quantification of RRM1 and CDA was calculated by delta–delta CT method and actual fold change in expression raised to power of \( 2 { }\left(2^{-\Delta \Delta \text{CT}} \right) \).
MicroRNA transfections
MiRNA-608 precursor (pre-miR-608) and scrambled miRNA were purchased from Hum Diagnostics Company (Tehran, Iran). About 3 × 106 of cells were transfected with 100 µl of vector containing mixture according to the transfection method of calcium phosphate. Transfection mixture included 8 μg of pre-miR-608 in 422 µl of 1× HBS and 16 µl TE1% buffer. Next 62 μl of CaCl2 (2.5 M) solution was added and mixed immediately. In the following step, 500 µl of 2× HBS (drop-wise) was added slowly to the above mixture. Thereafter, 1 ml of the final mix was dropped around the plate, and mixed gently. After 6 h, the transfection medium was substituted by fresh medium. The transfected cell lines were harvested 24, 48, and 72 h post-transfection.
Construction of luciferase reporter plasmids and evaluation of luciferase activity
3′UTR of rrm1 and cda containing the miR-608 recognition site were inserted into psiCHECK™2 vector (Promega) downstream to the stop codon of renilla luciferase gene confirmed by sequencing (primers are listed in Table 1).
To do luciferase assay, co-transfection of pre-miR-608 and psiCHECK–RRM1 and/or psiCHECK–CDA vectors into MIA PaCA, AsPC and 293-T was performed. Transfected solution was composed of 400 ng/µl psiCHECK–RRM1and/or psiCHECK–CDA and 100 ng/µl of each precursor miRs (608 or scrambled miRs). In the next day, luciferase activities were measured 24 and 48 h after transfection using Dual-Glo® Luciferase Assay System (Promega, E2940). Results were normalized to control samples. All tests were done in triplicate.
MTT proliferation assay
MTT assay (Sigma, M2128) was performed for assaying the cell proliferation. 2 × 104 of cells were added to each well and exposed to different concentrations (10 nM–1000 µM) of gemcitabine for 72 h. Based on the previous articles the least and the most reported IC50 for gemcitabine were chosen and MTT assays were carried out to find a proper inhibitory concentration of gemcitabine in MIA PaCa-2 and AsPC-1 cells [24]. MTT proliferation assay was done according to the previously mentioned protocols [22]. MTT chemosensitivity assays were carried out in triplicate which was repeated three times in the separate experiments. MTT proliferation assay on MIA PaCa-2 and AsPC-1 cells was performed 48 h after transfection with pre-miR-608 and miR-scrambled. The inhibitory effect of drugs on cell proliferation in each well was calculated by calculating the ([average absorbance of drug containing wells − average absorbance of blank wells]/[average absorbance of drug-free wells − average absorbance of blank wells]) × 100 (100% survival). Putting concentration and average percentage viability of the different clones in excel, a scatter plot of the data was made. We chose the trend line and equation on chart showing R-squared for each experiment. By solving the equation of each experiment the exact IC50% of the cells could be calculated.
Statistical methods
All the experiments were repeated at least three times. Results are expressed as mean ± standard error (SE). SPSS software (version 23, SPSS, Inc., Chicago, IL) was used for calculating Pearson correlation analysis. P value <0.05 was considered statistically significant.
Results
3′UTR of RRM1 and CDA have complementary sequence for miR-608
To identify the putative miRNAs targeting 3′UTR of RRM1 and CDA gene, an in silico approach was used, which was a combination of bioinformatics algorithms included TargetScan4.0 (http://www.targetscan.org/), miRwalk (http://zmf.umm.uni-heidelberg.de/apps/zmf/mirwalk2/), PicTar (http://www.pictar.mdc-berlin.de/), DIANA-microT (http://www.microrna.gr/ web Server) and FINDTAR3 (http://bio.sz.Tsinghua.edu.cn/) which are dedicated to miRNA target prediction and functional analysis. Due to four complementary sequences of miR-608 in 3′UTR of CDA gene and one in RRM1 (Fig. 1a) and good scores for miR-608 in binding to these genes, it was selected for further studies.
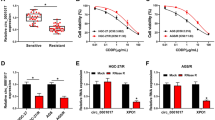
a Complementary sequences for miR-608 in 3′UTR of RRM1 and CDA genes; b luciferase assay for assessing the effect of pCDNA3.1-miR608 on 3′UTR of RRM1 and CDA. Data are mean ± SD from three independent experiments. Bars are indicative of standard deviations. Significant differences (p < 0.05) in the results are labeled with asterisks (Student T test)
RRM1 and CDA genes are imminent targets of miR-608
Co-transfection of premature-miR-608 with psiCHECK–RRM1 or psiCHECK–CDA into MIA PaCa-2 cell line resulted in reduced luciferase activity to 0.46 ± 0.05 and 0.48 ± 0.06%, respectively, compared to control group. Transfecting psiCHECK–RRM1 and/or psiCHECK–CDA together with premature-miR-608 into AsPC-1 cells decreased the luciferase activity to 0.45 ± 0.076 and 0.49 ± 0.072, respectively. To evaluate this phenomenon on a cell line other than pancreatic cancer cell lines, 293-T cells were subjected to co-transfection of premature-miR-608 in addition to psiCHECK–RRM1 or psiCHECK–CDA. Decreased luciferase activity to 0.42 ± 0.19 and 0.43 ± 0.17, respectively, were seen in transfected T-293 cell line. For negative control, a scrambled sequence was used. As shown in Fig. 1b, results showed that premature-miR-608 in comparison with scrambled miRNA downregulated the expression of luciferase in all transfected cells significantly (Student T test) which were in line with our bioinformatics and gene expression findings and validated that RRM1 and CDA as direct targets of miR-608.
Induction of Gemcitabine resistance in MIA PaCa-2 and AsPC-1 cell lines
IC50 of MIA PaCa-2 for gemcitabine was 20.95 ± 8.8 µM, while IC50 of AsPC-1 was 32.36 ± 7.9 µM. Initial incubation doses were below IC50 (AsPC-1 = 25 μM and MIA PaCa-2 = 10 μM) and exposure was continued by increasing the drug doses (Fig. 2a, b). Although during the process of resistance induction in MIA PaCa-2, IC50 level changed from 20.95 to 55.9 µM in four steps of resistance induction, in AsPC-1 cells IC50 level increased from 32.36 to 61.48 µM which only continued in two respective resistant cell lines. Finally the IC50 of resistant AsPC-1 and MIA PaCa-2 cells was near to each other. Resistant cell lines in this research were called resistant in comparison with the parental cell lines. In MIA PaCa-2 resistant cells, obtained IC50 s were 32.25 ± 5.1, 37.08 ± 4.7, 49.4 ± 7.5 and 55.9 ± 5.5 μM, while in AsPC-1 resistant cell lines, IC50 s were 41.41 ± 5.2, and 61.48 ± 3.8 μM, consecutively. IC50 of MIA PaCa-RG4 cells was 2.66 fold more than MIA PaCa-2 and IC50 of AsPC-RG2 cells was 1.89-fold more than non-resistant AsPC-1 cells. IC50 of the last resistant clones was significantly different from the parental cell lines.
MTT assay analyses (IC50) in consecutive resistant clones of a AsPC-1 and b MIA PaCa-2 cells showed significantly lower anti-proliferative effect of gemcitabine in MIA PaCa2-RGs than parental MiaPaCa-2 cells (MIA PaCa2-S) and in AsPC-1-RGs than in parental AsPC-1 cells (AsPC-1-S). As it is shown in the graph, viability of the resistant clones was higher in concentration below the respective IC50%. Values are representative of mean ± SD obtained from at least three independent tests
Compared to the parental cells, MIA PaCa-2 resistant cells changed to elongated spindle-shaped cells in which the proliferation time decreased so much that it took 4 weeks and higher to reach proper number of cells after each round of the drug exposition. On the contrary, AsPC-1 cell line had no significant morphologic changes during resistance induction.
Expression level of RRM1 and CDA was higher in gemcitabine-resistant clones compared to the parental cells
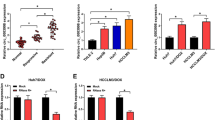
Interestingly, RRM1 and CDA expression was significantly higher in gemcitabine-resistant cells. As it can be seen in Fig. 2b, RRM1 and CDA expression changed about 1.6 ± 0.45 and 1.86 ± 0.79 fold, respectively, in AsPC-RG1 (p = 0.02 and 0.01), whilst they increased to 3.34 ± 0.35 and 4.14 ± 0.68-fold in AsPC-RG2 compared with the parental AsPC-1 cell line (p = 0.001 and 0.027). Although the first clones of resistant MIA PaCa-RG1 did not show significant alteration in both genes, CDA level showed 2.31-, 5.3- and 4-fold increase in MIA PaCa-RG2, MIA PaCa-RG3 and MIA PaCa-RG4, respectively, that they were all significantly different (p ≤ 0.05). On the other hand, RRM1 expression in MIA PaCa-RG1 was 0.2 fold in comparison with MIA PaCa-2, MIA PaCa-RG2, MIA PaCa-RG3 and MIA PaCa-RG4 showed the most gross upregulation (2.66, 4.5 and 3.73, respectively) compared to the sensitive parental cells (p ≤ 0.05) (Fig. 2a). Data demonstrated that the expressions of RRM1 and/or CDA in MIA PaCa-2 were related to IC50 (R = 0.948, p = 0.014 for RRM1), (R = 0.908, p = 0.033 for CDA), respectively. Also in AsPC-1 cell line, there was direct relationship between RRM1 expression and related IC50 (R = 0.956, p = 0.011 for RRM1), but not for CDA (R = 0.954, p = 0.193 for CDA) (Fig. 3c).
a Expression level of RRM1 and CDA in resistant clones of MIA PaCa-2 and b AsPC-1 cells, c MiR-608 expression in different clones of MIA PaCa-2 cells and d AsPC-1 cells. QRT–PCR demonstrated significantly lower miR-608 expression in MiaPaCa-2-RGs and AsPC-1-RGs clones than their parental cells. Values are means from triplicate experiments performed with each condition; bars indicate SD. Stars show the significant statistical difference (*p value <0.05)
MiR-608 was downregulated in gemcitabine-resistant clones
Expression level of miR-608 was measured in each clone. Interestingly compared to the sensitive cells, MIA PaCa-2 resistant clones showed decreased expression of miR-608 as below: MIA PaCa-RG1 0.75 ± 0.024, MIA PaCa-RG2 0.31 ± 0.056, MIA PaCa-RG3 0.54 ± 0.069 and MIA PaCa-RG4 0.65 ± 0.12. Whereas AsPC-1 resistant clones displayed lower level of miR-608 compared to the control cells as you see below: AsPC-RG1 0.53 ± 0.01, AsPC-RG2 0.68 ± 0.07 (p ≤ 0.05) (Fig. 3b, d). Moreover, there is a significant correlation between decreased expression level of miR-608 and IC50(R = −0.927; p = 0.023). Although there were no ordered sequences of reduction in miR-608 level in the resistant clones (Fig. 3), down-regulation of miR-608 is likely to be associated with resistance to gemcitabine.
MiR-608 increased sensitivity of MIA PaCa-2 and AsPC-1 to gemcitabine
To identify if miR-608 could impact the response of cells to gemcitabine, MIA PaCa-2 and AsPC-1 cell lines were transfected with premature-miR-608 and 24 h later they were incubated with different concentration of gemcitabine including IC50 of parental cell lines. MTT assay showed that in transfected MIA PaCa-2 cells IC50 value decreased to about 17.09 ± 4 µM while in AsPC-1 cell line this value decreased to 25.13 ± 7 µM (Fig. 4a, b).
a, b MTT assay analyses (IC50) in MIA PaCa and AsPC cell lines after transfection with pre-miR-608 and treatment with different concentration of gemcitabine. c, d MTT assay results (IC50) in MIA PaCa-RG4 and AsPC-RG-2 after transfection with pre-miR-608 transfection and treatment with different concentrations of gemcitabine. Data represents mean ± SD from three independent experiments. Significant differences in the results are labeled with stars
To study the effect of miR-608 on the drug sensitivity of the resistant cell lines, MIA-PaCa2-RG4 and AsPC-RG2 resistant clones were transfected with this miRNA. The day after, they were incubated with different concentrations of gemcitabine to determine any possible changes in the IC50. Pre-miR-608 transfection reduced IC50 from 61.48 to 27.14 µM in AsPC-RG2 while the IC50 of MIA PaCa-RG4 was decreased from 55.9 to 30.51 µM (Fig. 4c, d). These results suggest that miR-608 could be at least one of the gemcitabine resistance mediators in the MIA PaCa-2 and AsPC-1 cell lines that modulate their drug response. This miRNA sensitized AsPC-RG2 cell line more profoundly but it could not sensitize MIA PaCa-RG4 completely.
Expression level of RRM1 and CDA were decreased after transfection of cells with premature-miR-608
To investigate the association between miR-608 expression and RRM1 or CDA levels, a gain-of-function survey was carried out. As it is shown in Fig. 5a, c overexpression of miR-608 is significantly correlated with down-regulation of both target genes (p ≤ 0.05). In MIA PaCa-2 cells, increased expression of miR-608 decreased the expression of RRM1 to 0.39 (R = −0.968; p = 0.007) while the decreased expression of CDA was up to 0.42 (R = −0.936; p = 0.019). In AsPC-1 cells overexpression of miR-608 considerably reduced RRM1 to 0.43 (R = −0.908; p = 0.003) and CDA to 0.37 (R = −0.812; p = 0. 283).
a, c The effect of pCDNA3.1-miR-608 on expression of CDA and RRM1 genes in sensitive pancreatic cancer cell lines, and b, d resistant cells; e measuring miR-608 overexpression after transient transfection of MIA PaCa-2 and AsPC-1 cell lines. Data represent mean ± SD of three independent experiments. Asterisk show significant differences different groups (*p < 0.05)
It was proved that transfection of resistant cells with pre-miR-608 increased the level of miR-608 in the cells (Fig. 5e). After transfection with pre-miR-608, in resistant MIA PaCa-RG4 cells in comparison with non-transfected cells, RRM1 and CDA transcript levels were reduced to 0.45 and 0.47, respectively. In pre-miR-608 transfected AsPC-RG2 cells, reduction of expression in comparison with non-transfected cells were 0.40 and 0.42 for RRM1 and CDA, respectively (Fig. 5b, d). In transfected MIA PaCa-RG4 and AsPC-RG2 cells, RRM1 (R = −0.85 and −0.908) and CDA (R = −0.930 and −0.936) expressions were correlated with miR-608 expression. These results illustrated that miR-608 mediates regulation of RRM1 and CDA transcript expression in miR-608 transfected sensitive and resistant cells. Surprisingly CDA and RRM1 transcripts were affected in almost the same manner in all cell lines including sensitive and resistant cell lines.
Discussion
To the best of our knowledge, elevated expression of some genes like RRM1 and CDA is correlated with gemcitabine resistance and lower survival in patients with the pancreatic cancer [11, 12, 24]. In addition, some of the studies have considered a predictive value for RRM1 and CDA levels [11]. In vitro induction of resistance to gemcitabine in pancreatic cancer lines eventuated to 4.5-fold increase in RRM1 expression [9, 24]. The mechanism underlying gemcitabine resistance in RRM1 overexpressing cell lines may contribute to ‘‘molecular sink’’ property of RRM1 which inactivates the drug through binding of RRM1 subunit to the gemcitabine. To compensate lower ribonucleotide reductase activity of the cells incubated with gemcitabine, RRM1 expression in the cells is increased [27]. Overexpression of RRM1 might result in increased deoxynucleoside triphosphate (dNTP) concentration, which can compete with the incorporation of triphosphorylated gemcitabine into DNA [10]. In fact, resistant clones rectify lower level of dNTPs with higher RRM1 [10].
CDA catalyzes the deamination and salvaging of pyrimidine-containing compounds such as gemcitabine. Increased activity and expression level of CDA was associated with early progression of disease in pancreatic cancer patients treated with gemcitabine compared to whose CDA expression levels was low [28]. In the study of Davidson et al., two non-small cell lung cancer cell lines which have been exposed to increasing concentration of gemcitabine, demonstrated considerable increased expression of ribonucleotide reductase subunit 1 mRNA (RRM1). While in one study, CDA expression was slightly decreased in gemcitabine-resistant lung cancer cell lines [27]. Liu et al. [29] showed that CDA and RRM1 expressions were increased along with the gemcitabine resistance induction in these cell lines.
Cancer cells exhibit unique expression signature of specific miRNAs. Not only they act as cancer biomarkers, but also some of them have been connected with drug resistance phenotype in pancreatic cancer, including miR-15a, miR-21, miR-34, miR-200b, miR-200c, miR-214 and miR-221 [5, 30].
Present study uncovers the new role of miR-608 in gemcitabine-resistant clones derived from pancreatic cancer cell lines as the common regulator of RRM1 and CDA. In the previous studies, association between miR-608 polymorphism and recurrence of nasopharyngeal carcinoma has been investigated [31, 32]. Also Ryan et al. [33] showed that there is a significant association between rs4919510: C > G polymorphism and survival rate of colorectal cancer patients but not risk of this type of cancer. However, Huang et al. [34] ascertained the contribution of rs4919510: C > G to the risk of HER2-positive breast cancer rather than the other subtypes.
Through resistance induction in MIA PaCa-2 and AsPC-1 cells, RRM1 and CDA expression increased. This increase in the level of RRM1 and CDA expression was correlated with the increased level of IC50 in MIA PaCa-2 and AsPC-1.
We hypothesized that the favorite miRNA candidate was a miRNA that could target both genes with a good score. Luckily, in silico analysis showed that miR-608 have complementary sequences in 3′UTR of both genes. Luciferase assay validated that miR-608 was directly targeting 3′UTR of RRM1 and CDA. While downregulation of miR-608 was seen in resistant clones, MTT assay results after transferring pre-miR-608 to these cell lines, showed increased sensitivity of the cells to gemcitabine. Transfection with Pre-miR-608 in MIA PaCa-RG4 did not shift the IC50 to the original value of the sensitive parental cell line while in AsPC-RG2 the IC50 of transfected cell line was even less than parental cell line (AsPC). This suggest that there may be other mechanism of resistance induction in MIA PaCa-RG4 while in AsPC-RG2 miR-608 can neutralize all mechanisms of resistance induction and sensitizes the cell more than parental cell line. Sensitization of MIA PaCa and AsPC cell lines after transfection with pre-miR-608 was 18.42 and 22.34% while in MIA PaCa-RG4 and AsPC-RG2 these values were 45.42 and 55.85%, respectively, which might be due to different mechanisms of resistance induction in these two cell lines. While sensitization by miR-608 in induced resistant cell lines was more profound, this effect was also more significant in AsPC. This miRNA is targeting RRM1 and CDA, the sensitization effect may be through targeting RRM1 and CDA genes. It may be hypothesized that down-regulation of this miRNA play greater role in resistance induction of AsPC cell line.
However, in the last clones, increased expression of CDA and RRM1 had not linear correlation with decreased expression of miR-608. This phenomenon can be related to miRNA-mediated gene regulatory network including interactions between miRNA and their different targets or different transcriptional factors (TFs) [35]. For instance feedback loops (FBLs) that can fine-tune the gene expression, can be affected by internal and external factors [36]. Internal factors that may confer miRNA regulation network consist of randomized transcription and/or translation and molecular depravity [37], while external agents refer to environment fluctuations [38]. In highly resistant cell lines to gemcitabine other factors and gene mutations may confer the pancreatic cancer responsiveness; hence miR-608 may not be the only important factor for this resistance [39]. Also genetic and epigenetic changes may alter the expression of miR-608 in resistant cell lines [40]. So it can be concluded that after some period of time due to some unknown mechanisms, fluctuation in genes and miRNA was seen.
Although most cases of the long term exposure of the cells with the drugs resulted in obtaining high levels of resistance (100–10,000-fold increase in IC50), there are other reports showing lower increase in the IC50 value for gemcitabine in the case of resistance induction. Achiwa et al. [41] showed that in resistant lung cancer cell line, the IC50 was about 8.6 times higher than the parental cell line. Concordantly in our study, the IC50 of resistant cell lines did not increase exceedingly. In other studies gemcitabine resistance manifested a threshold level and there was no profound change in the IC50 s after prolonged drug exposure [42]. Moreover, since in our study the gemcitabine IC50 of parental cell lines was higher in comparison with the previous reports, IC50 in the resistant cell lines did not increase profoundly. Since altered expression of multiple individual genes might lead to resistance to gemcitabine in pancreatic cancer and in this study we just identified two of these genes and one miRNA. hence, variable response in the two transfected resistant cell lines may be the result of the molecular variability among these two gemcitabine-resistant models.
Our result is the first report revealing the direct repression role of miR-608 on RRM1 and CDA mediating responsiveness of pancreatic cancer cells to gemcitabine. Up to now, all studies on gemcitabine resistance are based on in vitro acquired resistance and no data are available in animal model systems showing a relationship between RRM1 and CDA expression and acquired gemcitabine resistance after gemcitabine administration. Also, miR-608 has not yet been reported to have implication in the pathology or prognosis of pancreatic cancer.
Tumor-suppressor-miRs have been indicated as ideal therapeutic tools in the battle against cancer [43], while our results showed it could be a potential target for sensitizing pancreatic cancer cells to chemotherapeutic drugs. Other types of cancer which are treated by gemcitabine would be good candidates for studying the expression of this miRNA and studying the genes targeted by this miRNA.
In vivo analysis of the effect of miR-608 in addition to gemcitabine in pancreatic cancer tissues should be done before human studies. Then therapeutic manipulation of miR-608 is ought to be explored in patients sample and if successful, might open opportunities to modify treatment strategies in pancreatic cancer and other types of cancer such as lung cancer as well.
In conclusion, it is suggested that to prescribe miR-608 along with gemcitabine to augment the effect of gemcitabine in pancreatic cancer patients. To achieve this goal, more assays such as in vivo analysis of the effect of miR-608 on the gemcitabine metabolism have to be done.
Abbreviations
- PDAC:
-
Pancreatic adenocarcinoma
- GEM:
-
Gemcitabine
- CDA:
-
Cytidine deaminase
- RRM1:
-
Ribonucleotide reductase M1
- dNTP:
-
Deoxynucleoside triphosphate
- miR:
-
miRNA, microRNA
- dFdU:
-
2′2′-Difluorodeoxyuridine
- IC:
-
Inhibitory concentration
- MTT:
-
3-(4,5-Di methylthiazole-2-yl)-2, 5-biphenyl tetrazolium bromide
References
Long J, Zhang Y, Yu X, Yang J, LeBrun DG, Chen C, Yao Q, Li M (2011) Overcoming drug resistance in pancreatic cancer. Expert Opin Ther Targets 15(7):817–828
Valsecchi ME, Holdbrook T, Leiby BE, Pequignot E, Littman SJ, Yeo CJ, Brody JR, Witkiewicz AK (2012) Is there a role for the quantification of RRM1 and ERCC1 expression in pancreatic ductal adenocarcinoma? BMC Cancer 12(1):1
Fujita H, Ohuchida K, Mizumoto K, Itaba S, Ito T, Nakata K, Yu J, Kayashima T, Souzaki R, Tajiri T (2010) Gene expression levels as predictive markers of outcome in pancreatic cancer after gemcitabine-based adjuvant chemotherapy. Neoplasia 12(10):807–808
Hagmann W, Jesnowski R, Löhr JM (2010) Interdependence of gemcitabine treatment, transporter expression, and resistance in human pancreatic carcinoma cells. Neoplasia 12(9):740–747
Rajabpour A, Rajaei F, Teimoori-Toolabi L (2016) Molecular alterations contributing to pancreatic cancer chemoresistance. Pancreatology 17(2):310–320
Rukov JL, Shomron N (2011) MicroRNA pharmacogenomics: post-transcriptional regulation of drug response. Trends Mol Med 17(8):412–423
Bhutia YD, Hung SW, Patel B, Lovin D, Govindarajan R (2011) CNT1 expression influences proliferation and chemosensitivity in drug-resistant pancreatic cancer cells. Can Res 71(5):1825–1835
Ueno H, Kiyosawa K, Kaniwa N (2007) Pharmacogenomics of gemcitabine: Can genetic studies lead to tailor-made therapy? Br J Cancer 97(2):145–151
Nakano Y, Tanno S, Koizumi K, Nishikawa T, Nakamura K, Minoguchi M, Izawa T, Mizukami Y, Okumura T, Kohgo Y (2007) Gemcitabine chemoresistance and molecular markers associated with gemcitabine transport and metabolism in human pancreatic cancer cells. Br J Cancer 96(3):457–463
Souglakos J, Boukovinas I, Taron M, Mendez P, Mavroudis D, Tripaki M, Hatzidaki D, Koutsopoulos A, Stathopoulos E, Georgoulias V (2008) Ribonucleotide reductase subunits M1 and M2 mRNA expression levels and clinical outcome of lung adenocarcinoma patients treated with docetaxel/gemcitabine. Br J Cancer 98(10):1710–1715
Jordheim LP, Dumontet C (2013) Do hENT1 and RRM1 predict the clinical benefit of gemcitabine in pancreatic cancer? Biomarkers 7(4):663–671
Jordheim LP, Sève P, Trédan O, Dumontet C (2011) The ribonucleotide reductase large subunit (RRM1) as a predictive factor in patients with cancer. Lancet Oncol 12(7):693–702
Elnaggar M, Giovannetti E, Peters GJ (2012) Molecular targets of gemcitabine action: rationale for development of novel drugs and drug combinations. Curr Pharm Des 18(19):2811–2829
Carpi FM, Vincenzetti S, Ubaldi J, Pucciarelli S, Polzonetti V, Micozzi D, Mignini F, Napolioni V (2013) CDA gene polymorphisms and enzyme activity: genotype–phenotype relationship in an Italian-Caucasian population. Pharmacogenomics 14(7):769–781
Sugiyama E, Kaniwa N, Kim S-R, Kikura-Hanajiri R, Hasegawa R, Maekawa K, Saito Y, Ozawa S, J-i Sawada, Kamatani N (2006) Pharmacokinetics of gemcitabine in Japanese cancer patients: the impact of a cytidine deaminase polymorphism. J Clin Oncol 25(1):32–42
Funamizu N, Okamoto A, Kamata Y, Misawa T, Uwagawa T, Gocho T, Yanaga K, Manome Y (2010) Is the resistance of gemcitabine for pancreatic cancer settled only by overexpression of deoxycytidine kinase? Oncol Rep 23(2):471
Rukov JL, Wilentzik R, Jaffe I, Vinther J, Shomron N (2013) Pharmaco-miR: linking microRNAs and drug effects. Brief Bioinform 15:648–659
Hashimoto Y, Akiyama Y, Yuasa Y (2013) Multiple-to-multiple relationships between microRNAs and target genes in gastric cancer. PLoS ONE 8(5):e62589
Zhang XJ, Ye H, Zeng CW, He B, Zhang H, Chen YQ (2010) Dysregulation of miR-15a and miR-214 in human pancreatic cancer. J Hematol Oncol 3(1):1
Hwang J-H, Voortman J, Giovannetti E, Steinberg SM, Leon LG, Kim Y-T, Funel N, Park JK, Kim MA, Kang GH (2010) Identification of microRNA-21 as a biomarker for chemoresistance and clinical outcome following adjuvant therapy in resectable pancreatic cancer. PLoS ONE 5(5):e10630
Ji Q, Hao X, Zhang M, Tang W, Yang M, Li L, Xiang D, DeSano JT, Bommer GT, Fan D (2009) MicroRNA miR-34 inhibits human pancreatic cancer tumor-initiating cells. PLoS ONE 4(8):e6816
Li Y, VandenBoom TG, Kong D, Wang Z, Ali S, Philip PA, Sarkar FH (2009) Up-regulation of miR-200 and let-7 by natural agents leads to the reversal of epithelial-to-mesenchymal transition in gemcitabine-resistant pancreatic cancer cells. Can Res 69(16):6704–6712
Park J-K, Lee EJ, Esau C, Schmittgen TD (2009) Antisense inhibition of microRNA-21 or-221 arrests cell cycle, induces apoptosis, and sensitizes the effects of gemcitabine in pancreatic adenocarcinoma. Pancreas 38(7):e190–e199
Nakahira S, Nakamori S, Tsujie M, Takahashi Y, Okami J, Yoshioka S, Yamasaki M, Marubashi S, Takemasa I, Miyamoto A (2007) Involvement of ribonucleotide reductase M1 subunit overexpression in gemcitabine resistance of human pancreatic cancer. Int J Cancer 120(6):1355–1363
Azizi M, Teimoori-Toolabi L, Arzanani MK, Azadmanesh K, Fard-Esfahani P, Zeinali S (2014) MicroRNA-148b and microRNA-152 reactivate tumor suppressor genes through suppression of DNA methyltransferase-1 gene in pancreatic cancer cell lines. Cancer Biol Ther 15(4):419–427
Gheidari F, Bakhshandeh B, Teimoori-Toolabi L, Mehrtash A, Ghadir M, Zeinali S (2014) TCF4 silencing sensitizes the colon cancer cell line to oxaliplatin as a common chemotherapeutic drug. Anticancer Drugs 25(8):908–916
Davidson JD, Ma L, Flagella M, Geeganage S, Gelbert LM, Slapak CA (2004) An increase in the expression of ribonucleotide reductase large subunit 1 is associated with gemcitabine resistance in non-small cell lung cancer cell lines. Can Res 64(11):3761–3766
Bengala C, Guarneri V, Giovannetti E, Lencioni M, Fontana E, Mey V, Fontana A, Boggi U, Del Chiaro M, Danesi R (2005) Prolonged fixed dose rate infusion of gemcitabine with autologous haemopoietic support in advanced pancreatic adenocarcinoma. Br J Cancer 93(1):35–40
Liu X, Wang W, Lin L, Song S (2010) Expression of Gemcitabine-resistance-related gene and polymorphism of ribonucleotide reductase M1 gene promoter in Gemcitabine-resistant A549/Gem and NCI-H460/Gem cell lines. Zhonghua Zhong Liu Za Zhi [Chin J Oncol] 32(1):17–21
Wang J, Chen J, Chang P, LeBlanc A, Li D, Abbruzzesse JL, Frazier ML, Killary AM, Sen S (2009) MicroRNAs in plasma of pancreatic ductal adenocarcinoma patients as novel blood-based biomarkers of disease. Cancer Prev Res 2(9):807–813
Zheng J, Deng J, Xiao M, Yang L, Zhang L, You Y, Hu M, Li N, Wu H, Li W (2013) A sequence polymorphism in miR-608 predicts recurrence after radiotherapy for nasopharyngeal carcinoma. Can Res 73(16):5151–5162
Ma X-P, Yu G, Chen X, Xiao Q, Shi Z, Zhang L-Y, Chen H, Zhang P, Ding D-L, Huang H-X (2016) MiR-608 rs4919510 is associated with prognosis of hepatocellular carcinoma. Tumor Biol 37(7):9931–9942
Ryan BM, McClary AC, Valeri N, Robinson D, Paone A, Bowman ED, Robles AI, Croce C, Harris CC (2012) rs4919510 in hsa-mir-608 is associated with outcome but not risk of colorectal cancer. PLoS ONE 7(5):e36306
Huang A-J, Yu K-D, Li J, Fan L, Shao Z-M (2012) Polymorphism rs4919510: C > G in mature sequence of human microRNA-608 contributes to the risk of HER2-positive breast cancer but not other subtypes. PLoS ONE 7(5):e35252
Alves R, Antunes F, Salvador A (2006) Tools for kinetic modeling of biochemical networks. Nat Biotechnol 24(6):667–672
Wang S, Raghavachari S (2011) Quantifying negative feedback regulation by micro-RNAs. Phys Biol 8(5):055002
Raj A, van Oudenaarden A (2008) Nature, nurture, or chance: stochastic gene expression and its consequences. Cell 135(2):216–226
Pedraza JM, van Oudenaarden A (2005) Noise propagation in gene networks. Science 307(5717):1965–1969
Bhardwaj V, Bhushan A, Lai JC, Tadinada SM (2012) Failure of pancreatic cancer chemotherapy: consequences of drug resistance mechanisms. INTECH Open Access Publisher, Rijeka
Azmi AS, Beck FW, Bao B, Mohammad RM, Sarkar FH (2011) Aberrant epigenetic grooming of miRNAs in pancreatic cancer:a systems biology perspective. Epigenomics 3(6):747–759
Achiwa H, Oguri T, Sato S, Maeda H, Niimi T, Ueda R (2004) Determinants of sensitivity and resistance to gemcitabine: the roles of human equilibrative nucleoside transporter 1 and deoxycytidine kinase in non-small cell lung cancer. Cancer Sci 95(9):753–757
Samulitis BK, Pond KW, Pond E, Cress AE, Patel H, Wisner L, Patel C, Dorr RT, Landowski TH (2015) Gemcitabine resistant pancreatic cancer cell lines acquire an invasive phenotype with collateral hypersensitivity to histone deacetylase inhibitors. Cancer Biol Ther 16(1):43–51
Kasinski AL, Slack FJ (2011) MicroRNAs en route to the clinic: progress in validating and targeting microRNAs for cancer therapy. Nat Rev Cancer 11(12):849–864
Funding
This research was funded by Qazvin University of Medical Sciences (Grant Number: 28/20/10563) and Pasteur Institute of Iran (Grant Number: 94/0201/1405).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
Azam Rajabpour declares that she has no conflict of interest. Ali Afgar declares that he has no conflict of interest. Habibollah Mahmoodzadeh declares that he has no conflict of interest. Jalal-e-Din Radfar declares that he has no conflict of interest. Farzad Rajaei declares that he has no conflict of interest. Ladan Teimoori-Toolabi declares that she has no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Rights and permissions
About this article
Cite this article
Rajabpour, A., Afgar, A., Mahmoodzadeh, H. et al. MiR-608 regulating the expression of ribonucleotide reductase M1 and cytidine deaminase is repressed through induced gemcitabine chemoresistance in pancreatic cancer cells. Cancer Chemother Pharmacol 80, 765–775 (2017). https://doi.org/10.1007/s00280-017-3418-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00280-017-3418-2