Abstract
The analysis of the cell concentration, volume concentration, and colony size of Microcystis is widely used to provide early warnings of the occurrence of blooms and to facilitate the development of predictive tools to mitigate their impact. This study developed a new approach for the analysis of the cell concentration, volume concentration, and colony size of Microcystis by applying a laser particle analyzer. Four types of Microcystis samples (55 samples in total) were analyzed by a laser particle analyzer and a microscope. By the application of the laser particle analyzer (1) when n = 1.40 and k = 0.1 (n is the intrinsic refractive index, whereas k is absorption of light by the particle), the results of the laser particle analyzer showed good agreement with the microscopic results for the obscuration indicator, volume concentration, and size distribution of Microcystis; (2) the Microcystis cell concentration can be calculated based on its linear relationship with obscuration; and (3) the volume concentration and size distribution of Microcystis particles (including single cells and colonies) can be obtained. The analytical processes involved in this new approach are simpler and faster compared to that by microscopic counting method. From the results, it was identified that the relationship between cell concentration and volume concentration depended on the colony size of Microcystis because the intercellular space was high when the colony size was high. Calculation of cell concentration and volume concentration may occur when the colony size information is sufficient.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Microcystis water bloom, one of the most alarming ecological and environmental issues around the world, seriously threatens the regional drinking water safety in China (Le et al. 2010; Otten et al. 2012). Analysis of Microcystis cell concentrationFootnote 1 and volume concentrationFootnote 2 is necessary to provide early warnings of the occurrence of blooms and to facilitate the development of predictive tools to mitigate their impact.
Microcystis bloom is generally understood to refer to the reproduction and accumulation of a large number of colonial Microcystis on the surfaces of lakes and reservoirs (Reynolds and Walsby 1975). Microcystis colony size dramatically varies in different developmental phases of water bloom. The colony size plays important roles in Microcystis bloom formation (Yamamoto et al. 2011) because of the ecological advantages of large colonies including fast vertical migration (Kromkamp and Walsby 1990; Nakamura et al. 1993), effective uptake of phosphorus (Shen and Song 2007), protection from predation by zooplankton (Cyr and Curtis 1999; Yang et al. 2009), as well as protection from toxic substances (Wu et al. 2007).
The cell concentration of algae is generally used to estimate algae biomass and can be measured by several methods such as microscopic counting (Fujimoto and Sudo 1997; Lee et al. 2010; Kim et al. 2010), spectrophotometry (Rohrlack and Hyenstrand 2007; Gremberghe et al. 2009), Casy method (Wiedner et al. 2003; Schober et al. 2007; Rohrlack and Hyenstrand 2007), and flow-cytometer method (Zhou et al. 2012). Among the above mentioned methods, the microscopic counting method is the most essential and frequently used method. However, cell concentration cannot be properly analyzed by microscopy because the internal cells would be shaded by peripheral cells in Microcystis colonies. Thus, the shaded cells in colonies cannot be counted accurately. Reynolds (Reynolds and Jaworski 1978) used ultrasonic disruption and alkaline hydrolysis process to pretreat Microcystis samples in order to estimate Microcystis cell concentration excluding the above problem. Through these pretreatments, Microcystis cell concentration can be analyzed more accurately.
Apart from cell concentration, analysis of volume concentration and colony size of Microcystis can provide deeper insight into the water bloom phenomenon (Kromkamp and Walsby 1990; Ghadouani et al. 2003; Yang et al. 2008; Shen and Song 2007). However, the methods mentioned above cannot be used to properly assess the situation. The spectrophotometry method is incapable of analyzing Microcystis colonies and cell diameters. Casy method cannot be used to analyze natural Microcystis colonies due to its measurement limit of 150 μm. The flow-cytometer method is so expensive that it was only used in some scientific studies (Zhou et al. 2012; Zucker and Fisher 2013). The upper measurement limit of 500–1,000 μm of the flow-cytometer method is also a weakness in analysis of large Microcystis colonies (bigger than 1,000 μm frequently, Yu et al. 2007). Although the microscopic counting method plays an important role in the taxonomy and biomass estimation of unicellular and small colonial algae, it has several limitations in the analysis of large, irregular colonies. For example, the microscopic counting method can analyze only a small quantity of samples (0.1 μL–0.1 mL). Large, irregular colony samples are normally diluted from one to ten times before analysis, which may result in misrepresentation of the original samples. Moreover, 400–600 particles have to be counted to ensure a highly accurate estimation of the Microcystis colonies, which is time consuming and labor intensive. Therefore, a new and simple but accurate approach for evaluating the cell and volume concentrations and the colony size of Microcystis is necessary.
In this paper, a laser particle analyzer (Mastersizer 2000 particle size analyzer, Malvern Instruments, Ltd.) was used to analyze Microcystis cell concentration, volume concentration, and colony size. This approach is based on the physical principle that particles can induce laser dispersion. The method has been used to study phytoplankton particles sizes. For example, Planner (Planner et al. 2000) evaluated the dimensions of Anabaena variabilis, Synechococcus, and Rhodospirillum rubrum by laser particle analyzer; Lukowski (Lukowski et al. 2008) also used a laser particle analyzer to determine the particle size distribution of emulsified microalgae. Lee et al. (2007) applied LISST-100 (laser in situ scattering and transmissometry, the measurement principle of which is similar to that of the laser particle analyzer) to measure phytoplankton particle size distribution. Most previous studies have focused only on the measurement of phytoplankton particle size distribution by LISST instruments (Anglès et al. 2008; Karp-Boss et al. 2007) and other optical systems (such as imaging-in-flow system; Olson and Sosik 2007). Although Microcystis is a dominant bloom species worldwide (Chen et al. 2003; Ozawa et al. 2005; Hambright and Zohary 2000; Calijuri et al. 2002), it has not been analyzed by laser particle analyzer, except when Andrews et al. (2010) assessed the effect of refractive index n Footnote 3 and absorption k on the results of a small particle size (<20 μm) distribution calculated by laser diffraction particle size analyzers (unicellular Microcystis monocultures were used in their research). However, the effects of n and k on the size distribution results of colonial Microcystis cultures and natural Microcystis colonies calculated by laser particle size analyzer were unknown. Hence, a new systematic approach for the analysis of cell concentration, volume concentration, and colony size of Microcystis by laser particle analyzer should be developed. The effects of n and k on the results should be evaluated as well.
One evident disadvantage of using a laser particle analyzer is its inability to distinguish between Microcystis particles and other phytoplankton particles. However, some previous studies have demonstrated that during the Microcystis bloom period (from July to October in the Northern Hemisphere and from December to July in the Southern Hemisphere), Microcystis is characterized as the dominant bloom-causing species, contributing between 80 and 98 % of the total biovolume in some lakes and reservoirs, such as Lake Taihu in China (Chen et al. 2003; Xu et al. 2010), Lake Biwa in Japan (Ozawa et al. 2005), Hartbeespoort Dam in South Africa (Hambright and Zohary 2000), and the Barra Bonita Reservoir in Brazil (Calijuri et al. 2002). Under these circumstances, the disturbance of other phytoplankton was negligible. Consequently, combined with the taxonomical observations by microscope, the cell concentration and colony characteristics of Microcystis can be analyzed by laser particle analyzer.
In this study, four types of Microcystis samples were analyzed by both microscopic counting method and laser particle analyzer approach. By comparing the results from these two different analysis methods, the laser particle analyzer approach was evaluated.
Materials and Procedures
Laser Particle Analyzer Description
A laser particle analyzer which works over the range of 0.02–2,000 μm (Mastersizer 2000 particle size analyzer, Malvern Instruments, Ltd.) and based on the technique of laser diffraction was used to measure the size of particles. Figure 1 shows the schema of optics and flow system of Mastersizer 2000 particle size analyzer. The collimated laser beams, with wavelengths of 632.8 nm (red laser) and 470 nm (blue laser), shine across a 1 cm flow cell containing particles from the flow system. The light scattered and diffracted is sensed by a focal plane detector and a wide angle detector system. With the software provided by the manufacturer, the intensities measured by the detector system were inverted mathematically to obtain the volume of different-sized particles assuming that particles were spheres. The sample was fully mixed by a churn dasher in the container and the rotated speed of the churn dasher was regulated. A syringe pump was used to draw the sample from the container through the flow cell of the instrument.
Samples preparation
Fifty-five samples (43 laboratory-cultured and 12 natural-bloom samples) were used to determine the parameters and verified by the complexity and variety of the Microcystis colonies.
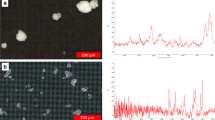
Four types of samples (named A, B, C, and D) with different colony characteristics were obtained through laboratory culture (samples A, B, and C) and natural-bloom collection (sample D). The M. aeruginosa strain (FACHB 469) used for laboratory culture was provided by the Freshwater Algae Culture Collection of the Institute of Hydrobiology (FACHB) of the Chinese Academy of Sciences. Afterward, the algae was batch-cultured axenically in liquid BG-11 medium in a 250 mL conical flask at 25 °C under a 12:12 h light–dark cycle with a fluorescent light intensity of 10–50 μE m−2 s−1 (a different light intensity was employed to obtain samples with different colony characteristics; Li et al. 2013). The natural-bloom samples were collected from a Microcystis water bloom area (coordinates: 31°25.31′N; 120°12.52′E) in the Meiliang Bay of Lake Taihu, Southeast China, in July 2010, during a heavy water bloom. The samples were collected at a depth of 30 cm below the lake surface into 500 mL plastic bottles by phytoplankton nets (64 μm mesh size; Choudhury and Pal 2010). These were used to get colony-dominated samples to assess the current method and then immediately fixed with formalin [2 % (v/v)] for the laboratory analysis. The microphotographs of the four types of samples are shown in Fig. 2.
Microphotographs of the four types of samples. a sample A, unicellular Microcystis samples (23 samples in total); b sample B, mixture of unicellular and large colonial Microcystis (8 samples in total); c sample C, colonial Microcystis (12 samples in total); d sample D, large Microcystis colonies (12 samples in total)
All the samples were divided into two groups (named Samples A-1, B-1, C-1, D-1 and Samples A-2, B-2, C-2, D-2) and analyzed by microscope and laser particle analyzer, respectively.
Observation by Microscope
The laser particle analyzer is unable to distinguish between Microcystis and other phytoplankton species. As a result, accurate results can only be achieved when the phytoplankton assemblage is dominated by a single species. In this study, the natural-bloom samples were also analyzed by microscopy to determine the composition of phytoplankton. Microcystis was identified by morphology monitoring and followed Yu et al. (2007) and Watanabe (1996). The observed results demonstrated that all the natural-bloom samples were dominated by the large Microcystis colonies (at least 95 % of the total biovolume).
Samples A-1, B-1, C-1, and D-1 were analyzed by microscope (Olympus CX31). One representative sample of Samples A-1, B-1, C-1, and D-1 was selected and separated into halves to measure the particle diameter and volume concentration by microscopy. The photomicrographs of the samples were taken using an Olympus C-5050 digital camera coupled to an Olympus CX31 optical microscope. The photomicrographs were analyzed using the UTHSCSA ImageTool v3.00 software (Department of Dental Diagnostic Science, University of Texas Health Science Center, San Antonio, TX, USA). The length and width of Microcystis colonies were obtained accordingly. The size of Microcystis colony was calculated as diameter = (length × width)1/2; and the volume was calculated as volume = π/6 (length × width)3/2 as it is almost impossible to measure the thickness of Microcystis colonies directly. A minimum of 200 colonies per sample were analyzed to determine the colony sizes of Microcystis colonies.
For the other half and the rest of the samples, Sample A-1 was counted directly by optical microscope at ×400 magnification. Samples B-1, C-1, and D-1 were also counted by optical microscope at ×400 magnification after the alkaline hydrolysis pretreatment (Reynolds and Jaworski 1978). After the pretreatment, most of the large colonies were fully disrupted to single cells (Fig. S1); and a fraction of large colonies were disrupted to small colonies composed dozens of cells which would not affect the cell counts. A hemocytometer with 100 fields was then used to count the cells. The cell concentration observed by the microscope was then represented by D m.
Analysis by Laser Particle Analyzer
The Mastersizer allows for the calculation of particle size distribution by specifying the n and k values, thus we tried several values of k and n for the representative samples of Samples A-2, B-2, C-2, and D-2 selected above to determine the optimal values. The speed of the churn dasher was set to 1,500 rpm to ensure intensive mixing without evident air bubbles. Afterward, all the samples of Samples A-2, B-2, C-2, and D-2 were analyzed by laser particle analyzer using the optimal n and k.
First, 600–700 mL (V1) of tap water was added into the container (volume of 800 mL) of the laser particle analyzer to measure the blank value. Microcystis samples were gradually added to the tap water by a 100-mL graduated cylinder. The obscuration increased as a function of the added Microcystis sample volume. The obscuration coefficientFootnote 4 is a property of the sample that needs to be kept within a certain range (10–20 %) for the instrument to function properly. According to the laser particle analyzer manual, the optimal measurements can be obtained when the obscuration is in the range of 10–20 %. It must not be arbitrarily changed for calibration. Therefore, when the obscuration was in this range, the added sample volume (V 2) and the related obscuration (O v) were registered. Finally, the Microcystis particle size distributions of the diluted samples (V 1 + V 2) were measured and their results were presented in logarithmic coordinates.
Results
Effects of n and k on Analyzed Results
The size distribution of different samples obtained from the microscope (gray rectangular) and laser particle analyzer with different n and k is presented in Figs. 3 and 4. Figure 3 reveals that the microscope results show no peak at sizes below 1 μm. However, clear peaks appear below 1 μm in the laser particle analyzer results when k = 0.01, 0.001, and 0, respectively (n = 1.40). The results processed by laser particle analyzer when k = 1 and 0.1 (n = 1.40) showed good agreement with the microscope results for size distribution.
Size distribution of the four types of samples in different k values (gray area is the result obtained by microscope). a Unicellular Microcystis samples; b mixture of unicellular and large colonial Microcystis; c colonial Microcystis; d large Microcystis colonies. Large differences between A to D reflected the differences in particle size of different types of samples
Size distribution of the four types of samples in different n values (gray area is the result obtained through microscopy). a Unicellular Microcystis samples; b mixture of unicellular and large colonial Microcystis; c colonial Microcystis; d large Microcystis colonies. Large differences between A to D reflected the differences in particle size of different types of samples
Figure 4 shows that the results obtained by laser particle analyzer and microscope differ greatly when n = 1.33 (k = 0.1). The laser particle analyzer results show good agreement with the microscope results when n ≥ 1.4 (k = 0.1).
Table 1 presents the obscuration, which could reflect the concentration of the sample, in different n and k values. Table 1 shows very small differences in obscuration with different n and k values.
Figures 5 and 6 show the volume concentration of different samples obtained from the microscope (shaded bars) and laser particle analyzer with different n and k values. The volume concentration obtained by laser particle analyzer shows good relationship with that of the microscopy results when n = 1.40 and k = 0.1, where all the relative deviations were below 10 %.
The variation of the results (including size distribution, obscuration, and volume concentration) in varying k looked similar given a different constant n and vice versa. Based on the comparison of the results of the representative samples by microscope observation and that of the laser particle analyzer method (Fig. 3, 4, 5, 6), the absorption coefficient and refractive index were set to 0.1 and 1.40, respectively.
Measurement of Cell Concentration by Laser Particle Analyzer
Microcystis cell concentration was measured by the laser particle analyzer based on the principle that obscuration is a function of the suspended particle concentration.
The obscuration of undiluted samples (O M) was calculated by the following formula (1):
Where O v is the obscuration of diluted samples, which must be in the range of 10–20 %; V 1 is the volume of tap water added into the container; and V 2 is the volume of added sample.
Figure 7 shows the relationship between the calculated obscuration of undiluted samples O M and their cell concentration D m observed with a microscope (35 samples were used: 13 from Sample A, 8 from Sample B, 5 from Sample C, and 9 from Sample D). Figure 7 shows significant linear relationships between D m and O M. Subsequently, Formula (2) was introduced to calculate the Microcystis cell concentration:
where D c is the calculated cell concentration of Microcystis and η is the coefficient of obscuration. In this study, the values of η were 22.98 and 9.03 for the laboratory-cultured samples and the natural-bloom samples, respectively.
The remaining samples (20 samples were used: 10 from Sample A, 4 from Sample B, 3 from Sample C, and 3 from Sample D) were used to test Formula (2). Figure 8 shows the relationship between D m and D c. The calculated results of D c based on the laser particle analyzer approach were very close to the observed results D m based on the microscopic counting method.
Size Distribution of Microcystis Particles
Apart from cell concentration and volume concentration, the size accumulation curve of Microcystis particles can also be acquired with a laser particle analyzer. The particle size values when the percentage volumes are 10, 50, and 90 are named as D 10, D 50, and D 90, respectively. These values indicate that the proportion of particles below D 10, D 50, and D 90 is 10, 50, and 90 %, respectively. These three parameters are always used to describe the size distribution of particles and refer to the smallest particle size, the mean particle size, and the largest particle size, respectively (Afoakwa et al. 2008). The particle size of Sample A is from 1.26 μm to 4.37 μm and the value of D 50 is 2.40 μm. The size distribution of Samples B and C is from 2 μm to 100 μm, but the D 10 of Samples B and C is 2.08 and 4.22 μm, respectively. The D 50 of Sample D is 197 μm and the interval distribution is between 10 and 1,000 μm (Table S1 and Fig. S2, see the supplementary material).
Relationship Between Cell Concentration and Volume Concentration
Both cell concentration and volume concentration are currently used to estimate the biomass of Microcystis. Undoubtedly, the biomass is high when cell concentration and volume concentration are high. However, thus far, no quantitative relationship of cell concentration and volume concentration has been established. All the cell concentration and volume concentration data of the samples used in this study are shown in Fig. 9. The cell concentration and volume concentration of all the four different types of samples show a positive correlation, but the positive correlations of the different types of samples differ greatly. Although the cell volume of Samples A, B, and C is the same, the positive correlations are very different.
Figure 10 shows the relationship between the ratio of volume concentration and cell concentration and D 50 of colony size. The analytic relation is:
where y is the ratio of volume concentration and cell concentration.
Based on the above content, the relationship between cell concentration and volume concentration is clear. The intercellular space is high when the colony size is high. Calculation of cell concentration and volume concentration could also be carried out even without sufficient colony size information.
Changes of the Particle Size Distribution During Analysis
A churn dasher was used to load samples into the instrument and the first thing we had to check was whether the instrument itself was likely to change the particle size distribution by breaking up the Microcystis colonies. Fig. S3 (see the supplementary material) shows the changes in the particle size distribution of colonial samples (sample C and D) mixed by a churn dasher in the container at a rotation speed of 1,500 rpm. Sample A was not discussed here because single cells were not deemed to be broken up. Sample B was considered as a mixture of Sample A and C, thus the changes of Sample B can be referenced to that of Sample C. It could be seen that 1-min length mixture could not affect the size distribution of both sample C and D but longer mixture (especially 10 min length) could affect the distribution significantly. Thus, the size distribution obtained with 1 min would be reliable at a rotation speed of 1,500 rpm.
Discussion
Assessment of the Approach
In this study, a new systematic approach combining laser particle analyzer and microscopic observation was proposed for the analyses of the cell concentration, volume concentration, and colony size of Microcystis.
Compared with other existing methods, this laser particle analyzer approach has several advantages. First, it allows simple and rapid measurements of cell concentration, volume concentration, and particle size distribution, especially when the sample volume is more than 50 mL. Second, it eliminates the influences of the randomicity of microscopic observation and gives a better representation of the samples. Furthermore, it eliminates the influences of the person-dependent estimation of equivalent spherical diameter of irregular colonies, which enables the comparison of different sample results. Finally, it provides more accurate measurements, which may reveal the tiny changes in Microcystis colony formation due to the different temperatures, light intensities, nutrient conditions, and other biological factors.
Remarkably, although the measurements of the size and distribution of Microcystis particles (single cells and colonies) can be significantly improved by applying the laser particle analyzer, the accuracy and reliability of the laser particle analyzer approach are dependent on the composition of phytoplankton, the refractive index n, and the absorption coefficient k. In addition, obtaining the field samples with a net will bias the results, as large colonies will be sampled, whereas small colonies and single cells will pass through the net. Thus, a wide-necked plastic or glass container could be employed to sample scum of Microcystis colonies and a suitable approach must be developed which allows for their observed heterogeneous density (Utkilen and Bartram 1999).
The cell concentration was calculated by obscuration based on significant linear relationships. A proper obscuration coefficient k should be determined to gain accurate results. In this study, the η values for the laboratory-cultured samples and the natural-bloom samples were quite different. The possible reason for this difference is that the cell volume of the natural-bloom samples was significantly bigger than that of laboratory-cultured samples (Fig. S4). Thus, the cell obscuration of natural-bloom samples was bigger. The average cell diameter of the laboratory-cultured samples and the natural-bloom samples was 2.43 and 3.47 μm, respectively, and the cell volume of the natural-bloom samples was 2.9 times as much as that of the laboratory-cultured samples. In this study, the η value of the laboratory-cultured samples was approximately 2.6 times as much as that of natural samples. The above values of 2.9 and 2.6 were so similar that the η value could be considered to be dependent on the cell volume of Microcystis.
It was also interesting that the K values in Fig. 9 are representative of the biovolume per cell. And since K A < K B < K C, it seems sparser colonies have smaller average cell biovolumes. Our previous study (Li et al. 2013) showed that smaller colony size was associated with higher specific growth rate of Microcystis. Recently, we found that the growth of cell biomass cannot keep pace with growth in cell numbers when the specific growth rate was high. Thereby, the mean cell diameter of Microcystis decreased with the increase of specific growth rate. It was thus clear that both colony size and cell diameter of Microcystis decreased with the increase of the specific growth rate. The differences in growth rate of biomass, biovolume, cell number, and biochemical components (especially polysaccharide) would be the cause that sparser colonies having smaller average cell biovolumes.
There are dozens of morphotypes of Microcystis colonies, such as Microcystis wesenbergii and Microcystis viridis. Our approach would certainly work on these colony types, including analysis of volume concentration and colony size. However, the varying morphotypes would affect the relationship between cell number and colony size. Thus, the obscuration coefficient η should be calibrated when new colonial types or sample locations are applied.
The values of the refractive index n and the absorption coefficient k were calculated as 1.40 (relative to the air) and 0.1, respectively. The n value was the same as that (1.05 relative to the water) calculated by Andrews et al. (2010). The absorption coefficient k was calculated as 0.1, which was much higher than that used in a previous study (Andrews et al. 2010). However, Microcystis cells have high absorption because of the mass of the light-absorbing chlorophyll pigments they contain.
Pretreatment and Robustness of the Method
This study shows the application of the laser particle analyzer for the analyses of cell concentration, volume concentration, and colony size distribution of Microcystis.
In this study, no pretreatment was carried out because Microcystis only dominated the natural sample and no inorganic particles were observed. However, further studies are required to develop a more effective pretreatment to utilize fully the potential of the laser particle analyzer. In some cases, some coarse particles, such as phytoplankton, zooplankton, and inorganic particles, are dominant in the natural-bloom samples. However, Microcystis colonies are easily separated from these particles. Zooplankton such as Daphnia, can easily be picked out. Some small protozoa and phytoplankton, which cannot be picked out, could be eliminated by centrifugal separation effectively. Inorganic particles can also be separated from Microcystis colonies by precipitation as Microcystis colonies always float on the surface of the water.
Our results showed that the measurements of cell concentration, volume concentration, and colony size of Microcystis are robust. However, there are outstanding points (circled) in Figs. 9 and 10, both of which were the measurement results of natural-bloom samples (Sample D). These disunited points were probably caused by the differences in morphology of Microcystis colonies. The most colonies of sample D were identified as Microcystis aeruginosa usually with distinct holes. When the porosity (ratio of the volume of holes and the colony volume) was higher, the biovolume of colony was higher, but the cell number was lower as a large amount of biovolume just reflected the holes in the colony. Thus, these outstanding points in the figures resulted from the higher porosity of two special samples. These outstanding points were far away from most of the points and they would not be consistent with other points. Thus, they were eliminated during the regression analysis to obtain significant fittings. Moreover, these figures are just propositional quantitative relationships of cell concentration and volume concentration and some difference would exist among different samples. But, these differences would not affect the robustness of the measurements of cell concentration, volume concentration, and colony size of Microcystis.
Notes
Cell concentration was defined as cells number of Microcystis per unit water sample.
Volume concentration was defined as biovolume of Microcystis colonies per unit water sample.
This is a parameter of device settings, with the meaning close to that of the corresponding physical value of the intrinsic refractive index. It is used to quantify changes in light propagation that occurs when light passes in between the particle and its surrounding medium.
Obscuration O is the intensity of the scattered and diffracted light that can be used as an indicator, quantifying the concentration of particles, volume concentration and size distribution of Microcystis.
References
Afoakwa EO, Paterson A, Fowler M (2008) Effects of particle size distribution and composition on rheological properties of dark chocolate. Eur Food Res Technol 226:1259–1268
Andrews S, Nover D, Schladow SG (2010) Using laser diffraction data to obtain accurate particle size distributions: the role of particle composition. Limnol Oceanogr Methods 8:507–526
Anglès S, Jordi A, Garcés E, Masó M, Basterretxea G (2008) High-resolution spatio-temporal distribution of a coastal phytoplankton bloom using laser in situ scattering and transmissometry (LISST). Harmful Algae 7:808–816
Calijuri MC, Santos ACAD, Jati S (2002) Temporal changes in the phytoplankton community structure in a tropical and eutrophic reservoir (Barra Bonita, S.P.-Brazil). J Plankton Res 24:617–634
Chen Y, Qin B, Teubner K, Dokulil MT (2003) Long-term dynamics of phytoplankton assemblages: Microcystis-domination in Lake Taihu, a large shallow lake in China. J Plankton Res 25:445–453
Choudhury AK, Pal R (2010) Phytoplankton and nutrient dynamics of shallow coastal stations at Bay of Bengal, Eastern Indian coast. Aquat Ecol 44:55–71
Cyr H, Curtis JM (1999) Zooplankton community size structure and taxonomic composition affects size-selective grazing in natural communities. Oecologia 118:306–315
Fujimoto N, Sudo R (1997) Nutrient-limited growth of Microcystis aeruginosa and Phormidium tenue and competition under various N:P supply ratios and temperatures. Limnol Oceanogr 42:250–256
Ghadouani A, Bernadette P-A, Prepas EE (2003) Effects of experimentally induced cyanobacterial blooms on crustacean zooplankton communities. Freshw Biol 48:363–381
Gremberghe IV, Vanormelingen P, Gucht KV, Mancheva A, D’hondt S, Meester LD, Vyverman W (2009) Influence of Daphnia infochemicals on functional traits of Microcystis strains (Cyanobacteria). Hydrobiologia 635:147–155
Hambright KD, Zohary T (2000) Phytoplankton species diversity control through competitive exclusion and physical disturbances. Limnol Oceanogr 45:110–122
Karp-Boss L, Azevedo L, Boss E (2007) LISST-100 measurements of phytoplankton size distribution: evaluation of the effects of cell shape. Limnol Oceanogr Methods 5:396–406
Kim S-G, Joung S-H, Ahn C-Y, Ko S-R, Boo SM, Oh H-M (2010) Annual variation of Microcystis genotypes and their potentia toxicity in water and sediment from a eutrophic reservoir. FEMS Microbiol Ecol 74:93–102
Kromkamp J, Walsby AE (1990) A computer model of buoyancy and vertical migration in cyanobacteria. J Plankton Res 12:161–183
Le C, Zha Y, Li Y, Sun D, Lu H, Yin B (2010) Eutrophication of lake waters in China: cost, causes, and control. Environ Manag 45:662–668
Lee K-B, Azevedo L, Boss E (2007) LISST-100 measurements of phytoplankton size distribution: evaluation of the effects of cell shape. Limnol Oceanogr Methods 5:396–406
Lee Y-K, Ahn C-Y, Kim H-S, Oh H-M (2010) Cyanobactericidal effect of Rhodococcus sp. isolated from eutrophic lake on Microcystis sp. Biotechnol Lett 32:1673–1678
Li M, Zhu W, Gao L, Lu L (2013) Changes in extracellular polysaccharide content and morphology of Microcystis aeruginosa at different specific growth rates. J Appl Phycol 25:1023–1030
Lukowski G, Lindequist U, Mundt S, Kramer A, Jülich W-D (2008) Inhibition of dermal MRSA colonization by microalgal micro- and nano-particles. Skin Pharmacol Phys 21:98–105
Nakamura T, Adachi Y, Suzuki M (1993) Flotation and sedimentation of a single Microcystis floc collected from surface bloom. Water Res 27:979–983
Olson RJ, Sosik HM (2007) A submersible imaging-in-flow instrument to analyze nano-and microplankton: imaging FlowCytobot. Limnol Oceanogr Methods 5:195–203
Otten TG, Xu H, Qin B, Zhu G, Paerl HW (2012) Spatiotemporal patterns and ecophysiology of toxigenic Microcystis blooms in Lake Taihu, China: implications for water quality management. Environ Sci Technol 46:3480–3488
Ozawa K, Fujioka H, Muranaka M, Yokoyama A, Katagami Y, Homma T, Ishikawa K, Tsujimura S, Kumagai M, Watanabe MF, Park H-D (2005) Spatial distribution and temporal variation of Microcystis species composition and microcystin concentration in Lake Biwa. Environ Toxicol 20:270–276
Planner A, Hara M, Stachowiak Z, Miyake J (2000) Properties of photosynthetic bacteria in anisotropic rigid matrix and in suspension. Photosynthetica 38:251–258
Reynolds CS, Jaworski GHM (1978) Enumeration of natural Microcystis populations. Eur J Phycol 13:269–277
Reynolds CS, Walsby AE (1975) Water-blooms. Biol Rev 50:437–481
Rohrlack T, Hyenstrand P (2007) Fate of intracellular microcystins in the cyanobacterium Microcystis aeruginosa (Chroococcales, Cyanophyceae). Phycologia 46:277–283
Schober E, Wernd M, Laakso K, Korschineck I, Sivonen K, Kurmayer R (2007) Interlaboratory comparison of Taq nuclease assays for the quantification of the toxic cyanobacteria Microcystis sp. J Microbiol Methods 69:122–128
Shen H, Song L (2007) Comparative studies on physiological responses to phosphorus in two phenoltypes of bloom-forming Microcystis. Hydrobiologia 592:475–486
Utkilen JF, Bartram J (1999) Fieldwork: site inspection and sampling. In: Chorus I, Bartram J (eds) Toxic cyanobacteria in water: a guide to public health significance, monitoring and management. E&FN Spon, London
Watanabe M (1996) Isolation, cultivation and classification of bloom-forming Microcystis in Japan. In: Watanabe MF, Harada K, Carmichael WW, Fujiki H (eds) Toxic Microcystis. CRC Press, Boca Raton, pp 13–34
Wiedner C, Visser PM, Fastner J, Metcalf JS, Codd GA, Mur LR (2003) Effects of light on the microcystin content of Microcystis strain PCC 7806. Appl Environ Microbiol 69:1475–1481
Wu Z, Gan N, Huang Q, Song L (2007) Response of Microcystis to copper stress: do phenotypes of Microcystis make a difference in stress tolerance? Environ Pollut 147:324–330
Xu H, Paerl HW, Qin B, Zhu G, Gao G (2010) Nitrogen and phosphorus inputs control phytoplankton growth in eutrophic Lake Taihu, China. Limnol Oceanogr 55:420–432
Yamamoto Y, Shiah FK, Chen Y-L (2011) Importance of large colony formation in bloom-forming cyanobacteria to dominate in eutrophic ponds. Ann Limnol Int J Limnol 47:167–173
Yang Z, Kong F, Zhang M, Yu Y, Qian S (2009) Benefits and costs of the grazer-induced colony formation in Microcystis aeruginosa. Ann Limnol Int J Lim 45:203–208
Yang Z, Kong F, Shi X, Zhang M, Xing P, Cao H (2008) Changes in the morphology and polysaccharide content of Microcystis aeruginosa (Cyanobacteria) during flagellate grazing. J Phycol 44:716–720
Yu GL, Song LR, Li RH (2007) Taxonomic notes on water bloom forming Microcystis species (cyanophyta) from China: an example from samples of the Dianchi Lake. J Syst Evol 45:727–741
Zhou Q, Chen W, Zhang H, Peng L, Liu L, Han Z, Wan N, Li L, Song L (2012) A flow cytometer based protocol for quantitative analysis of bloom-forming cyanobacteria (Microcystis) in lake sediments. J Environ Sci China 24:1709–1716
Zucker RM, Fisher NC (2013) Evaluation and purchase of an analytical flow cytometer: some of the numerous factors to consider. Curr Protoc Cytom. doi:10.1002/0471142956.cy0128s63
Acknowledgments
We would like to thank the Taihu Laboratory for Lake Ecosystem Research (TLLER) for their assistance during field sampling. This study was sponsored by the National Program on Key Basic Research Project of China (2012CB719804), the Natural Science Foundation of Jiangsu Province (BK2011025), and the Hydraulic Science & Technology Project of Jiangsu Province (2012012).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Li, M., Zhu, W. & Gao, L. Analysis of Cell Concentration, Volume Concentration, and Colony Size of Microcystis Via Laser Particle Analyzer. Environmental Management 53, 947–958 (2014). https://doi.org/10.1007/s00267-014-0252-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00267-014-0252-8