Abstract
Purpose
Neutropenia is a life-threatening side effect of irinotecan, and uridine diphosphate glucuronosyltransferases (UGTs) gene polymorphisms are considered to be one of the predictive markers of irinotecan-related toxicities. Many studies have demonstrated that patients bearing UGT1A1*28 have a higher risk of severe neutropenia on toxicity of irinotecan. However, UGT1A1 (TA7/TA7) was very rare in Asian populations. Some researches reported that UGT1A1*28 and/or UGT1A1*6 could predict irinotecan-induced toxicities in Asian populations, but controversial conclusions still remained. This study aims to investigate the association between UGT1A1 gene polymorphisms *6, *6/*28 and irinotecan-related neutropenia in Asian cancer patients receiving irinotecan regimen chemotherapy.
Experimental design
Meta-analyses were done to assess the relationship between UGT1A1*6 or UGT1A1*6/*28 and irinotecan-induced neutropenia.
Results
The risk of neutropenia was significantly higher among patients with a UGT1A1*6 genotype than among those carrying the UGT1A1*1 allele(s) [odds ratio (OR) 3.276; 95 % confidence interval (CI) 1.887–5.688; P = 0.000 (*6/*6 vs. *1/*6 or *1/*1)], [OR 1.542; 95 % CI 1.180–2.041; P = 0.001 (*6/*6 or *1/*6 vs. *1/*1)]. Also, the risk was significantly higher among patients with a UGT1A1*6/*28 than among those carrying the UGT1A1*1 allele(s) [OR 3.275; 95 % CI 2.152–4.983; P = 0.000 (*6/*6 or *28/*28 or *6/*28 vs. *1/*6 or *1/*28 or *1/*1)].
Conclusions
In conclusion, the UGT1A1*6 and UGT1A1*6/*28 genotypes were associated with an increased risk of irinotecan-induced neutropenia in Asian cancer patients.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Camptothecin is a topoisomerase I inhibitor that causes cancer cell death by forming cleavable complexes with cell DNA, resulting in DNA strand breaks. Irinotecan hydrochloride (CPT-11, CAMPTOSAR) is a prodrug derivative of camptothecin, an alkaloid obtained from plants such as the Camptotheca acuminata tree. Irinotecan is a valuable drug in the treatment for several types of solid tumor, especially colon, small cell lung [1–3], and gynecological cancers [4, 5]. First-line treatment with irinotecan plus fluorouracil and leucovorin had led to improved survival in patients with advanced colon cancer [6]. However, irinotecan has significant side effects, including neutropenia and delayed-type diarrhea. The risk of life-threatening neutropenia of irinotecan has been consistently associated with genetic variations in UGT1A1 [7, 8]. Irinotecan requires metabolic activation by carboxylesterase 2 to form the active metabolite SN-38, which is further cleared via formation of SN-38 glucuronide (SN-38G) by UGT1A1 isoforms in human liver [9]. Meta-analyses have shown that individuals with reduced UGT1A1 activity, as detected by the presence of the UGT1A1*28 allele, have increased risk of the two major adverse effects of irinotecan: neutropenia and diarrhea.
The US Food and Drug Administration (FDA) has informed that individuals who are homozygous for the UGT1A1*28 allele (UGT1A1 7/7 genotype) are at increased risk for neutropenia following initiation of irinotecan treatment. FDA amended the label of irinotecan in November 2004 and approved the diagnostic UGT1A1 test (Invader UGT1A1 Molecular Assay: Third Wave Technologies Inc., Madison, WI, USA) in August 2005 [10]. The frequency of UGT1A1*28 was higher in whites than in Asians (40–50 vs. 15–20 % for heterozygosity; about 10 vs. 4–6 % for homozygosity) [9, 11–15]. In contrast, the polymorphism of UGT1A1*6 characterized by a single-nucleotide substitution in exon 1 of UGT1A1 (211 G > A; GG, GA, and AA genotypes; G71R) and related with reduced SN-38 glucuronidation activity [16, 17] occurred at a higher frequency in Asians [18, 19]. Studies reported that the polymorphism of UGT1A1*6 was associated with irinotecan-induced neutropenia in Asians [16, 20].
Although many studies had been conducted to investigate the associations between UGT1A1*6 or UGT1A1*28/*6 polymorphisms and irinotecan-induced neutropenia, the conclusions were still controversial [21–36]. Our study was conducted to elucidate whether UGT1A1*6 and UGT1A1*6/*28 polymorphisms could predict irinotecan-induced neutropenia in Asian cancer patients on a large scale.
Materials and methods
Bioinformatics analysis of ugt1A1 G71R protein
The characteristics of wild-type ugt1A1 and the G71R mutation were calculated by CLC Protein Workbench software: hydrophobicity, protein charge, antigenicity, and secondary structure of wild-type and mutated ugt1A1.
Search strategy and selection criteria
Two investigators (Fei-fei Han and Chang-long Guo) independently searched PubMed and Embase (from 1980 until July 16, 2013) database using the terms “irinotecan” and “UGT1A1” and “neutropenia”. Furthermore, we reviewed citations in the retrieved articles to search for additional relevant studies. Articles included in meta-analysis were in English, with human subjects, published in the primary literature and with no obvious overlap of subjects with other studies. The retrieved literatures were then read in their entirety to assess their appropriateness for the inclusion in this meta-analysis. Conference abstracts, case reports, editorials, review articles, and letters were excluded. A priori we defined strict criteria for the inclusion of studies. Studies were included if (a) they could be defined as clinical trials, (b) the exposure of interest was the UGT1A1*6 or *6/*28 genotype, (c) the outcome of interest was irinotecan-induced neutropenia (grade III–IV/IV), and (d) the numbers of patients with and without neutropenia were provided. We excluded studies that were not published in English, studies that included <20 patients, and studies that included children patients.
Data extraction
The following information was abstracted from included publications: study design, year, race, irinotecan dose, number of patients with neutropenia (grade III–IV/IV) in each genotype group (UG T1A1*1/*1, UGT1A1*1/*6, and UGT1A 1*6/*6), (UGT1A1*6/*6, UGT1A1*28/*28, UGT 1A1*6/*28, UGT1A1*1/*1, UGT1A1*1/*6, and UGT1A1*1/*28), mutation detection method, and potential confounders.
Statistical analysis
Statistical analyses were performed using Review Manager (Review Manager 5.0 software) and Stata/MP 11.0. Cochran’s w 2 test and the inconsistency index (I 2) were used to evaluate heterogeneity across the included studies. P values of <0.1 for the w 2 test indicated a lack of heterogeneity, and the fixed-effects model was then used to calculate the summary odds ratio (OR). Otherwise, a random-effects model was applied. OR and their corresponding 95 % confidence intervals (CI) were estimated. Z test was performed to determine the statistical significance of pooled OR and was considered significant when P < 0.05. We also conducted sensitive analyses, where each study was excluded one at a time to determine the magnitude of influence on the overall summary estimate. We assessed potential publication bias by using a funnel plot and Egger’s test [37].
Results
Characteristics of ugt1A1 G71R protein
To assess the influence of G71R mutation to naive protein characteristics, CLC Protein Workbench software was used to test the alteration and the results turned (Fig. 1a, b): site 71 glycine is neutral amino acid and arginine is alkaline amino acid, the mutation of glycine 71 to arginine did not alter the protein charge and antigenicity of ugt1A1 greatly; however, the hydrophobicity seemed be different in ugt1A1 G71R (Fig. 1a). And the protein secondary structure prediction showed that the amino acid change also changed the seventh second structure of this protein, from alpha-strand to beta-helix (Fig. 1b).
Characteristics of wild and mutated UGT1A1 protein. a The hydrophobicity analysis of wild and mutated UGT1A1 protein. The red box indicates the hydrophobicity alteration of Gly to Arg at 71 site, b the second structure of wild and mutated UGT1A1 protein the red box indicates the changed second structure, and the red ellipse indicates the position of G71R
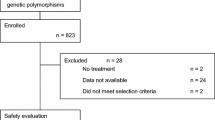
Identification of studies, data extraction, and assessment of study quality
Of the 389 possibly relevant reports identified, 18 studies matched the eligibility criteria. The results of the stages of identifying relevant studies (with reasons for study exclusion) are depicted in Fig. 2. The major reason for exclusion was missing data required to estimate the OR (including the standard error) for the OR between UGT1A1 genotypes. Characteristics of the studies included in meta-analyses are shown in Table 1. Data comparison of OR between *6/*6 and *1/*6 plus *1/*1 genotype patients were available in 8 of the included studies for the analysis [24–27, 30–33]. For the comparison of OR between *6/*6 plus *1/*6 and *1/*1 genotype groups, nine studies had relevant data [23–28, 30, 31, 34]. For the comparison of OR between *6/*28 groups, nine studies had data [23, 30, 31, 38–42]. All studies included in the meta-analyses were generally of comparable methodological quality. Table 2 shows the quality assessment results.
Association between UGT1A1*6 and irinotecan-induced neutropenia
Analysis of pooled data from all samples indicated that UGT1A1*6 allele was significantly associated with irinotecan-induced neutropenia in Asian cancer patients. In the homozygous model, the OR was 3.276 [95 % CI 1.887–5.688; P = 0.000 (*6/*6 vs. *1/*6 or *1/*1)] (Fig. 3a). For UGT1A1*1/*6 or *6/*6 versus UGT1A1*1/*1, the OR was 1.542 [95 % CI 1.180–2.041; P = 0.001 (*6/*6 or *1/*6 vs. *1/*1)] (Fig. 3b). The heterogeneity across all studies was not statistically significant for any model. I 2 values were 3.83 % (P = 0.799) and 30.6 % (P = 0.173), respectively, for homozygous and heterozygous models (Table 2). No publication bias was detected by either the funnel plot (Fig. 3a, b) or Egger’s tests (P > 0.05, each comparison) (Figure S). However, the sensitivity analysis conducted by removed and cumulative statistics have showed that combined ORs of UGT1A1*6 polymorphism was influenced by individual study under *6/*6 and *1/*6 plus *1/*1 genotype comparison.
Summary odds ratio (OR) of irinotecan-induced neutropenia and the Begg’s funnel plot for the publication bias test for UGT1A1*6 or UGT1A1*6/*28. a *6/*6 versus *1/*6 or *1/*1, b *6/*6 or *1/*6 versus *1/*1, c *6/*6 or *28/*28 or *6/*28 versus *1/*6 or *1/*28 or *1/*1. A fixed-effects model was used for all analyses. Squares represent study-specific estimates (size of the square reflects the study-specific statistical weight); horizontal lines represent 95 % confidence intervals (CIs); diamonds represent summary estimates with corresponding 95 % CIs
Association between UGT1A1*6/*28 and irinotecan-induced neutropenia
The results showed that UGT1A1*6/*28 allele was associated with irinotecan-induced neutropenia in Asian cancer patients. In the homozygous model, the OR was 3.275 [95 % CI 2.152–4.983; P = 0.000 (*6/*6 or *28/*28 or *6/*28 vs. *1/*6 or *1/*28 or *1/*1)] (Fig. 3c). The heterogeneity across all study was not statistically significant. I 2 value was 10.3 % (P = 0.65) (Table 2). No publication bias was detected by either the funnel plot (Fig. 3c) or Egger’s tests (P > 0.05, each comparison) (Figure S).
Discussion
UGT1A1*6 polymorphism is characterized by a single amino acid substitution (G71R). Based on the bioinformatic analysis, the alteration of Gly to Arg at this site changed the hydrophobicity and second structure of this protein and these might be the decreased efficiency of SN-38 glucuronidation activity. Also, the protein expression levels were found to vary among the UGT1A1s, from a low of approximately 40 %. Some researches suggested that G71R could be critical in combination with other polymorphisms in the UGT1A1 gene [17].
Personal treatment has been a popular field in recent years, and consequently, the molecules involved in the targeting and metabolism of drugs were highlighted in many studies to predict the efficacy and toxicity of treatment. The role of UGT1A1*28 polymorphism in the development of irinotecan-induced neutropenia had been documented in many studies from western countries, the US FDA claimed in 2005 that UGT1A1*28 testing should be included in the label of irinotecan as a risk factor for severe toxicity. However, many studies of Asian cancer patients showed that UGT1A1*28 was not significantly associated with irinotecan-induced neutropenia [25, 27, 28] and this might be because of the lower allelic frequency of UGT1A1*28 than whites and individuals of African descent. In Asian population, UGT1A1*6 is an important mutation variant. Some researches pointed out the importance of UGT1A1*6 in combination with UGT1A1*28 for predicting irinotecan-related neutropenia [25, 39, 43, 44].
A total of 20 publications were selected in this meta-analysis, and different data were extracted from publications for different analysis. In the UGT1A1*6 genotype analysis, significant effects were observed in both recessive genetic model (*6/*6 vs. *1/*6 and *1/*1) and dominant genetic model (*6/*6 and *1/*6 vs. *1/*1) on irinotecan-induced neutropenia in Asian. For the UGT1A1*6/*28 analysis, significant effects were observed in recessive genetic model (*6/*6, *28/*28, and *6/*28 vs. *1/*6, *1/*28 and *1/*1). Taken together, the above results provided strong evidence that the combination of UGT1A1*6 and *28 can be more clinically important than that of UGT1A1*28 in predicting the risk of irinotecan-related neutropenia in Asian.
There are limitations to this analysis. First, not all studies included adequate data for all comparison analyses. Further, there is inherent heterogeneity to all meta-analyses, and in the analyzed studies, different combinations of chemotherapy regimens were used and patients were of varied performance status. Second, there were differences in study design, doses, polymorphism detection methods, toxicity grade criteria, kinds of cancer, and pretreatment with other regimens. Especially, different combinations of chemotherapy regimens (different types and doses) could have impacted on treatment tolerability, these regimes, such as combination with 5-FU, S1, gemcitabine, with diverse doses will lead to neutropenia with different degrees, but most references involved in this study did not provide detailed data to analyze, and it should be emphasized in future researches. Especially, in most researches, grade 3 (1,000–500/mm3) or 4 (<500/mm3) neutropenia was used as the criteria. Grade 3 neutropenia is quite an acceptable toxicity from cytotoxic treatment. So, in the future studies UGT1A1*28 and UGT1A1*6, grade 4 neutropenia should be the criteria used, which is reasonable, as clinicians would wish to either reduce dose or consider colony-stimulating factors in this circumstance, but certainly not so with grade 3 neutropenia without fever or sepsis.
In summary, this meta-analysis provided evidence for the association between the UGT1A1*6 and UGT1A1*6/*28 polymorphism and an increased risk of irinotecan-induced neutropenia in Asian cancer patients. The combination of UGT1A1*6 and *28 might be a hypothetical biomarker for irinotecan in Asian. However, clinical validity is only the first step of several that determine whether a biomarker is applied into clinical. The clinical significance of this last finding requires replication and further research. More larger studies on patients of studies stratified for interactions between tumor stage, genotyping method, and clinical outcome should be conducted to confirm the predictive roles of UGT1A1*6/*28 for irinotecan-induced neutropenia.
References
Meyerhardt JA, Mayer RJ (2005) Systemic therapy for colorectal cancer. N Engl J Med 352:476–487
Noda K, Nishiwaki Y, Kawahara M, Negoro S, Sugiura T, Yokoyama A, Fukuoka M, Mori K, Watanabe K, Tamura T et al (2002) Irinotecan plus cisplatin compared with etoposide plus cisplatin for extensive small-cell lung cancer. N Engl J Med 346:85–91
Rivera F, Vega-Villegas ME, Lopez-Brea MF (2007) Chemotherapy of advanced gastric cancer. Cancer Treat Rev 33:315–324
Yamamoto K, Kokawa K, Umesaki N, Nishimura R, Hasegawa K, Konishi I, Saji F, Nishida M, Noguchi H, Takizawa K (2009) Phase I study of combination chemotherapy with irinotecan hydrochloride and nedaplatin for cervical squamous cell carcinoma: Japanese gynecologic oncology group study. Oncol Rep 21:1005–1009
Takakura S, Takano M, Takahashi F, Saito T, Aoki D, Inaba N, Noda K, Sugiyama T, Ochiai K (2010) Randomized phase II trial of paclitaxel plus carboplatin therapy versus irinotecan plus cisplatin therapy as first-line chemotherapy for clear cell adenocarcinoma of the ovary: a JGOG study. Int J Gynecol Cancer 20:240–247
Saltz LB, Cox JV, Blanke C, Rosen LS, Fehrenbacher L, Moore MJ, Maroun JA, Ackland SP, Locker PK, Pirotta N et al (2000) Irinotecan plus fluorouracil and leucovorin for metastatic colorectal cancer. Irinotecan Study Group. N Engl J Med 343:905–914
Innocenti F, Undevia SD, Iyer L, Chen PX, Das S, Kocherginsky M, Karrison T, Janisch L, Ramirez J, Rudin CM et al (2004) Genetic variants in the UDP-glucuronosyltransferase 1A1 gene predict the risk of severe neutropenia of irinotecan. J Clin Oncol 22:1382–1388
Hoskins JM, Goldberg RM, Qu P, Ibrahim JG, McLeod HL (2007) UGT1A1*28 genotype and irinotecan-induced neutropenia: dose matters. J Natl Cancer Inst 99:1290–1295
Inoue K, Sonobe M, Kawamura Y, Etoh T, Takagi M, Matsumura T, Kikuyama M, Kimura M, Minami S, Utsuki H et al (2013) Polymorphisms of the UDP-glucuronosyl transferase 1A genes are associated with adverse events in cancer patients receiving irinotecan-based chemotherapy. Tohoku J Exp Med 229:107–114
Kim TW, Innocenti F (2007) Insights, challenges, and future directions in irinogenetics. Ther Drug Monit 29:265–270
Martinez-Balibrea E, Abad A, Martinez-Cardus A, Gines A, Valladares M, Navarro M, Aranda E, Marcuello E, Benavides M, Massuti B et al (2010) UGT1A and TYMS genetic variants predict toxicity and response of colorectal cancer patients treated with first-line irinotecan and fluorouracil combination therapy. Br J Cancer 103:581–589
Liu CY, Chen PM, Chiou TJ, Liu JH, Lin JK, Lin TC, Chen WS, Jiang JK, Wang HS, Wang WS (2008) UGT1A1*28 polymorphism predicts irinotecan-induced severe toxicities without affecting treatment outcome and survival in patients with metastatic colorectal carcinoma. Cancer 112:1932–1940
Ferraldeschi R, Minchell LJ, Roberts SA, Tobi S, Hadfield KD, Blackhall FH, Mullamitha S, Wilson G, Valle J, Saunders M, Newman WG (2009) UGT1A1*28 genotype predicts gastrointestinal toxicity in patients treated with intermediate-dose irinotecan. Pharmacogenomics 10:733–739
Schulz C, Heinemann V, Schalhorn A, Moosmann N, Zwingers T, Boeck S, Giessen C, Stemmler HJ (2009) UGT1A1 gene polymorphism: impact on toxicity and efficacy of irinotecan-based regimens in metastatic colorectal cancer. World J Gastroenterol 15:5058–5066
Glimelius B, Garmo H, Berglund A, Fredriksson LA, Berglund M, Kohnke H, Bystrom P, Sorbye H, Wadelius M (2011) Prediction of irinotecan and 5-fluorouracil toxicity and response in patients with advanced colorectal cancer. Pharmacogenomics J 11:61–71
Takano M, Kato M, Yoshikawa T, Sasaki N, Hirata J, Furuya K, Takahashi M, Yokota H, Kino N, Horie K et al (2009) Clinical significance of UDP-glucuronosyltransferase 1A1*6 for toxicities of combination chemotherapy with irinotecan and cisplatin in gynecologic cancers: a prospective multi-institutional study. Oncology 76:315–321
Jinno H, Tanaka-Kagawa T, Hanioka N, Saeki M, Ishida S, Nishimura T, Ando M, Saito Y, Ozawa S, Sawada J (2003) Glucuronidation of 7-ethyl-10-hydroxycamptothecin (SN-38), an active metabolite of irinotecan (CPT-11), by human UGT1A1 variants, G71R, P229Q, and Y486D. Drug Metab Dispos 31:108–113
Desai AA, Innocenti F, Ratain MJ (2003) Pharmacogenomics: road to anticancer therapeutics nirvana? Oncogene 22:6621–6628
Park SR, Kong SY, Rhee J, Park YI, Ryu KW, Lee JH, Kim YW, Choi IJ, Kim CG, Lee JY et al (2011) Phase II study of a triplet regimen of S-1 combined with irinotecan and oxaliplatin in patients with metastatic gastric cancer: clinical and pharmacogenetic results. Ann Oncol 22:890–896
Minami H, Sai K, Saeki M, Saito Y, Ozawa S, Suzuki K, Kaniwa N, Sawada J, Hamaguchi T, Yamamoto N et al (2007) Irinotecan pharmacokinetics/pharmacodynamics and UGT1A genetic polymorphisms in Japanese: roles of UGT1A1*6 and *28. Pharmacogenet Genomics 17:497–504
Jo JC, Lee JL, Ryu MH, Chang HM, Kim M, Lee HJ, Kim HS, Shin JG, Kim TW, Kang YK (2012) Phase II and UGT1A1 genotype study of irinotecan dose escalation as salvage therapy for advanced gastric cancer. Br J Cancer 106:1591–1597
Seo BG, Kwon HC, Oh SY, Lee S, Kim SG, Kim SH, Han H, Kim HJ (2009) Comprehensive analysis of excision repair complementation group 1, glutathione S-transferase, thymidylate synthase and uridine diphosphate glucuronosyl transferase 1A1 polymorphisms predictive for treatment outcome in patients with advanced gastric cancer treated with FOLFOX or FOLFIRI. Oncol Rep 22:127–136
Zhou CF, Ma T, Su Y, Ye ZB, Ji J, Yu YY, Zhang J, Liu BY, Zhu ZG (2013) UGT1A1 gene polymorphisms and the toxicities of FOLFIRI in Chinese Han patients with gastrointestinal cancer. Anticancer Agents Med Chem 13:235–241
Ando Y, Saka H, Ando M, Sawa T, Muro K, Ueoka H, Yokoyama A, Saitoh S, Shimokata K, Hasegawa Y (2000) Polymorphisms of UDP-glucuronosyltransferase gene and irinotecan toxicity: a pharmacogenetic analysis. Cancer Res 60:6921–6926
Onoue M, Terada T, Kobayashi M, Katsura T, Matsumoto S, Yanagihara K, Nishimura T, Kanai M, Teramukai S, Shimizu A et al (2009) UGT1A1*6 polymorphism is most predictive of severe neutropenia induced by irinotecan in Japanese cancer patients. Int J Clin Oncol 14:136–142
Okuyama Y, Hazama S, Nozawa H, Kobayashi M, Takahashi K, Fujikawa K, Kato T, Nagata N, Kimura H, Oba K et al (2011) Prospective phase II study of FOLFIRI for mCRC in Japan, including the analysis of UGT1A1 28/6 polymorphisms. Jpn J Clin Oncol 41:477–482
Wang Y, Shen L, Xu N, Wang JW, Jiao SC, Liu ZY, Xu JM (2012) UGT1A1 predicts outcome in colorectal cancer treated with irinotecan and fluorouracil. World J Gastroenterol 18:6635–6644
Park SR, Kong SY, Rhee J, Park YI, Ryu KW, Lee JH, Kim YW, Choi IJ, Kim CG, Lee JY et al (2010) Phase II study of a triplet regimen of S-1 combined with irinotecan and oxaliplatin in patients with metastatic gastric cancer: clinical and pharmacogenetic results. Ann Oncol 22:890–896
Sunakawa Y, Ichikawa W, Fujita K, Nagashima F, Ishida H, Yamashita K, Mizuno K, Miwa K, Kawara K, Akiyama Y et al (2011) UGT1A1*1/*28 and *1/*6 genotypes have no effects on the efficacy and toxicity of FOLFIRI in Japanese patients with advanced colorectal cancer. Cancer Chemother Pharmacol 68:279–284
Gao J, Zhou J, Li Y, Lu M, Jia R, Shen L (2013) UGT1A1 6/28 polymorphisms could predict irinotecan-induced severe neutropenia not diarrhea in Chinese colorectal cancer patients. Med Oncol 30:604
Gao J, Zhou J, Li Y, Peng Z, Li Y, Wang X, Shen L (2013) Associations between UGT1A1*6/*28 polymorphisms and irinotecan-induced severe toxicity in Chinese gastric or esophageal cancer patients. Med Oncol 30:630
Han JY, Lim HS, Shin ES, Yoo YK, Park YH, Lee JE, Jang IJ, Lee DH, Lee JS (2006) Comprehensive analysis of UGT1A polymorphisms predictive for pharmacokinetics and treatment outcome in patients with non-small-cell lung cancer treated with irinotecan and cisplatin. J Clin Oncol 24:2237–2244
Han JY, Lim HS, Park YH, Lee SY, Lee JS (2007) Integrated pharmacogenetic prediction of irinotecan pharmacokinetics and toxicity in patients with advanced non-small cell lung cancer. Lung Cancer 63:115–120
Nakamura Y, Soda H, Oka M, Kinoshita A, Fukuda M, Fukuda M, Takatani H, Nagashima S, Soejima Y, Kasai T et al (2011) Randomized phase II trial of irinotecan with paclitaxel or gemcitabine for non-small cell lung cancer: association of UGT1A1*6 and UGT1A1*27 with severe neutropenia. J Thorac Oncol 6:121–127
Shulman K, Cohen I, Barnett-Griness O, Kuten A, Gruber SB, Lejbkowicz F, Rennert G (2011) Clinical implications of UGT1A1*28 genotype testing in colorectal cancer patients. Cancer 117:3156–3162
Yamamoto N, Takahashi T, Kunikane H, Masuda N, Eguchi K, Shibuya M, Takeda Y, Isobe H, Ogura T, Yokoyama A et al (2009) Phase I/II pharmacokinetic and pharmacogenomic study of UGT1A1 polymorphism in elderly patients with advanced non-small cell lung cancer treated with irinotecan. Clin Pharmacol Ther 85:149–154
Egger M, Smith GD, Schneider M, Minder C (1997) Bias in meta-analysis detected by a simple, graphical test. BMJ 315:629–634
Choi YH, Kim TW, Kim KP, Lee SS, Hong YS, Ryu MH, Lee JL, Chang HM, Ryoo BY, Kim HS et al (2012) A Phase II study of clinical outcomes of 3-week cycles of irinotecan and S-1 in patients with previously untreated metastatic colorectal cancer: influence of the UGT1A1 and CYP2A6 polymorphisms on clinical activity. Oncology 82:290–297
Minami H, Sai K, Saeki M, Saito Y, Ozawa S, Suzuki K, Kaniwa N, Sawada J, Hamaguchi T, Yamamoto N et al (2007) Irinotecan pharmacokinetics/pharmacodynamics and UGT1A genetic polymorphisms in Japanese: roles of UGT1A1*6 and *28. Pharmacogenet Genomics 17:497–504
Sai K, Saito Y, Sakamoto H, Shirao K, Kurose K, Saeki M, Ozawa S, Kaniwa N, Hirohashi S, Saijo N et al (2008) Importance of UDP-glucuronosyltransferase 1A1*6 for irinotecan toxicities in Japanese cancer patients. Cancer Lett 261:165–171
Satoh T, Ura T, Yamada Y, Yamazaki K, Tsujinaka T, Munakata M, Nishina T, Okamura S, Esaki T, Sasaki Y et al (2011) Genotype-directed, dose-finding study of irinotecan in cancer patients with UGT1A1*28 and/or UGT1A1*6 polymorphisms. Cancer Sci 102:1868–1873
Takano M, Kato M, Yoshikawa T, Sasaki N, Hirata J, Furuya K, Takahashi M, Yokota H, Kino N, Horie K et al (2009) Clinical significance of UDP-glucuronosyltransferase 1A1*6 for toxicities of combination chemotherapy with irinotecan and cisplatin in gynecologic cancers: a prospective multi-institutional study. Oncology 76:315–321
Pharmaceutical and Food Safety Bureau MoH, Labour and Welfare (2008) Pharmaceuticals and medical devices safety information. Pharmaceutical and food safety Bureau MoH, labour and welfare, Japan ed., vol. No. 248 Pharmaceutical and food safety Bureau, ministry of health, labour and welfare
Association between UGT1A1 variant alleles and irinotecan-induced severe neutropenia (2010) Authority HS ed., vol. 12: Health Products Regulation Group, HSA and the HSA Pharmacovigilance Advisory Committee
Han JY, Lim HS, Park YH, Lee SY, Lee JS (2009) Integrated pharmacogenetic prediction of irinotecan pharmacokinetics and toxicity in patients with advanced non-small cell lung cancer. Lung Cancer 63:115–120
Takahara N, Nakai Y, Isayama H, Sasaki T, Satoh Y, Takai D, Hamada T, Uchino R, Mizuno S, Miyabayashi K et al (2013) Uridine diphosphate glucuronosyl transferase 1 family polypeptide A1 gene (UGT1A1) polymorphisms are associated with toxicity and efficacy in irinotecan monotherapy for refractory pancreatic cancer. Cancer Chemother Pharmacol 71:85–92
Acknowledgments
This research was financially supported by 81172986 National Natural Science Foundation.
Conflict of interest
All authors declare no conflicts of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
Fei-fei Han and Chang-long Guo have contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
280_2014_2405_MOESM1_ESM.tif
The Egger’s funnel plot for the publication bias test. A) *6/*6 versus *1/*6 or *1/*1, B) *6/*6 or *1/*6 versus *1/*1, C) *6/*6 or *28/*28 or *6/*28 versus *1/*6 or *1/*28 or *1/*1 (TIFF 1063 kb)
Rights and permissions
About this article
Cite this article
Han, Ff., Guo, Cl., Yu, D. et al. Associations between UGT1A1*6 or UGT1A1*6/*28 polymorphisms and irinotecan-induced neutropenia in Asian cancer patients. Cancer Chemother Pharmacol 73, 779–788 (2014). https://doi.org/10.1007/s00280-014-2405-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00280-014-2405-0