Abstract
Deficient mismatch repair (dMMR)/microsatellite instability (MSI) colorectal cancer (CRC) has high immunogenicity and better prognosis compared with proficient MMR (pMMR)/microsatellite stable (MSS) CRC. Although the activation of the cyclic GMP-AMP synthase (cGAS)-stimulator of interferon genes (STING) pathway has been considered to contribute to the high number of CD8+ TILs, its role in dMMR/MSI CRC is largely unknown. In this study, to examine the role of the cGAS-STING pathway on the recruitment of CD8+ TILs in dMMR/MSI CRC, we used public datasets and clinical tissue samples in our cohorts to evaluate the expression of cGAS, STING, and CD8+ TILs in pMMR/MSS and dMMR/MSI CRCs. According to the analysis of public datasets, the expression of cGAS-STING, CD8 effector gene signature, and CXCL10-CCL5, chemoattractants for CD8+ TILs which regulated by the cGAS-STING pathway, was significantly upregulated in dMMR/MSI CRC, and the expression of cGAS-STING was significantly associated with the expression of CD8 effector gene signature. Immunohistochemistry staining of the clinical tissue samples (n = 283) revealed that cGAS-STING was highly expressed in tumor cells of dMMR CRC, and higher expression of cGAS-STING in tumor cells was significantly associated with the increased number of CD8+ TILs. Moreover, we demonstrated that the downregulation of MMR gene in human CRC cell lines enhanced the activation of the cGAS-STING pathway. Taken together, for the first time, we found that dMMR/MSI CRC has maintained a high level of cGAS-STING expression in tumor cells, which might contribute to abundant CD8+ TILs and immune-active TME.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Colorectal cancer (CRC) remains one of the most common causes of cancer death worldwide [1, 2]. Although the advances in primary and adjuvant treatments have improved the prognosis of CRC, the cases with advanced CRC still have a poor prognosis [3], and thus further therapeutic strategies should be required. CRC is biologically divided into two groups according to microsatellite stability classification: DNA mismatch repair deficient (dMMR) CRC tumors generate microsatellite instability (MSI) phenotype; MMR proficient (pMMR) CRC tumors are defined as microsatellite stable (MSS) but chromosomal unstable. The germline mutation in MMR genes including MutL homolog 1 (MLH1) (30–40%), MutS homolog 2 (MSH2) (50%), MutS homolog 6 (MSH6) (7–10%), and PMS1 homolog 2 (PMS2) (< 5%), and epithelial cell adhesion molecule gene (EPCAM) (1–3%) causes Lynch syndrome [4, 5], whereas sporadic dMMR/MSI CRCs most commonly arise from hypermethylation of MLH1 promoter [3, 6]. dMMR/MSI CRC accounts for 15–20% of all CRC cases [3, 6, 7] and is reported to be heavily infiltrated by immune cells such as CD8+ TILs, T helper 1 CD4+ TILs, and antigen-presenting cells such as dendritic cells and macrophages due to a high number of tumor mutational burden and neoantigen load [6,7,8,9,10,11,12,13]. dMMR/MSI CRC might therefore show a better prognosis and response to immunotherapy using immune checkpoint inhibitors (ICI). However, the regulatory mechanisms of the immune-active TME in dMMR/MSI CRC are not yet fully understood.
The cyclic GMP-AMP synthase (cGAS) is the direct cytosolic DNA sensor and innate immune response initiator, which binds to cytosolic DNA and activates stimulator of interferon genes (STING) as a downstream adaptor through the generation of the second messenger cyclic GMP-AMP (cGAMP) [14]. The activated STING subsequently recruits and activates TANK-binding kinase 1 and IFN regulatory factor 3 (IRF3) via a phosphorylation-dependent manner, leading to the production of type I IFN and other inflammatory mediators including C–C chemokine ligand-5 (CCL5) and C-X-C motif ligand 10 (CXCL10) [15, 16]. It has been thought that the cGAS-STING pathway in tumor cells is a critical component for regulations of anti-tumor adaptive immunity within the TME in several cancers including CRC [17,18,19,20,21,22]. Indeed, Chon et al. reported that patients with STING-high CRC showed increasing the number of CD8+ TILs and better prognosis when compared to those with STING-low CRC, and intra-tumoral injections of the STING agonist, 3′3’-cGAMP, effectively enhanced infiltration of CD8+ effector T cells and suppressed colon cancer progression in mice [21]. Moreover, it has been reported that dysregulation of the cGAS-STING pathway in colorectal carcinoma might escape the tumor from immune recognition [23].
The majority of CRC (~ 85%) exhibit chromosomal instability (CIN) which promotes chromosome segregation errors leading to the formation of micronuclei and rupture spills genomic DNA into the cytosol [7, 24, 25]. CIN, therefore, activates inflammatory signaling downstream of cGAS-STING in response to cytoplasmic DNA and triggers immune surveillance of malignancies during the early steps of tumorigenesis [26]. Santaguida et al. also demonstrated that the cells with abnormal karyotypes were eliminated by cGAS-STING-mediated activation of the immune system that may serve as cancer cell immunosurveillance [27]. On the other hand, recent studies reported that dMMR-mediated cytosolic DNA sensing by the cGAS-STING pathway also contributed to anti-tumor immunity [28]. MLH1-deficiency triggered DNA hyper-resection by loss of MutLα-specific regulation of exonuclease 1, which activates the cGAS-STING pathway in dMMR tumors [29]. Mowat et al. also demonstrated that genomic instability induced by dMMR stimulated the cGAS-STING pathway and type I IFN signaling in murine CRC [30]. Given the accumulating evidence described above, both CIN CRC and MSI CRC might have the potential to activate the cGAS-STING pathway through the generation of cytosolic dsDNA in tumor cells, resulting in immune activation during tumorigenesis.
In this study, to investigate the role of the cGAS-STING pathway in the immune-active TME of dMMR/MSI CRC especially, we investigated the expression of cGAS and STING between pMMR/MSS and dMMR/MSI CRCs by analyzing public datasets and IHC of our cohort and evaluated the association of the expression of cGAS and STING with CD8+ TILs between pMMR and dMMR CRCs. We also examined whether the downregulation of MMR genes led to activating the cGAS-STING pathway in human CRC cells in vitro.
Materials and methods
Data analyses of the cancer genome atlas (TCGA) and the gene expression omnibus (GEO) database
Publicly accessible datasets of mRNA expression of genes for colon and rectal adenocarcinoma were obtained from cBioPortal (http://www.cbioportal.org/) [31] or the GEO database in April 2021. To compare mRNA expression levels of cGAS (MB21D1) and STING (TMEM173) between MSI (MSI-H) and MSS (MSI-low and microsatellite stable) CRCs, we obtained the expression z-score from TCGA (COADREAD) or the log2 signal intensity from GSE39582. We also calculated multi-gene expression signatures including CD8 effector genes (GZMK, CD3E, CD3G, CXCR3, CD3D, BCL11B, CD28, KLRG1, IL7R), CD4 mature genes (IGFBP4, ITM2A, AMIGO2, TRAT1, CD40LG, ICOS, RCAN3), regulatory T (Treg) genes (IL2RA, FOXP3, CTLA4, SLC35D1, GDPD3, CISH) [32], and myeloid-derived suppressor cell (MDSC) genes (ITGAM, CD14, FUT4, CD33) [33] using TCGA and GSE39582 samples, and compared each score between MSS and MSI CRCs.
Patient samples
We enrolled 283 patients with primary CRC (MSS, n = 243; MSI, n = 40) who underwent surgical resection at Fukushima Medical University Hospital between 2002 and 2013 (Table 1). The available formalin-fixed paraffin-embedded tissue samples were used for IHC. Clinical and pathological information were retrospectively obtained from reviewing medical records. The study was conducted in accordance with the Declaration of Helsinki and was approved by the Institutional Review Board of Fukushima Medical University (Fukushima, Japan).
IHC
Paraffin-embedded 4-μm CRC tissue sections were deparaffinized in xylene and rehydrated in ethanol. Endogenous peroxidases were blocked with 0.3% hydrogen peroxide in methanol. Antigens were retrieved by autoclave with Target Retrieval Solution pH6.0 or pH9.0 (Dako/Agilent Technologies, Santa Clara, CA, USA). After washing with PBS, the sections were incubated at 4 ℃ overnight with the following primary antibodies: anti-cGAS mAb (#79,978; Cell Signaling Technology, Danvers, MA, USA), anti-STING mAb (#13,647; Cell Signaling Technology), anti-three-prime repair exonuclease 1 (TREX1) mAb (ab185228; Cambridge, UK), PD-L1 mAb (#13,684, Cell Signaling Technology), anti-CD4 mAb (M7310; Dako/Agilent Technologies), anti-CD8 mAb (M7103; Dako/Agilent Technologies), and anti-Foxp3 mAb (ab20034; Abcam, Cambridge). The sections were then incubated with horseradish peroxidase-conjugated anti-mouse or anti-rabbit secondary antibodies (K4003 or K4001; Dako/Agilent Technologies). Peroxidase was visualized with diaminobenzidine (Dojindo Molecular Technology, Kumamoto, Japan), and nuclei were counterstained with Mayer Hematoxylin Solution (FUJIFILM Wako Pure Chemical Corporation, Osaka, Japan).
The expression of cGAS, STING, and TREX1 in tumor cells was evaluated using IHC score (H-score; 0–300) calculated by multiplying intensity score and extent score. The intensity score was graded by staining in the cytoplasm as follows: 0 (none), 1 + (weak), 2 + (moderate), or 3 + (strong), and the extent score was defined as the percentage of stained cytoplasm (0%–100%). The medians were adopted for estimating cGAS-low and cGAS-high, and STING-low and STING-high. PD-L1 expression was evaluated by assessing membranous staining without cytoplasmic staining and tumor specimens were considered to be PD-L1-positive when > 1% of the tumor cells exhibited membranous staining of any intensity [34]. For the assessment of CD4+ and CD8+ TILs, the invasive front region of the tumor was reviewed in four independent areas, and counted the number of lymphocytes at a magnification of × 400, as previously described [35]. IHC analyses were evaluated by two observers (A.K and S.N) who were blinded to all of the clinical and pathological information. Samples with contradictory scores between the observers were discussed and reevaluated jointly until they reached an agreement.
Determination of MMR status
IHC for MMR proteins was performed using primary antibodies for MLH1 (M3640; Dako/Agilent Technologies), MSH2 (M3639; Dako/Agilent Technologies), MSH6 (M3646; Dako/Agilent Technologies), and PMS2 (M3647; Dako/Agilent Technologies) as previously described by us [36]. Loss of at least one MMR protein was defined as dMMR and tumors with intact MMR protein expression as pMMR.
CRC cell lines
Human colorectal cancer cell lines were purchased from the Korean Cell Line Bank (SNU81) and the American Type Culture Collection (SW480), and the cell lines have been authenticated using STR analysis (Promega Japan, Tokyo, Japan). The cells were maintained with RPMI-1640 (Merck Sigma-Aldrich, St-Louis, MO, USA) containing 10% heat-inactivated FBS and 1% penicillin/streptomycin at 37 ℃ in an atmosphere of 5% CO2. The cells were seeded into 12-well plates at 2.0 × 105 cells/well 24 h before the stimulation and then treated with 2’,3’-cGAMP (#35,573; Cell Signaling Technology) at a final concentration of 5 μg/ml for 18 h.
Knockdown experiments were conducted using siRNA oligonucleotides for MLH1 (s297; Thermo Fisher Scientific, Waltham, MA, USA), MSH2 (s534362; Thermo Fisher Scientific), TMEM173 (s50646; Thermo Fisher Scientific) genes, or scramble control (negative control#1; Thermo Fisher Scientific) with Lipofectamine RNAiMAX Transfection Reagent (Thermo Fisher Scientific), according to manufacturer’s instructions. Knockdown of MLH1 and MSH2 was confirmed by immunoblot analysis.
CD8+ cell isolation from human peripheral blood mononuclear cells (PBMC)
PBMC were isolated from the peripheral blood of healthy volunteers using Ficoll-Paque (GE Healthcare, Little Chalfont, UK). Human CD8+ cells were isolated from PBMC using the MACS magnetic cell separation system (CD8 MicroBeads, human; Miltenyie Biotec, Bergisch Gladbach, Germany).
Cell migration assay
Human CRC cell line SW480 cells were stimulated with or without 2’,3’-cGAMP, a STING agonist, for 18 h, and the conditioned medium (CM) was collected and added to the bottom wells of the migration assay chamber (CytoSelect; CELL BIOLABS, San Diego, CA, USA). After setting a membrane chamber into the migration assay chamber containing the CM, isolated CD8+ cells from PBMCs were added to the membrane chamber. After 24 h of incubation, the level of migrated cells was measured according to the manufacturer's protocol.
Quantitative real-time PCR (qPCR)
Total RNA was extracted using TRIzol Reagent (QIAGEN, Valencia, CA, USA). Obtained RNA was quantified on a NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific). Total RNA was reverse transcribed to cDNA using ReverTra Ace qPCR RT Master Mix with gDNA remover (TOYOBO, Osaka, Japan) according to the manufacturer’s instructions. qPCR reaction was performed on the QuantStudio3 real-time PCR system (Applied Biosystems, Carlsbad, CA, USA) using PrimeTime Gene Expression Master Mix (Integrated DNA Technologies, Coral Ville, IA, USA) with specific primers and probes against CXCL10 (Accession No: NM_001565.1), CCL5 (Accession No: NM_002985.1), MLH1(Accession No: NM_001258271), and GAPDH (Accession No: NM_002046.1) (Integrated DNA Technologies). The levels of gene expression were normalized to GAPDH.
Immunoblot analysis
Total cell proteins were extracted using RIPA buffer supplemented with protease inhibitor and phosphatase inhibitor cocktail. The protein concentration was measured using the XL-Bradford kit (Aproscience, Tokushima, Japan). Tris–Glycine SDS sample buffer (Thermo Fisher Scientific) was added to protein samples and boiled at 100 ℃ for 5 min. The equal amount of protein was loaded onto the 4–20% or 10% Tris–Glycine gels (Thermo Fisher Scientific) and electrophoresed at 125 V for 60 min using Invitrogen XCell SureLock electrophoresis system with Tris–Glycine SDS Running buffer (Thermo Fisher Scientific). Then, the proteins were transferred onto the PVDF membrane using iBlot2 Dry Blotting System (Thermo Fisher Scientific). The membranes were blocked with 5% non-fat dried skimmed milk and incubated with primary antibodies including anti-phospho-IRF3 mAb (#37,829; Cell Signaling Technology), anti-STING mAb (#13,647; Cell Signaling Technology), anti-MLH1 mAb (#3515; Cell Signaling Technology), anti-MSH2 mAb (#2017; Cell Signaling Technology), and anti-β-actin mAb (sc-69879; Santa Cruz Biotechnology, Dallas, TX, USA) at 4 ℃ overnight. The membranes were then incubated with HRP-linked anti-mouse IgG or anti-rabbit IgG antibodies (Cell Signaling Technology) for 1 h at room temperature. Immunoreactive proteins were visualized using ImageQuant LAS 4000 mini (Fuji Film, Tokyo, Japan) with ECL prime western blot detection reagent (GE Healthcare, Chicago, IL, USA).
Detection of intracellular dsDNA in flow cytometry
Cells were fixed in 4% paraformaldehyde for 15 min at room temperature, and then, the cell membranes were disrupted by 0.1% Triton-X in PBS with 3% donkey serum (blocking solution) at room temperature for 30 min. After washing with PBS, the fixed cells were stained with anti-dsDNA mAb (ab270732; Abcam) or mouse IgG3 isotype control (14–4742-82; Thermo Fisher Scientific) at 4 ℃ for 2 h, followed by incubation in the blocking solution with Alexa Fluor 488-conjugated anti-mouse secondary antibody (A-21202; Thermo Fisher Scientific) at 4 ℃ for 1 h. The stained cells were analyzed on a BD FACSCanto II flow cytometer (BD Biosciences, San Jose, CA, USA), and flow cytometry data were analyzed using FlowJo software (FlowJo, Ashland, OR, USA).
Statistical analysis
Statistical analyses were performed using STATA15 (STATA Corp., College Station, TX, USA) or Graph pad Prism 6 (Graph Pad Software, San Diego, CA, USA). Fisher’s exact test, Welch’s t-test, or Mann–Whitney U test were used to determine differences between two variables. For multigroup comparisons, we applied a one-way ANOVA with post hoc Tukey–Kramer test or Kruskal–Wallis test with post hoc Dunn test after analyzing the raw data to check whether they were normally distributed. P-values less than 0.05 were considered statistically significant.
Results
Higher expression of cGAS-STING and CD8 effector gene signature in MSI CRCs
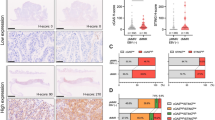
To first examine the expression of cGAS-STING between dMMR/MSI and pMMR/MSS CRCs, we analyzed their expressions in data obtained from TCGA COADREAD and GSE39582 contained 526 (450 MSS and 76 MSI) and 536 (459 MSS and 77 MSI) CRC samples, respectively. mRNA expression levels of MB21D1 (cGAS) and TMEM173 (STING) were significantly higher in MSI CRC than those in their MSS counterparts (Fig. 1a, b). We also examined the expression of CD8 effector, CD4 mature, and Treg gene signatures between MSI and MSS CRCs using these datasets, and analyzed the association between the expression of those gene signatures and cGAS-STING. CD8 effector gene signature was highly expressed in MSI CRC compared with that in MSS CRC (Fig. 1c, d), while the expression of CD4 mature and Treg gene signatures was comparable between MSI and MSS CRCs (Supplementary Fig. S1a–S1d). When dividing all CRC cases into cGAS-high or cGAS-low, and STING-high or STING-low, the expression of CD8 effector gene signature was significantly higher in cGAS-high and STING-high CRCs than those in cGAS-low and STING-low CRCs (Fig. 1c, d), suggesting that the infiltration of CD8+ effector T cells might be associated with the expression of cGAS and STING in CRC. CCL5 and CXCL10 are well known as STING-dependent genes and function as chemoattractants for CD8+ TILs in CRC [37, 38]. We found that both chemokines were highly expressed in MSI CRC and significantly associated with the expression of cGAS and STING (Fig. 1e–h).
MSI CRC has a higher expression of cGAS-STING and CD8 effector gene signature a, b mRNA expression levels of MB21D1 (cGAS) and TMEM173 (STING) in 450 MSS CRCs and 76 MSI CRCs in TCGA (a), and in 459 MSS CRCs and 77 MSI CRCs in GSE39582 (b). c–h The expression of CD8 effector gene signature (c and d), CXCL10 (e and f), and CCL5 g and h in CRC samples in TCGA and GSE39582, respectively (left). Comparison of the expression of CD8 effector gene (c and d), CXCL10 (e and f), and CCL5 g and h between cGAS-low and cGAS-high CRCs (middle), or STING-low and STING-high CRCs (right). Median values were used to differentiate cGAS-low and cGAS-high CRCs, and STING-low and STING-high CRCs. Values are shown as means ± SD. **p < 0.01, ***p < 0.001, ****p < 0.0001, n.s. not significant
CCL5 has been reported to function as a chemo-attractant not only for CD8+ T cells but also for MDSCs [39, 40]. We, therefore, examined the presence of MDSC in pMMR/MSS CRC and dMMR/MSI CRC using the public data. The analyses of the gene expression data of TCGA and GSE39582 revealed that the expression of a gene signature of MDSC was significantly higher in dMMR/MSI CRC than pMMR/MSS CRC (Supplementary Fig. S1e), suggesting that the presence of MDSC might be higher in dMMR/MSI CRC than pMMR/MSS CRC.
Taken together, these results suggest that the expression of cGAS-STING is upregulated in MSI CRC, and the activation of the cGAS-STING pathway might be involved in the recruitment of CD8+ TILs in MSI CRC.
Higher expression of cGAS-STING in tumor cells in dMMR/MSI CRC
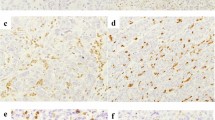
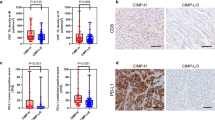
Analysis of such public datasets is based on whole tissue transcript (Fig. 1), and thus, it remains unclear whether cGAS-STING is upregulated in tumor cells but not in stromal cells of dMMR/MSI CRC. We, therefore, conducted IHC for cGAS-STING using surgically resected CRC specimens including 243 pMMR CRCs and 40 dMMR CRCs to evaluate the expression of cGAS-STING in tumor cells. As shown in Table 1, dMMR CRCs were significantly associated with proximal tumor location (p < 0.001), poorly histological differentiation (p < 0.001), and higher PD-L1 expression (p < 0.001) compared with pMMR CRCs. Figure 2a shows representative IHC images for cGAS and STING staining in each intensity (0–3 +) based on tumor cells, and the percentage of moderate and strong staining intensity (2 + and 3 +) of cGAS and STING were increased in dMMR CRC (Fig. 2b). We found that H-scores of cGAS and STING were significantly higher in dMMR CRC compared with those in pMMR CRC (Fig. 2c, d), suggesting that cGAS-STING is highly expressed in tumor cells of dMMR CRC. We also evaluated the number of CD8+ and CD4+ TILs by IHC. As shown in Fig. 2e, f, the number of CD8+ TILs was significantly higher in dMMR CRC than that in pMMR CRC, while the number of CD4+ TILs did not differ between the two groups. In addition, significant associations were observed between the number of CD8+ TILs and expression of cGAS and STING in CRC (Fig. 2g). On the other hand, neither expression of cGAS nor STING had a significant impact on the number of CD4+ or Foxp3+ TILs (Treg cells) (Supplementary Fig. S2a and S2b).
The expression of cGAS-STING is upregulated in tumor cells of dMMR CRC and associated with the number of CD8+ TILs a Representative IHC staining images for cGAS (upper) and STING (lower) in surgically resected CRC specimens. Scale bars, 100 μm; original magnification × 200. b The percentage of IHC staining intensity of cGAS (left) and STING (right) in pMMR CRC and dMMR CRC tissues. c and d Comparison of H-scores of cGAS (c) and STING (d) between pMMR CRC and dMMR CRC. e Representative IHC staining images of CD8 and CD4 in pMMR and dMMR CRCs. Scale bars, 100 μm; original magnification × 200. f Comparison of the number of CD8+ or CD4+ TILs between pMMR and dMMR CRCs. g Comparison of the number of CD8+ TILs between cGAS-low and cGAS-high CRCs (left), or STING-low and STING-high CRCs (right). Median values were used to differentiate cGAS-low and cGAS-high CRCs, and STING-low and STING-high CRCs. Values are shown as means ± SD. Blue dots; pMMR, red dots; dMMR. **p < 0.01, ****p < 0.0001, n.s. not significant
TREX1 is a DNA 3’ → 5’ exonuclease that degrades DNA and prevents aberrant nucleic acid-sensing. TREX1 is known as a negative regulator of cGAS-STING, and TREX1 dysfunction was reported to activate the cGAS-STING pathway, leading to the activation of IFN response [41]. mRNA expression of TREX1 was significantly higher in MSI CRC than that in MSS CRC according to the analysis of TCGA dataset (Supplementary Fig. S3a). Consistent with this result, the proportion of high staining intensity (2 + and 3 +) and H-score of TREX1 was increased in dMMR CRC (Supplementary Fig. S3b and S3c). Although significant associations between the expression of TREX1 and STING were observed in both pMMR and dMMR CRCs, the correlation value in dMMR CRC was higher than that in pMMR CRC (Supplementary Fig. S3d), suggesting that TREX1 might negatively regulate the activation of STING signaling especially in dMMR CRC.
Taken together, these results suggest that dMMR CRC shows higher expression of cGAS-STING in tumor cells, resulting in highly infiltration of CD8+ TILs.
The activation of STING signaling by downregulation of MMR genes in human CRC cell lines
We finally investigated whether (i) the activation of the cGAS-STING pathway in CRC cells contributed to CD8+ T cell infiltration, and (ii) the downregulation of MMR genes could affect the activation of the cGAS-STING pathway in CRC cells. CD8+ cells were isolated from PBMC (Fig. 3a, left) and subjected to cell migration assay using CM of human CRC cell line SW480 cells or CM of STING-activated SW480 cells. The migration level of CD8+ cells was significantly increased by the CM of STING-activated SW480 cells compared with the CM of STING-non-activated SW480 cells (Fig. 3a, right), suggesting that the activation of STING signaling in CRC cells contributes to CD8+ T cell infiltration. We also investigated the effect of the downregulation of MMR genes on the activation of the cGAS-STING pathway in CRC cells. mRNA expression of MLH1 was markedly decreased by MLH1-specific siRNA in human CRC cell line SNU81 and SW480 cells, which are corresponding to MSS CRC cells (Fig. 3b). Please note that siRNA by itself might induce IFN response [42], we used scrambled non-targeting siRNA as a negative control, which has the same lengths (21-bp) as other targeted siRNAs. 2’,3’-cGAMP markedly phosphorylated IRF3, an activation marker of the cGAS-STING pathway, and this phosphorylation was increased by the knockdown of MLH1 in SNU81 and SW480 cells (Fig. 3c). Consistent with this result, the downregulation of MLH1 significantly enhanced 2’,3’-cGAMP-induced expressions of CXCL10 and CCL5, STING-dependent genes and crucial chemoattractants of CD8+ T cells, in both CRC cell lines (Fig. 3d). We also found that the downregulation of another MMR gene, MSH2, reinforced the phosphorylation of IRF3 and the expressions of CXCL10 and CCL5 by 2’,3’-cGAMP in SW480 cells (Supplementary Fig. S4a and S4b). Moreover, the downregulation of STING significantly blunted the enhancement of the expression of CXCL10 and CCL5 by the knockdown of MLH1 in SNU81 and SW480 cells (Fig. 3d), suggesting that high expression of STING contributes to high productions of CXCL10 and CCL5 in MLH1-defective CRC cells.
The downregulation of MMR genes induces the activation of the cGAS-STING pathway in human CRC cell lines a Flow cytometry analysis of PBMC and CD8+ cells isolated from PBMC (left). Cell migration analysis of CD8+ cells using CM of SW480 cells or CM of STIGN-activated (2’,3’-cGAMP-stimulated) SW480 cells (n = 3) (right). b qPCR analysis of MLH1 in SNU81 and SW480 cells transfected with MLH1-specific siRNA (n = 3). c Immunoblot analysis of MLH1 and phosphorylated IRF3 (p-IRF3) in SNU81 and SW480 cells transfected with siRNA for MLH1 and stimulated with 5 μg/ml 2’,3’-cGAMP for 6 h. β-actin was used as a loading control. d Immunoblot analysis of STING and MLH1 in SNU81 and SW480 cells transfected with MLH1-specific siRNA together with or without STING-specific siRNA was shown (left). qPCR analysis of CXCL10 and CCL5 in SNU81 and SW480 cells transfected with MLH1-specific siRNA together with or without STING-specific siRNA and stimulated with 5 μg/ml 2’,3’-cGAMP for 18 h (n = 3) (right). The mRNA expression level was normalized to GAPDH. Values are shown as means ± SD. **p < 0.01, ***p < 0.001, ****p < 0.0001
Because the downregulation of MLH1 by siRNA did not affect the expression of STING in SNU81 and SW480 cells (Supplementary Fig. S4c), another mechanism might be involved in the enhancement of the activation of STING signaling by the knockdown of MLH1 in CRC cells. Guan et al. recently demonstrated that MLH1-deficient tumors exhibited cytosolic DNA accumulation by DNA breaks in the cells, resulting in the activation of the cGAS-STING pathway [29]. We, therefore, tested whether the downregulation of MLH1 and MSH2 increased cytosolic dsDNA in CRC cells. The cytosolic dsDNA levels were found to be significantly increased in SW480 cells when the cells were treated with siRNA for MLH1 and MSH2 (Supplementary Fig. S4d), suggesting that increased expression of cytosolic dsDNA by the downregulation of MMR genes might contribute to the enhancement of the activation of STING signaling by 2’,3’-cGAMP in CRC cell lines.
Taken together, our findings suggest that high expression and activation of cGAS-STING in tumor cells contributes to the high infiltration of CD8+ T cells in dMMR/MSI CRC.
Discussion
It has been widely accepted that dMMR/MSI is associated with a high tumor mutation burden, leading to immune cell infiltration including CD8+ TILs, and therefore, the treatment with ICIs results in improved survival in patients with dMMR/MSI tumors [12]. In this study, as a novel finding, we found that cGAS and STING are highly upregulated in dMMR/MSI CRC compared with pMMR/MSS CRC by analyzing public datasets and our IHC cohort, and the expression of cGAS and STING was significantly associated with the number of CD8+ TILs in CRC. Our in vitro experiments also revealed that the downregulation of MMR genes significantly enhanced the activation of STING signaling by the treatment with 2’,3’-cGAMP in CRC cell lines. Therefore, our current findings suggest that the cGAS-STING pathway in tumor cells could be one of the critical components that activate the tumor-immune microenvironment in dMMR/MSI CRC.
Although we found that the expression of cGAS-STING in tumor cells was markedly higher in dMMR/MSI CRC than pMMR/MSS CRC (Fig. 2), the regulatory mechanisms of higher expressions of cGAS and STING in dMMR/MSI CRC are still unclear. Because sporadic dMMR/MSI CRCs are most commonly caused by MLH1 deficiency, we examined the association between the expressions of MLH1 and cGAS-STING in CRC. However, TCGA dataset revealed that the expression of MLH1 was not inversely correlated with the expression of MB21D1 (cGAS) and TMEM173 (STING) in CRCs (Supplementary Fig. S5), suggesting that defective or lower expressions of MLH1 might not affect the higher expressions of cGAS and STING in dMMR/MSI CRC. In a previous report, cGAS and STING were commonly suppressed in a wide variety of cancer by epigenetic silencing, and promoter methylations of both cGAS and STING genes in CRC were higher than those in normal tissues [43]. When comparing methylation levels of cGAS and STING genes between MSS and MSI CRCs, the methylation level of the STING gene was significantly higher in MSS CRC than MSI CRC (Supplementary Fig. S6a). It should be noted that the methylation level of STING was inversely correlated with the mRNA expression level of STING (Supplementary Fig. S6b). Importantly, our IHC data also revealed that the percentage of loss or lower expressions of cGAS and STING (intensity 0–1 +) was markedly decreased in dMMR CRC compared with pMMR CRC. Thus, higher expressions of cGAS and STING in dMMR/MSI CRC might be partially mediated by the lower frequency of their epigenetic silencing.
TREX1 is a known IFN-stimulated gene and might be upregulated by the activation of the cGAS-STING pathway through a negative feedback mechanism [44, 45]. Indeed, in MSI CRC, the expression of TREX1 significantly correlated with the expressions of several IFN-stimulated genes which are mainly regulated by STING signaling (Supplementary Fig. S3e). Based on the previous reports and our current correlation data between the expressions of STING and TREX1 in CRC (Supplementary Fig. S3d and S3e), the high expression of TREX1 might be modulated by the high activation of the cGAS-STING pathway in dMMR/MSI CRC. Because the blockade of enzymatic activity of TREX1 could stimulate cGAS-STING and type I IFN signaling, TREX1 inhibition in tumor cells should potentiate the anti-tumor effect by activating immune cells in the TME [41]. Indeed, Silson et al. reported that microRNA-based TREX1-knockdown successfully repressed tumor growth through the immune activation of the TME [46]. Therefore, inhibition of TREX1 might become a novel therapeutic strategy for ICI-based immunotherapy of dMMR/MSI CRC by enhancing cGAS-STING-mediated anti-tumor immune responses.
Recent studies suggested that the cGAS-STING pathway is necessary for the anti-cancer immune responses in many types of cancer, and STING agonists showed a favorable effect in promoting CD8+ TILs into the TME [47]. Several clinical studies targeting solid tumors using STING agonists such as 5,6-dimethylxanthenone-4-acetic acid and ADU-S100 have been ongoing [48]. Our current findings suggest that the cGAS-STING pathway is important for the recruitment of CD8+ TILs in CRC tumors including pMMR/MSS CRC, and therefore STING agonists might be applicable in the treatment of CRC, which has a low activity of cGAS-STING signaling.
In conclusion, we, for the first time, showed that cGAS/STING axis is highly upregulated in dMMR/MSI CRC compared with pMMR/MSS CRC, and targeting for cGAS/STING axis may become a novel treatment strategy, when ICI treatment is considered.
Data availability
The datasets generated during and/or analyzed during the current study are available from corresponding author on reasonable request.
Abbreviations
- CCL5:
-
C–C chemokine ligand-5
- cGAMP:
-
Cyclic GMP-AMP
- cGAS:
-
Cyclic GMP-AMP synthase,
- CIN:
-
Chromosomal instability
- CRC:
-
Colorectal cancer
- CXCL10:
-
C-X-C motif ligand 10
- dMMR:
-
Mismatch repair deficient
- ICI:
-
Immune checkpoint inhibitor
- IRF3:
-
Interferon regulatory factor 3
- MLH1:
-
MutL homolog 1
- MSH2:
-
MutS homolog 2
- MSH6:
-
MutS homolog 6
- MSI:
-
Microsatellite instability
- MSS:
-
Microsatellite stable
- pMMR:
-
Mismatch repair proficient
- PMS2:
-
PMS1 homolog 2
- STING:
-
Stimulator of interferon genes
- TREX1:
-
Three-prime repair exonuclease 1
References
Siegel RL, Miller KD, Goding Sauer A, Fedewa SA, Butterly LF, Anderson JC, Cercek A, Smith RA, Jemal A (2020) Colorectal cancer statistics, 2020. CA Cancer J Clin 70:145–164. https://doi.org/10.3322/caac.21601
Keum N, Giovannucci E (2019) Global burden of colorectal cancer: emerging trends, risk factors and prevention strategies. Nat Rev Gastroenterol Hepatol 16:713–732. https://doi.org/10.1038/s41575-019-0189-8
Xie YH, Chen YX, Fang JY (2020) Comprehensive review of targeted therapy for colorectal cancer. Signal Transduct Target Ther 5:22. https://doi.org/10.1038/s41392-020-0116-z
Tutlewska K, Lubinski J, Kurzawski G (2013) Germline deletions in the EPCAM gene as a cause of Lynch syndrome - literature review. Hered Cancer Clin Pract 11:9. https://doi.org/10.1186/1897-4287-11-9
Kuiper RP, Vissers LE, Venkatachalam R et al (2011) Recurrence and variability of germline EPCAM deletions in Lynch syndrome. Hum Mutat 32:407–414. https://doi.org/10.1002/humu.21446
Amodio V, Mauri G, Reilly NM, Sartore-Bianchi A, Siena S, Bardelli A, Germano G (2021) Mechanisms of immune escape and resistance to checkpoint inhibitor therapies in mismatch repair deficient metastatic colorectal cancers. Cancers (Basel). https://doi.org/10.3390/cancers13112638
Randrian V, Evrard C, Tougeron D (2021) Microsatellite instability in colorectal cancers: carcinogenesis, neo-antigens immuno-resistance and emerging therapies. Cancers (Basel). https://doi.org/10.3390/cancers13123063
Boland CR, Goel A (2010) Microsatellite instability in colorectal cancer. Gastroenterology 138:2073-2087.e2073. https://doi.org/10.1053/j.gastro.2009.12.064
Deshpande M, Romanski PA, Rosenwaks Z, Gerhardt J (2020) Gynecological cancers caused by deficient mismatch repair and microsatellite instability. Cancers (Basel) 12:3319. https://doi.org/10.3390/cancers12113319
Pritchard CC, Grady WM (2011) Colorectal cancer molecular biology moves into clinical practice. Gut 60:116–129. https://doi.org/10.1136/gut.2009.206250
Phillips SM, Banerjea A, Feakins R, Li SR, Bustin SA, Dorudi S (2004) Tumour-infiltrating lymphocytes in colorectal cancer with microsatellite instability are activated and cytotoxic. Br J Surg 91:469–475. https://doi.org/10.1002/bjs.4472
Ganesh K, Stadler ZK, Cercek A, Mendelsohn RB, Shia J, Segal NH, Diaz LA Jr (2019) Immunotherapy in colorectal cancer: rationale, challenges and potential. Nat Rev Gastroenterol Hepatol 16:361–375. https://doi.org/10.1038/s41575-019-0126-x
Le DT, Durham JN, Smith KN et al (2017) Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 357:409–413. https://doi.org/10.1126/science.aan6733
Wu J, Sun L, Chen X, Du F, Shi H, Chen C, Chen ZJ (2013) Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science 339:826–830. https://doi.org/10.1126/science.1229963
Gao M, He Y, Tang H, Chen X, Liu S, Tao Y (2020) cGAS/STING: novel perspectives of the classic pathway. Mol Biomed. https://doi.org/10.1186/s43556-020-00006-z
Li T, Chen ZJ (2018) The cGAS-cGAMP-STING pathway connects DNA damage to inflammation, senescence, and cancer. J Exp Med 215:1287–1299. https://doi.org/10.1084/jem.20180139
Jiang M, Chen P, Wang L et al (2020) cGAS-STING, an important pathway in cancer immunotherapy. J Hematol Oncol 13:81. https://doi.org/10.1186/s13045-020-00916-z
Tokunaga R, Zhang W, Naseem M, Puccini A, Berger MD, Soni S, McSkane M, Baba H, Lenz H-J (2018) CXCL9, CXCL10, CXCL11/CXCR3 axis for immune activation–a target for novel cancer therapy. Cancer Treat Rev 63:40–47. https://doi.org/10.1016/j.ctrv.2017.11.007
Sokolowska O, Nowis D (2018) STING signaling in cancer cells: Important or not? Arch Immunol Ther Exp (Warsz) 66:125–132. https://doi.org/10.1007/s00005-017-0481-7
Song S, Peng P, Tang Z et al (2017) Decreased expression of STING predicts poor prognosis in patients with gastric cancer. Sci Rep 7:39858. https://doi.org/10.1038/srep39858
Chon HJ, Kim H, Noh JH et al (2019) STING signaling is a potential immunotherapeutic target in colorectal cancer. J Cancer 10:4932–4938. https://doi.org/10.7150/jca.32806
Bu Y, Liu F, Jia Q-A, Yu S-N (2016) Decreased expression of TMEM173 predicts poor prognosis in patients with hepatocellular carcinoma. PLoS One 11:e0165681. https://doi.org/10.1371/journal.pone.0165681
Xia T, Konno H, Ahn J, Barber GN (2016) Deregulation of STING signaling in colorectal carcinoma constrains DNA damage responses and correlates with tumorigenesis. Cell Rep 14:282–297. https://doi.org/10.1016/j.celrep.2015.12.029
Bakhoum SF, Cantley LC (2018) The multifaceted role of chromosomal instability in cancer and its microenvironment. Cell 174:1347–1360. https://doi.org/10.1016/j.cell.2018.08.027
de la Chapelle A, Hampel H (2010) Clinical relevance of microsatellite instability in colorectal cancer. J Clin Oncol 28:3380–3387. https://doi.org/10.1200/jco.2009.27.0652
Dou Z, Ghosh K, Vizioli MG et al (2017) Cytoplasmic chromatin triggers inflammation in senescence and cancer. Nature 550:402–406. https://doi.org/10.1038/nature24050
Santaguida S, Richardson A, Iyer DR et al (2017) Chromosome mis-segregation generates cell-cycle-arrested cells with complex karyotypes that are eliminated by the immune system. Dev Cell 41:638-651.e635. https://doi.org/10.1016/j.devcel.2017.05.022
Lu C, Guan J, Lu S et al (2021) DNA sensing in mismatch repair-deficient tumor cells is essential for anti-tumor immunity. Cancer Cell 39(96–108):e106. https://doi.org/10.1016/j.ccell.2020.11.006
Guan J, Lu C, Jin Q et al (2021) MLH1 deficiency-triggered DNA hyperexcision by exonuclease 1 activates the cGAS-STING pathway. Cancer Cell 39(109–121):e105. https://doi.org/10.1016/j.ccell.2020.11.004
Mowat C, Mosley SR, Namdar A, Schiller D, Baker K (2021) Anti-tumor immunity in mismatch repair-deficient colorectal cancers requires type I IFN–driven CCL5 and CXCL10. J Exp Med. https://doi.org/10.1084/jem.20210108
Gao J, Aksoy BA, Dogrusoz U et al (2013) Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. https://doi.org/10.1126/scisignal.2004088
Davoli T, Uno H, Wooten EC, Elledge SJ (2017) Tumor aneuploidy correlates with markers of immune evasion and with reduced response to immunotherapy. Science. https://doi.org/10.1126/science.aaf8399
Zhang C, Li J, Wang H, Song SW (2016) Identification of a five B cell-associated gene prognostic and predictive signature for advanced glioma patients harboring immunosuppressive subtype preference. Oncotarget 7:73971–73983. https://doi.org/10.18632/oncotarget.12605
Herbst RS, Baas P, Kim DW et al (2016) Pembrolizumab versus docetaxel for previously treated, PD-L1-positive, advanced non-small-cell lung cancer (KEYNOTE-010): a randomised controlled trial. Lancet 387:1540–1550. https://doi.org/10.1016/s0140-6736(15)01281-7
Kikuchi T, Mimura K, Okayama H et al (2019) A subset of patients with MSS/MSI-low-colorectal cancer showed increased CD8(+) TILs together with up-regulated IFN-γ. Oncol Lett 18:5977–5985. https://doi.org/10.3892/ol.2019.10953
Noda M, Okayama H, Tachibana K et al (2018) glycosyltransferase gene expression identifies a poor prognostic colorectal cancer subtype associated with mismatch repair deficiency and incomplete glycan synthesis. Clin Cancer Res 24:4468–4481. https://doi.org/10.1158/1078-0432.ccr-17-3533
Abe T, Harashima A, Xia T, Konno H, Konno K, Morales A, Ahn J, Gutman D, Barber GN (2013) STING recognition of cytoplasmic DNA instigates cellular defense. Mol Cell 50:5–15. https://doi.org/10.1016/j.molcel.2013.01.039
Zumwalt TJ, Arnold M, Goel A, Boland CR (2015) Active secretion of CXCL10 and CCL5 from colorectal cancer microenvironments associates with GranzymeB+ CD8+ T-cell infiltration. Oncotarget 6:2981–2991. https://doi.org/10.18632/oncotarget.3205
Aldinucci D, Colombatti A (2014) The inflammatory chemokine CCL5 and cancer progression. Med Inflamm 2014:292376. https://doi.org/10.1155/2014/292376
Yang L, Wang B, Qin J, Zhou H, Majumdar AP, Peng F (2018) Blockade of CCR5-mediated myeloid derived suppressor cell accumulation enhances anti-PD1 efficacy in gastric cancer. Immunopharmacol Immunotoxicol 40:91–97
Hemphill WO, Simpson SR, Liu M, Salsbury FR Jr, Hollis T, Grayson JM, Perrino FW (2021) TREX1 as a novel immunotherapeutic target. Front Immunol 12:660184. https://doi.org/10.3389/fimmu.2021.660184
Sledz CA, Holko M, de Veer MJ, Silverman RH, Williams BR (2003) Activation of the interferon system by short-interfering RNAs. Nat Cell Biol 5:834–839. https://doi.org/10.1038/ncb1038
Konno H, Yamauchi S, Berglund A, Putney RM, Mulé JJ, Barber GN (2018) Suppression of STING signaling through epigenetic silencing and missense mutation impedes DNA damage mediated cytokine production. Oncogene 37:2037–2051. https://doi.org/10.1038/s41388-017-0120-0
Schoggins JW, Wilson SJ, Panis M, Murphy MY, Jones CT, Bieniasz P, Rice CM (2011) A diverse range of gene products are effectors of the type I interferon antiviral response. Nature 472:481–485. https://doi.org/10.1038/nature09907
Flood BA, Higgs EF, Li S, Luke JJ, Gajewski TF (2019) STING pathway agonism as a cancer therapeutic. Immunol Rev 290:24–38. https://doi.org/10.1111/imr.12765
Wilson R, Espinosa-Diez C, Kanner N et al (2016) MicroRNA regulation of endothelial TREX1 reprograms the tumour microenvironment. Nat Commun 7:13597. https://doi.org/10.1038/ncomms13597
Demaria O, De Gassart A, Coso S et al (2015) STING activation of tumor endothelial cells initiates spontaneous and therapeutic antitumor immunity. Proc Natl Acad Sci U S A 112:15408–15413. https://doi.org/10.1073/pnas.1512832112
Zhu Y, An X, Zhang X, Qiao Y, Zheng T, Li X (2019) STING: a master regulator in the cancer-immunity cycle. Mol Cancer 18:152. https://doi.org/10.1186/s12943-019-1087-y
Acknowledgements
This work was supported by the grants from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (to S. Nakajima and K. Kono). We thank Masayo Sugeno, Sakino Arai, Eri Takahashi, and Saori Naruse for excellent technical assistant and helpful secretarial assistance.
Author information
Authors and Affiliations
Contributions
SN and KK contributed to the study conception. SN, HO, KM, and KK contributed to the supervision of the study. Material preparation was performed by AK, SN, and KS. Data curation and analysis were performed by SN, AK, and HO. Data collection and investigation were performed by AK, SN, TMa, KS, TK, EE. Patient recruitment was carried out by AK, HO, TMa, TK, EE, MI, YK, MS, ZS, SF, WS, HO, TMo, and SO. The first draft and the revised version of the manuscript were written by AK, SN, and KK. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Conflict of Interest
No potential conflicts of interest relevant to this article were reported.
Ethics approval
This study was conducted in accordance with the principles of the Declaration of Helsinki and was approved by the Institutional Review Board of Fukushima Medical University (Fukushima, Japan). Cell line authentication: Human colorectal cancer cell lines were purchased from the Korean Cell Line Bank (SNU81) and the American Type Culture Collection (SW480), and the cell lines have been authenticated using STR analysis (Promega Japan, Tokyo, Japan).
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kaneta, A., Nakajima, S., Okayama, H. et al. Role of the cGAS-STING pathway in regulating the tumor-immune microenvironment in dMMR/MSI colorectal cancer. Cancer Immunol Immunother 71, 2765–2776 (2022). https://doi.org/10.1007/s00262-022-03200-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00262-022-03200-w