Abstract
Multiple mating by honeybee queens results in colonies of genotypically diverse workers. Recent studies have demonstrated that increased genetic diversity within a honeybee colony increases the variation in the frequency of tasks performed by workers. We show that genotypically diverse colonies, each composed of 20 subfamilies, collect more pollen than do genotypically similar colonies, each composed of a single subfamily. However, genotypically similar colonies collect greater varieties of pollen than do genotypically diverse colonies. Further, the composition of collected pollen types is less similar among genotypically similar colonies than among genotypically diverse colonies. The response threshold model predicts that genotypic subsets of workers vary in their response to task stimuli. Consistent with this model, our findings suggest that genotypically diverse colonies likely send out fewer numbers of foragers that independently search for pollen sources (scouts) in response to protein demand by the colony, resulting in a lower variety of collected pollen types. The cooperative foraging strategy of honeybees involves a limited number of scouts monitoring the environment that then guide the majority of foragers to high quality food sources. The genetic composition of the colony appears to play an important role in the efficiency of this behavior.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Honeybee (Apis mellifera) queens mate with multiple males, a reproductive strategy known as polyandry. Evolutionary reasons for the behavior are not entirely understood given the potential risks of multiple mating, which include exposure to predation and disease (Sherman et al. 1988), as well as energetic costs. Further, the reduction in nestmate relatedness with increasing mating frequency (Palmer and Oldroyd 2000) seems counter to eusociality, as it negates the advantage of kin selection, a well accepted explanation for eusocial evolution (Hamilton 1964). Interestingly, a recent hypothesis based on a comprehensive phylogenetic analysis suggests that polyandry is likely a derived trait that evolved either after or concurrently with eusociality (Hughes et al. 2008). While uncommon, polyandry occurs repeatedly among the eusocial Hymenoptera (Boomsma and Ratnieks 1996; Strassmann 2001), suggesting the net effect of this mating strategy is adaptive. In particular, all species of the genus Apis exhibit extremely high levels of polyandry (Oldroyd et al. 1998; Strassmann 2001). Among the many hypotheses put forward to explain the adaptive significance of polyandry (e.g., Ridley 1988; Keller and Hudson 1994; Cole and Wiernasz 1999; Jennions and Petrie 2000; Crozier and Fjerdingstad 2001; Strassmann 2001; Tarpy and Page 2002; Kraus et al. 2004; Schlüns et al. 2005), the most credible hypotheses for social insects address improved fitness from the increased genetic diversity among nestmates (Crozier and Fjerdingstad 2001; Crozier and Page 1985; Palmer and Oldroyd 2000; Tarpy and Page 2002). Because honeybee queens can be instrumentally inseminated (Laidlaw 1977), it is possible to explore in a controlled manner how the genotypic diversity of a honeybee colony affects its performance. For example, a honeybee colony with reduced genetic diversity can be created by inseminating the queen with a reduced number of males. Conversely, a queen inseminated with a high number of males increases the genetic diversity of the colony population. When compared to genotypically similar (GS) colonies in which queens were mated with a reduced number of males, genotypically diverse (GD) colonies exhibit greater disease resistance (Hamilton 1987; Seeley and Tarpy 2007; Tarpy and Seeley 2006), more stable nest thermoregulation (Jones et al. 2004), and increased communication regarding foraging conditions (Mattila et al. 2008). There is also enhanced productivity and fitness after colony founding among GD colonies compared to GS colonies (Mattila and Seeley 2007).
The response threshold model for division of labor in honeybees (Robinson and Page 1989) offers a plausible explanation for enhanced performance among GD colonies. The model predicts that genotypic subsets of workers (i.e., patrilines) vary in their propensities to perform certain tasks. As such, a worker will only engage in a particular task when the intensity of the task stimulus meets or exceeds her genetically based response threshold. A low stimulus level affects only those patrilines with the lowest thresholds. As individuals perform a task, the stimulus for that task is reduced, thereby lowering the probability that additional individuals will perform the task. However, if the stimulus level continues to increase, additional patrilines perform the task until the overall stimulus level in the colony is reduced (Beshers and Fewell 2001; Fewell and Page 1993; Robinson and Page 1989; Oldroyd and Thompson 2007). Differences in worker response thresholds create genetic polyethism, whereby workers of each genotype differentially allocate their labor among available tasks (Calderone and Page 1988; Fewell and Page 1993; Robinson and Huang 1998). Indeed, the link between worker genotype and the probability of task performance has been demonstrated repeatedly in honeybees (reviewed by Crozier and Fjerdingstad 2001).
The influence of genotype has been demonstrated for several honeybee traits, including variability in foraging distance and the ability to switch between nectar and pollen resources (see Page et al. 1995). The propensity to collect pollen is a highly heritable trait in honeybees (Hellmich et al. 1985), as is the tendency to scout independently for food sources (Dreller 1998). Further, there is some evidence that bees exhibit preferences among pollens in choice tests (Campana and Moeller 1977; Levin and Bohart 1955; Schmidt 1984). However, such preferences are apparently not based upon inherent differences in pollen quality (Pernal and Currie 2002). Rather, bees use pollen cues such as odor (Cook et al. 2005) and pollen grain size, which may serve to increase collection efficiency and/or nestmate recruitment (Pernal and Currie 2002). Regardless, the extent to which such behaviors are genetically based is unknown. While much is known about the effect of genotype on individual worker behaviors, the effect of extreme polyandry on the breadth of pollen types collected by the foraging population has not been explored. Because pollen types vary in their nutritional content (Somerville 2001; Standifer 1967), the ability to collect a variety of pollen types may be important for colony health and fitness. However, floral resources tend to be patchy and ephemeral, making it difficult to locate the most profitable sites. Rather than foraging independently, the scout-recruit division of labor within a honeybee colony is a highly efficient foraging strategy whereby most foragers are instead guided to high quality resources (Visscher and Seeley 1982). For maximum efficiency, the colony foraging strategy must strike a balance between an optimal number of scouts to scan the environment and a substantial number of recruits to collect food from those profitable resources (Dreller 1998).
In our study, we considered how the genotypic diversity within a honeybee colony affects pollen foraging efficiency. To the extent that pollen foraging traits such as pollen preference and foraging distance are genetically based, increased genotypic diversity within the colony may lead to greater variety of pollen types collected. However, the response threshold model predicts an alternative outcome, consistent with the efficient foraging strategy. A genotypically diverse (GD) colony composed of multiple patrilines should, on average, maintain greater variability in response thresholds for any particular task, including specialized foraging behaviors such as scouting. Response threshold variability among patrilines suggests that fewer workers will initially engage in task performance. For example, response to the scouting stimulus would be performed first by the patriline(s) with the lowest threshold for scouting. In a multi-patriline colony, this cohort represents a small proportion of the colony population. As pollen sources are discovered and collected, the scouting stimulus is reduced, as is the probability of engaging additional scouts. In a genotypically similar (GS) colony composed of a single patriline, response threshold variability is greatly reduced. As such, the stimulus to scout for pollen would be detected by a much larger proportion of the colony population. Thus, the reduced genetic variability of a GS colony could produce an overreaction to the scouting stimulus, sending significantly more scouts into the field than a GD colony. To investigate this question, we compared total weight of collected pollen as well as richness of collected pollen taxa between GS colonies, each composed of a single patriline, and GD colonies, each composed of 20 patrilines. We then assessed the uniformity of collected pollen taxa among colonies of each treatment group to assess similarity in foraging patterns, and measured adult population, brood area, and pollen foraging rate.
Methods
Instrumentally inseminated queens
Honeybee queens were produced by a commercial queen breeder (Glenn Apiaries, Fallbrook, CA, USA) following standard queen rearing practices (Laidlaw and Page 1997). To minimize phenotypic variability due to individual queen genetics, queens reared for this study were supersisters, having a relatedness factor of 0.75. Twenty queens were instrumentally inseminated in a manner similar to that of Tarpy (2003). Ten of the queens were each inseminated with 1.0 μl of semen collected from a single drone (i.e., one unique drone per queen). The remaining ten queens were each inseminated with a 1.0-μl blend of semen collected from 20 different drones (i.e., 20 unique drones per queen). This technique produced one treatment group with genotypically diverse (multi-patriline) worker populations and another group with genotypically similar (single patriline) worker populations, based on the genetic contributions of the males (Haberl and Moritz 1994). Drones were selected from among five lines of breeder stock maintained at Glenn Apiaries: Minnesota Hygienic, USDA Russian, Cordovan Italian, Carniolan, and Varroa Sensitive Hygiene (VSH). Prior to shipping, inseminated queens were confined under push-in cages within mating nucs until the breeder confirmed that oviposition had begun.
Colony establishment and monitoring
Queens arrived in Tucson, AZ, USA on 10 June 2008 and were introduced into queenless colonies of equal strength in nine-frame, single-story hives. To minimize drift—particularly between treatment groups—colonies with polyandrous queens were co-located approximately 50 m from colonies with monandrous queens. Colonies within each treatment group were arranged in a horseshoe pattern. During the following 8 weeks, workers in each colony were gradually replaced by each inseminated queen’s offspring. Colonies were examined weekly during this period to remove any developing queen cells and to assess overall colony health; colonies that replaced their queens were removed from the study. Once colonies with unacceptable queens were eliminated, nine GD colonies and seven GS colonies remained.
Pollen trapping
On 10 Aug 2008, each colony was fitted with a Sundance™ pollen trap. By design, the traps can be readily engaged or disengaged. Installed traps were initially disengaged for 1 week to allow the bees to adjust to the presence of the equipment. On 17 Aug 2008, traps were engaged, and bees were allowed to acclimatize to the engaged configuration for 48 h. The first sampling period commenced on 19 Aug 2008 and continued for 10 days. Two additional 10-day sampling periods immediately followed. Traps were emptied several times during each sampling period. Trapped pollen was labeled by sampling period and by source colony, weighed, and then stored frozen until processed.
Colony strength assessment
The adult population of each colony was estimated at the beginning of each sampling period by estimating the percentage of each frame covered by adult bees (Waller et al. 1985). Each frame was examined using a wire grid held over the comb as a visual guide to estimate the coverage area. Percent coverage by bees was estimated to the nearest 25% (i.e., 0%, 25%, 50%, 75%, or 100% coverage). Coverage estimates for each frame side were then summed and the sum divided in half to calculate the total number of frames (NOF) of adult bees in each hive. Population estimates were compared between GD and GS colonies for each sampling period.
Brood area measurements
Brood area and pollen foraging are positively correlated (Dreller and Tarpy 2000; Fewell and Winston 1992), and estimates of brood area for each colony were determined at the end of each sampling period. Brood area on both sides of every frame in each hive was measured with a wire grid as above to estimate the coverage area to a resolution of 0.5 cm2. Square centimeters of brood for each frame were then summed to determine total brood area for each hive. Brood area estimates were compared between GD and GS colonies for each sampling period.
Foraging activity
GD colonies have been shown to maintain higher foraging rates than GS colonies (Mattila and Seeley 2007), which could potentially account for variation in the types and amounts of pollen collected. We assessed foraging activity of colonies several times during each of the three sampling periods. At the beginning, middle, and end of each sampling period, we counted the inbound forager population between 08:00:00 and 10:00:00, when pollen foraging activity was high. GD and GS colonies were paired randomly. Paired colonies were then observed concurrently for 1 min. An observer at each colony recorded numbers of pollen and non-pollen foragers entering the hive. The entire survey was repeated (with unique colony pairings) four times. Differences in foraging activity between GD and GS colonies were compared.
Pollen processing and identification
At the end of each sampling period, each colony’s trapped pollen was thoroughly mixed, and then a subsample was collected in a 15-ml centrifuge tube and sent to the Palynology Laboratory at Washington State University (Pullman, WA, USA). Samples were processed using a standard laboratory procedure known as acetolysis, which dissolves unwanted organic material, exposing the exine of the pollen grain for identification (Kapp et al. 2000). Slides were prepared from each processed sample, representative of each hive’s foraging effort per sampling period, and then examined by light microscopy. A 500 pollen grain count identifying types and frequencies was used to indicate the pollen foraging patterns of each colony during the study period. Few pollen types can be identified to species with this approach, and available identification keys often resolve only to plant family or genus (Kapp et al. 2000). Therefore, it was often only possible to associate pollen grains with higher taxonomic levels.
Statistical analysis
Data from each colony within each treatment group were sampled repeatedly over time and evaluated using repeated measures analysis of variance (ANOVA; 2 genetic groups × 3 time periods; Zar 1999). The Greenhouse–Geisser correction to the degrees of freedom was applied in cases where assumptions of sphericity were violated. To assess the degree of similarity in pollen types collected among colonies within each group, we calculated the Bray–Curtis similarity index among colony pairs (1-Bray–Curtis distance). The Bray–Curtis similarity index varies from 0 to 1, where 1 represents identical pollen foraging patterns (Bray and Curtis 1957). We then assessed the results using the same repeated measures ANOVA approach. The criterion for statistical significance was set at α = 0.05.
Variable amounts of pollen collected by colonies could bias the number of identified pollen taxa. Therefore, in order to validate species richness differences between GD and GS colonies, pollen counts from each colony were aggregated for each treatment group over the entire study period. We then constructed rarefaction curves by repeatedly sampling at random each of the aggregated counts (Gotelli and Graves 1996). Data for the rarefaction curves were generated using EcoSim software (Gotelli and Entsminger 2009).
Results
Brood area
The difference in average brood area of GD colonies (M = 1099.9 ± 102.1 cm2) compared to GS colonies (M = 685.7 ± 115.8 cm2) was significant (F 1, 14 = 7.20; p = 0.018; η 2 = 0.3; Table 1). A significant temporal effect was obtained (F 2, 28 = 5.021, p = 0.014), though this was mild (η 2 = 0.22). A significant time × diversity interaction was also obtained (F 2, 28 = 3.72, p = 0.037), though this effect was even milder (η 2 = 0.16). There was an inexplicable decrease in average brood area among GD colonies from 28 Aug 2008 (M = 1245.2 ± 128.0 cm2) to 7 Sep 2008 (M = 901.1 ± 95.4 cm2), whereas the change in brood area for GS colonies from 28 Aug 2008 (M = 671.0 ± 145.1 cm2) to 7 Sep 2008 (M = 660.8 ± 108.1 cm2) was negligible.
Adult population
The average number of frames (NOF) of adult bees of GD colonies (M = 3.77 ± 0.22 frames) was slightly greater than for GS colonies (M = 3.06 ± 0.253 frames). However, this difference was not significant (F 1, 14 = 4.37, p = 0.055, η 2 = 0.24; Table 1). A temporal effect was not significant (F 2, 28 = 0.333, p = 0.720) nor was the time × diversity interaction (F 2, 28 = 2.02, p = 0.152).
Pollen foragers and collected pollen weight
A greater number of pollen foragers from GD colonies returned to the hive each minute (M = 18.3 ± 1.5 bees/min) compared to GS colonies (M = 14.0 ± 1.7 bees/min); however, this difference was not significant (F 1, 14 = 3.59, p = 0.079, η 2 = 0.20; Table 1). As a measure of the total foraging activity, the percentage of returning foragers carrying pollen was higher in GD colonies (M = 51.6 ± 2.5%) compared to GS colonies (M = 45.8 ± 2.8%), but this difference was not significant (F 1, 14 = 2.40, p = 0.144, η 2 = 0.15; Table 1). A significant temporal effect was obtained (F 8, 14 = 21.03, p < 0.001) likely due to temporal availability of pollen sources or changes in brood load. A time × diversity interaction was not significant (F 8, 14 = 0.90, p = 0.521).
GD colonies collected nearly twice as much pollen (M = 259.4 ± 23.3 g) as did GS colonies (M = 140.0 ± 26.4 g), and this difference was significant (F 1, 14 = 11.5, p = 0.004, η 2 = 0.45). A significant temporal effect was obtained both for pollen foraging rate (F 2, 28 = 22.2, p < 0.001, η 2 = 0.50) and for pollen weight (F 2, 28 = 42.0, p < 0.001, η 2 = 0.66; Table 1). This effect was again likely due to the temporal availability of various pollen sources and/or changes in brood load. A significant time × diversity interaction was also obtained for both pollen foraging rate (F 2, 28 = 8.61, p = 0.001, η 2 = 0.19) and for pollen weight (F 2, 28 = 8.03, p = 0.002, η 2 = 0.13).
Pollen foraging breadth
Among the 52 pollen taxa identified, 46 types were collected by GS colonies, whereas only 30 types were collected by GD colonies. Only 24 pollen types collected were found in common between both treatment groups, whereas 22 pollen types were unique to the GS colonies, and only six pollen types were unique to the GD colonies (see Fig. 1). There was a significant difference in the richness of pollen taxa collected by genotypically diverse colonies compared to genotypically similar ones (F 1, 14 = 10.66, p = 0.006, η 2 = 0.43; Table 1). Despite a tendency for higher population levels and greater numbers of returning foragers among genotypically diverse colonies, the average number of pollen taxa collected by genotypically diverse colonies (M = 9.48 ± 0.56 types) was lower than for genotypically similar colonies (M = 12.24 ± 0.63 types). A significant temporal effect was also obtained (F 2, 28 = 6.29, p = 0.006, η 2 = 0.22), attributable to pollen availability. The interaction of time × diversity was not significant (F 2, 28 = 2.13, p = 0.138, η 2 = 0.13).
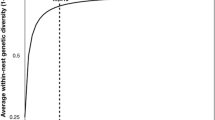
Rarefaction curves comparing pollen diversity between GD and GS colonies are shown in Fig. 2. Curves are displayed with their 95% confidence intervals. While curves were not yet asymptotic, the rate of pollen type accumulation for GS colonies was clearly higher than that of GD colonies.
Pollen foraging similarity
Pair-wise comparisons of Bray–Curtis similarity indices indicated that the pollen collected by GD colonies was significantly more uniform (M = 79 ± 1.1% similar) compared with the pollen collected by the GS colonies (M = 63 ± 1.4% similar; F 1, 55 = 89.52, p < 0.001, η 2 = 0.62; Table 1). A significant temporal effect was also obtained (F 1.5, 81.6 = 76.10, p < 0.001, η 2 = 0.49) as was the interaction of time × diversity (F 1.5, 81.6 = 5.54, p = 0.011, η 2 = 0.05), although this interaction was weak. Of note, from 29 Aug 2008 to 7 Sep 2008, there was a marked increase in foraging similarity in both groups. This was likely due to an abundance of Chenopodium pollen, which accounted for 87% of the pollen taxa collected by GD colonies and 80% of the pollen taxa collected by GS colonies during that period.
Discussion
For polyandrous social insects, mating frequency establishes the genetic architecture of a colony, which in turn affects individual worker behavior. Several hypotheses have been put forth to explain why increased genetic diversity among nestmates is adaptive, including enhanced division of labor, resistance to pathogens, and increased tolerance to environmental stress, among others (reviewed by Boomsma and Ratnieks 1996). Such colony level traits, however, occur widely among social Hymenoptera, including monandrous species. Polyandry is not therefore a strict requirement for fitness among social insects. For honeybees, however, decreased genotypic variability among nestmates does appear to be deleterious to colony performance.
In our study, we found that GD colonies outperformed GS colonies in nearly every colony level measure, although not every difference was significant (see Table 1). Our findings were generally consistent with those of Fuchs and Schade (1994) as well as Mattila and Seeley (2007) in studies comparing colony performance between treatment groups similar to ours. However, we also found that GD colonies collected significantly fewer pollen types than did GS colonies, suggesting that pollen foraging breadth is affected by the colony’s genetic composition. We ascribed the greater number of pollen taxa occurring among GS colonies to a higher frequency of “rare” pollen types (i.e., pollens represented by only one or a few grains out of a 500 grain count (see Online Resource 1). Our results were counter to those of Woyciechowski and Warakomska (1994) who found no correlation between pollen diversity and colony genetics. Our study, however, compared GS colonies against colonies with a known and very high number of patrilines (i.e., 1 vs. 20), rather than comparing low and moderate diversity colonies against those headed by “open-mated” queens. Further, our pollen was subsampled from pollen traps that continuously trapped pollen over the duration of the 30-day study period, rather than simply collecting the first 200 pollen loads on seven different days. Of note, the six most common pollen taxa identified in our study (Chenopodium, Leucophyllum, Asteraceae, Tamarisk, Poaceae, and Solanaceae) represented the vast majority of collected pollen in both GD and GS colonies (see Online Resource 1), suggesting a comparable foraging pattern between both groups. However, based on our Bray–Curtis similarity index, we found the overall composition of collected pollen types to be less similar among GS colonies than among GD colonies (see Table 1).
Because scouts search independently for food sources, the variety of pollen types brought back to the colony may serve as a proxy for overall pollen scouting effort. One possible interpretation of our findings, therefore, is that the number of pollen scouts is negatively associated with intracolonial genetic diversity. This association might exist because some foragers will act as scouts when they fail to find a dance to follow (Beekman et al. 2007). GD colonies produce greater numbers of waggle dances than GS colonies (Mattila et al. 2008), perhaps increasing the chances that a forager will find a dance. Thus, a high frequency of recruitment dances may reduce scouting more rapidly. However, because scouting is also affected by an individual’s genotype (Dreller 1998), numbers of waggle dances alone may not fully regulate a colony’s scouting effort.
Genetic polyethism predicted by the response threshold model affords an additional (and not mutually exclusive) hypothesis for differences in pollen variety between our treatment groups. Genetic polyethism is an evolutionary strategy that contributes to colony level behavioral diversity by exploiting the genetically based tendencies of individual workers to perform particular tasks. Specifically, behavioral variability among patrilines suggests that those patrilines with lower response thresholds likely respond to behavioral cues sooner than other patrilines in the colony. As such, patrilines with higher response thresholds may never perform certain tasks, while patrilines with the lowest thresholds may appear as behavioral specialists. Subsequently, the allocation of tasks across patrilines suggests fewer bees on average may engage in any particular behavior, rendering more workers available for other tasks, which increases overall colony efficiency. If scouting behavior is allocated across patrilines, scouting effort is predicted to vary with intracolonial genetic diversity.
Pollen foragers are stimulated to collect pollen when either brood area increases (Fewell and Page 1993) or stored pollen decreases (Camazine 1993; Fewell and Winston 1992). A proportion of these foragers will exhibit scouting behavior, seeking out new pollen sources (Seeley 1983). If only patrilines with the lowest thresholds for pollen scouting undertake the task, then GD colonies may send fewer numbers of pollen scouts in response to colony protein demand. In a colony composed entirely of a single patriline, however, the genetic diversity that underlies pollen foraging behavior is reduced, requiring each individual to perform a greater array of tasks, including scouting behavior. Thus, relatively greater numbers of bees from GS colonies will likely respond to cues for pollen scouting. In this sense, an increased number of scouts represents a breakdown in polyethism; more foragers engaged in scouting reduces the number of bees available for other tasks, including being recruited to previously discovered, high value pollen sources. This explanation is also consistent with the significantly greater amounts of pollen collected by GD colonies (Table 1). A more detailed study to identify and genotype scout bees could lend support for such a mechanism, particularly if any patrilineal bias for scouting was demonstrated.
Despite the larger variety of pollen types collected by GS colonies, we found that GD colonies collected significantly greater amounts of pollen. However, because GD colonies had more brood, it was not surprising that they also had more pollen foragers and collected more pollen than GS colonies. Measurements of pollen collection and brood area are not independent; the presence of brood stimulates pollen foraging (Pankiw et al. 1998), and large amounts of incoming pollen enable colonies with strong queens to maximize brood rearing.
If the overall foraging strategy of a honeybee colony is to put an optimal number of scouts into the environment and maximize recruiting to the high value resources, then a colony must efficiently regulate its foraging effort. Combined with their greater foraging rates (Mattila and Seeley 2007), GD colonies are likely able to reduce the overall stimulus level for pollen foraging more rapidly and with fewer scouts than GS colonies. By contrast, GS colonies may be losing task allocation efficiency by sending out far more scouts than necessary. Recently, Mattila and Seeley (2010) reported that, contrary to GD colonies, initial foragers in GS colonies often would not dance when returning to the hive, and thus would not mobilize their nestmates to a food source. Our results and interpretation are an extension of these findings in that the diversity of pollen types in GS colonies is consistent with a foraging force comprised largely of individuals that independently search for pollen sources but do not recruit sufficient numbers of nestmates to fully exploit identified resources.
References
Beekman M, Gilchrist AL, Duncan M, Sumpter DJT (2007) What makes a honeybee scout? Behav Ecol Sociobiol 61:985–995
Beshers SN, Fewell JH (2001) Models of division of labor in social insects. Annu Rev Entomol 46:413–440
Boomsma JJ, Ratnieks FLW (1996) Paternity in eusocial hymenoptera. Philos Trans R Soc B Biol Sci 351:947–975
Bray JR, Curtis JT (1957) An ordination of the upland forest communities of Southern Wisconsin. Ecol Monogr 27(4):325–349
Calderone NW, Page RE (1988) Genotypic variability in age polyethism and task specialization in the honey bee, Apis mellifera (Hymenoptera: Apidae). Behav Ecol Sociobiol 22:17–25
Camazine S (1993) The regulation of pollen foraging by honey bees: how foragers assess the colony’s need for pollen. Behav Ecol Sociobiol 32:265–272
Campana BJ, Moeller FE (1977) Honey bees: preference for and nutritive value of pollen from five plant sources. J Econ Entomol 70:39–41
Cole BJ, Wiernasz DC (1999) The selective advantage of low relatedness. Science 285:891–893
Cook SM, Sandoz JC, Martin AP, Murray DA, Poppy GM, Williams IH (2005) Could learning of pollen odours by honey bees (Apis mellifera) play a role in their foraging behaviour? Physiol Entomol 30:164–174
Crozier RH, Fjerdingstad EJ (2001) Polyandry in social Hymenoptera—disunity or diversity? Ann Zool Fenn 38:267–285
Crozier RH, Page RE (1985) On being the right size: male contributions and multiple mating in social Hymenoptera. Behav Ecol Sociobiol 18:105–115
Dreller C (1998) Division of labor between scouts and recruits: genetic influence and mechanisms. Behav Ecol Sociobiol 43:191–196
Dreller C, Tarpy DR (2000) Perception of the pollen need by foragers in a honeybee colony. Anim Behav 59:91–96
Fewell JH, Page RE (1993) Genotypic variation in foraging responses to environmental stimuli by honey bees, Apis mellifera. Experientia 49:1106–1112
Fewell JH, Winston ML (1992) Colony state and regulation of pollen foraging in the honey bee, Apis mellifera L. Behav Ecol Sociobiol 30:387–393
Fuchs S, Schade V (1994) Lower performance in honeybee colonies of uniform paternity. Apidologie 25:155–168
Gotelli NJ, Entsminger GL (2009) EcoSim: null models software for ecology. Version 7. Acquired Intelligence Inc., Jericho. Available at http://garyentsminger.com/ecosim.htm
Gotelli NJ, Graves GR (1996) Null models in ecology. Smithsonian Books, Washington
Haberl M, Moritz RFA (1994) Estimation of intracolonial worker relationship in a honey bee colony (Apis mellifera L.) using DNA fingerprinting. Insect Soc 41:263–272
Hamilton WD (1964) The genetical evolution of social behaviour. J Theor Biol 7(1):1–52
Hamilton WD (1987) Kinship, recognition, disease, and intelligence: constraints of social evolution. In: Ito Y, Brown JL, Kikkawa J (eds) Animal societies: theory and facts. Japanese Scientific Society Press, Tokyo, pp 81–102
Hellmich RL, Kulincevic JM, Rothenbuhler WC (1985) Selection for high and low pollen-hoarding honey bees. J Hered 76:155–158
Hughes WO, Ratnieks FL, Oldroyd BP (2008) Multiple paternity or multiple queens: two routes to greater intracolonial genetic diversity in the eusocial Hymenoptera. Journal of Evolutionary Biology 21:1090–1095
Jennions MD, Petrie M (2000) Why do females mate multiply? A review of the genetic benefits. Biol Rev 75:21–64
Jones JC, Myerscough MR, Graham S, Oldroyd BP (2004) Honey bee nest thermoregulation: diversity promotes stability. Science 305:402–404
Kapp RO, Davis OK, King JE (2000) Guide to pollen and spores. American Association of Stratigraphic Palynologists, College Station
Keller L, Hudson KR (1994) Genetic variability, queen number, and polyandry in social Hymenoptera. Evolution 48:694–704
Kraus FB, Neumann P, Praagh J, Moritz RFA (2004) Sperm limitation and the evolution of extreme polyandry in honeybees (Apis mellifera L.). Behav Ecol Sociobiol 55(5):494–501
Laidlaw HH (1977) Instrumental insemination of honey bee queens. Dadant, Hamilton
Laidlaw HH, Page RE (1997) Queen rearing and bee breeding. Wicwas, Cheshire
Levin MD, Bohart GE (1955) Selection of pollens by honey bees. Am Bee J 95:392–393
Mattila HR, Seeley TD (2007) Genetic diversity in honey bee colonies enhances productivity and fitness. Science 317:362–364
Mattila HR, Seeley TD (2010) Promiscuous honeybee queens generate colonies with a critical minority of waggle-dancing foragers. Behav Ecol Sociobiol 64(5):875–889
Mattila HR, Burke KM, Seeley TD (2008) Genetic diversity within honeybee colonies increases signal production by waggle-dancing foragers. Proc R Soc B Biol Sci 275:809–816
Oldroyd BP, Thompson GJ (2007) Behavioural genetics of the honey bee, Apis mellifera. Adv Insect Physiol 33:1–49
Oldroyd BP, Clifton MJ, Parker K, Wongsiri S, Rinderer TE, Crozier RH (1998) Evolution of mating behavior in the genus Apis and an estimate of mating frequency in Apis cerana (Hymenoptera: Apidae). Ann Entomol Soc Am 91(5):700–709
Page RE, Robinson GE, Fondrk MK, Nasr ME (1995) Effects of worker genotypic diversity on honey bee colony development and behavior (Apis mellifera L.). Behav Ecol Sociobiol 36:387–396
Palmer KA, Oldroyd BP (2000) Evolution of multiple mating in the genus Apis. Apidologie 31:235–248
Pankiw T, Page RE, Fondrk KM (1998) Brood pheromone stimulates pollen foraging in honey bees (Apis mellifera). Behav Ecol Sociobiol 44(3):193–198
Pernal SF, Currie RW (2002) Discrimination and preferences for pollen-based cues by foraging honeybees, Apis mellifera L. Anim Behav 63:369–390
Ridley M (1988) Mating frequency and fecundity in insects. Biol Rev 63:509–549
Robinson GE, Huang ZY (1998) Colony integration in honey bees: genetic, endocrine, and social control of division of labor. Apidologie 29:159–170
Robinson GE, Page RE (1989) Genetic basis for division of labor in an insect society. In: Breed MD, Page RE (eds) The genetics of social evolution. Westview, Boulder, pp 61–80
Schlüns H, Moritz RFA, Neumann P, Kryger P, Koeniger G (2005) Multiple nuptial flights, sperm transfer and the evolution of extreme polyandry in honeybee queens. Anim Behav 70(1):125–131
Schmidt JO (1984) Feeding preferences of Apis mellifera L. (Hymenoptera: Apidae): individual versus mixed pollen species. J Kansas Entomol Soc 57:323–327
Seeley TD (1983) Division of labor between scouts and recruits in honeybee foraging. Behav Ecol Sociobiol 12:253–259
Seeley TD, Tarpy DR (2007) Queen promiscuity lowers disease within honeybee colonies. Proc R Soc B Biol Sci 274:67–72
Sherman PW, Seeley TD, Reeve HK (1988) Parasites, pathogens, and polyandry in social Hymenoptera. Am Nat 151:392–396
Somerville DC (2001) Nutritional value of bee collected pollens. Report DAN 134A. Rural Industries Research and Development Corporation
Standifer L (1967) A comparison of the protein quality of pollens for growth-stimulation of the hypopharyngeal glands and longevity of honey bees, Apis mellifera L. (Hymenoptera: Apidae). Insect Soc 14:415–425
Strassmann J (2001) The rarity of multiple mating by females in the social Hymenoptera. Insect Soc 48:1–13
Tarpy DR (2003) Genetic diversity within honeybee colonies prevents severe infections and promotes colony growth. Proc R Soc B Biol Sci 270(1510):99–103
Tarpy DR, Page RE (2002) Sex determination and the evolution of polyandry in honey bees (Apis mellifera). Behav Ecol Sociobiol 52:143–150
Tarpy DR, Seeley TD (2006) Lower disease infections in honeybee (Apis mellifera) colonies headed by polyandrous vs. monandrous queens. Naturwissenschaften 93:195–199
Visscher PK, Seeley TD (1982) Foraging strategy of honeybee colonies in a temperate deciduous forest. Ecology 63:1790–1801
Waller GD, Gary NE, Chester ST Jr, Karim MS, Martin JH, Vaissiere BE (1985) Honeybee colony populations and foraging rates on onion seed-fields in Arizona. J Apic Res 24:93–101
Woyciechowski M, Warakomska Z (1994) Worker’s genetic diversity has no relation to pollen diversity in a honey bee colony (Apis mellifera L.). J Ethol 12:163–167
Zar JH (1999) Biostatistical analysis. Prentice Hall, Inc., Upper Saddle River
Acknowledgements
We are grateful to Tom Glenn for producing the instrumentally inseminated queens and to John Jones for identification of pollen grains. Thanks also to Ruben Alarcon for statistical guidance and to Mona Chambers, Morgan Caseman, Delayne Caseman, and Maurisa Two Two for help with the bees. Linda Eckholm, Tim Griffith, and two anonymous reviewers provided helpful comments that greatly improved the manuscript. This experiment complied with current guidelines and regulations concerning subject handling. The authors declare that they have no conflicts of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by R. Moritz
Electronic supplementary material
Below is the link to the electronic supplementary material.
Online Resource 1
Identification and frequencies of pollen types collected by honey bee colonies between 19-Aug-2008 and 17-Sep-2008 in Tucson, Arizona USA. Sampling periods 1, 2, and 3 occurred from 19-Aug-2008 to 28-Aug-2008, 29-Aug-2008 to 07-Sep-2008, and 08-Sep-2008 to 17-Sep-2008, respectively. Pollen types were identified via 500-grain pollen count using light microscopy. Samples were collected from each colony at the end of each sampling period.(XLS 46 kb)
Rights and permissions
About this article
Cite this article
Eckholm, B.J., Anderson, K.E., Weiss, M. et al. Intracolonial genetic diversity in honeybee (Apis mellifera) colonies increases pollen foraging efficiency. Behav Ecol Sociobiol 65, 1037–1044 (2011). https://doi.org/10.1007/s00265-010-1108-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00265-010-1108-8