Abstract
The natural cytotoxicity receptors (NCRs; NKp30, NKp44, and NKp46) were first defined as activating receptors on human NK cells that are important in recognition of and response to tumors. A flurry of recent research, however, has revealed that differential splicing can occur during transcription of each of the NCR genes, resulting in some transcripts that encode receptor isoforms with inhibitory functions. These alternative transcripts can arise in certain tissue microenvironments and appear to be induced by cytokines. Evidence indicates that some of the inhibitory NCRs are triggered by specific ligands, such as the interaction of the inhibitory isoform of NKp44 with PCNA on the surface of tumor cells. Here, we review the different NCR splice variants, cytokines that modulate their expression, their functional impacts on innate immune cells, and their differential expression in the contexts of cancer, pregnancy, and infections. The recent discovery of these inhibitory NCR isoforms has revealed novel innate immune checkpoints, many of which still lack defined ligands and clear mechanisms driving their expression. These NCR checkpoint pathways offer exciting potential therapeutic targets to manipulate innate immune functions under defined pathological conditions, such as cancer, pregnancy disorders, and pathogen exposure.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The family of natural cytotoxicity receptors (NCRs) is comprised of NKp46 (NCR1, CD335), NKp44 (NCR2, CD336), and NKp30 (NCR3, CD337) [1]. These three receptors were first characterized on human natural killer (NK) cells. NCR-mediated NK cell activation results in lysis of target cells and secretion of cytokines, mainly IFNγ and TNFα [2,3,4,5]. NKp46 and NKp30 are expressed constitutively on the cell membrane of human peripheral blood NK (pNK) cells, while NKp44 protein expression requires stimulation with IL-2, IL-15, or IL-1β, primarily on the CD56bright subset [6, 7]. An exception for NKp44 protein expression is decidual NK (dNK) cells, which constitutively express NKp44 and can secrete placenta-associated cytokines, such as VEGF, IL-10, and PLGF that promote tissue remodeling [8, 9]. NKp44 protein expression was also observed on intratumoral NK cells, innate lymphoid cell type 3 (ILC3), plasmacytoid dendritic cells (pDCs), and Natural Killer T (NKT) cells [10,11,12,13,14]. Since the discovery of NCRs in the late 1990s, the role of those receptors in immune surveillance has been intensely studied in cancer, viral and bacterial infections, pregnancy, and autoimmunity [1, 15, 16].
NKp30 and NKp44 are encoded by the NCR3 and NCR2 genes, respectively, which are both located on chromosome 6 in the human MHC class III locus [17]. NKp46 is encoded by the NCR1 gene located on chromosome 19 near the leukocyte regulatory complex [18]. The NKp30 and NKp44 proteins are characterized by a single V-type Ig-like extracellular domain, while NKp46 protein has two extracellular C2-type Ig-like domains (domain 1 “D1” domain 2 “D2”, respectively) [1, 19,20,21]. The transmembrane domain of the NCRs contains a positive amino acid, either arginine (for NKp30 and NKp46) or lysine for NKp44. The transmembrane positive amino acid facilitates the activating signal from each receptor by facilitating association with a corresponding negative charged residue in a transmembrane adaptor protein that contains an immunoreceptor tyrosine-based activation motif (ITAM). In this way, both NKp30 and NKp46 associate with CD3ζ and FcεRI-γ, while NKp44 is associated with DAP12, which is essential for the membrane expression of NKp44 (Fig. 1) [22].
NKp46 was the first natural cytotoxic receptor to be discovered in the NCR family and is the only receptor that has a homologous gene in Mus musculus (ncr1/Ly94) [5]. An NKp30 pseudogene is found in 12 mouse strains, however, and a functional ncr3 gene was found in Mus caroli [23]. The presence of the NKp44 gene (NCR2) was first shown only in H. sapiens, but later was found also in non-human primates that also express the NKp44 protein in their NK cells [24]. Thus, NKp44 is the newest member in the evolution of the NCR family, and its function may be relevant only in primates. This should be taken into account when studying NCR-mediated NK function using mouse models [25].
NKp30 splice variant profiles
The mRNA of the NCR3 gene is transcribed as four exons, and these can be differentially spliced (Fig. 2a). NKp30 splice variants that have been studied to date are NKp30a, NKp30b, and NKp30c (NCBI; Accession: NM_147130.2, NM_001145466.1, and NM_001145467.1, respectively). NKp30a-c splice variants differ from each other at exon no. 4 due to splicing events, resulting in a variable NKp30 intracellular domain. Other NKp30 splice variants that have been described are NKp30d, NKp30e, and NKp30f that encode a C-type Ig domain with the corresponding intracellular domains of NKp30b, NKp30a, and NKp30c, respectively (Fig. 1a) [23]. The NKp30 splice variants can only be detected at the mRNA level, as there is no available specific antibody for each of the NKp30 isoform proteins. Both NKp30a and NKp30b lead to NK cell activation that is mediated by association with CD3ζ. On the other hand, NKp30c has a weak association to CD3ζ and leads to the activation of p38 MAP kinase, resulting in secretion of IL-10 by NK cells and immunosuppression [26]. However, NK cells express all three NKp30 splice variants, yet the functional outcome depends on the expression pattern of NKp30a, NKp30b, and NKp30c [26]. Therefore, NKp30 splice variant profiles have been classified based on the relative abundance of the splice variants; namely: the activating NKp30a/b profile and the inhibitory NKp30c profile [26]. The impact of NKp30 splice variant profiles in cancer is yet to be fully understood and was tested also in viral infection [27, 28]. NKp30 splice variant profiles were described in tumor infiltrating lymphocytes (TILs) from gastrointestinal stromal tumor (GIST) patients that were treated with Imatinib [26]. GIST patients with a predominant NKp30c profile had lower survival rate. Furthermore, in 50% of the patients, the NKp30c profile was associated with single-nucleotide polymorphism (SNP) in the NCR3 gene, resulting in two haplotype groups (Table 1; [26]). Subsequently, NKp30 splice variant profiles were also described to have an effect in neuroblastoma, melanoma, and NSCLC patients (Table 1; [29,30,31,32]). Moreover, NKp30 splice variant profiles from PBMC were recently suggested to be prognostic predictive biomarkers in patients with metastatic GIST (Table 1; [33]).
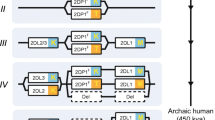
Exon map of NCR splice variants; a NKp30, b NKp44, and c NKp46. Regions representing the various exons are colored differently, and in accordance with the colors of the exon-encoded protein domains in Fig. 1. Teardrops mark locations of charged residues in the transmembrane domains (R = arginine, K = lysine). Yellow arrows mark premature stop codons caused by frame shifts during differential splicing. Location of the ITIM-like sequence is marked with hashed shading in NKp44-1. The ΔA teardrop in NKp46b marks the location of an alanine (A) deletion
The influence of the microenvironment on NKp30 splice variant profiles was also examined [34, 35]. NKp30 is expressed in dNK cells and human primary trophoblasts cells express NKp30 ligands [8]. The expression profile of NKp30 splice variants in PBMC and matched placenta tissue in women that undergo elective, sporadic or recurrent first trimester abortion, showed matched cases with different NKp30 splice variant profiles, indicating that the placenta microenvironment can influence the NKp30 splice variants expression pattern (Table 1; [35]). Decidua-enriched cytokines have been shown to influence NKp30 splice variant profiles [34]. IL-15 and IL-18 induce the expression of both NKp30a and NKp30b relative to NKp30c. The presence of TGF-β, with IL-15 and/or IL-18 combinations, resulted in higher expression of NKp30c relative to NKp30a (Table 1; [34]). TGF-β is abundant in the placenta and tumor microenvironment [34]. Intratumoral NK cells share phonotypical characterization with dNK cells (CD3−, CD56bright and CD16−) [36]. Analysis of NKp30 splice variant profiles in tumor and matched normal tissues in different organs revealed higher incidence of NKp30a/b profile relative to NKp30c profile [37]. The NKp30a/b profile was more abundant both in the tumor and in the normal tissues. However, in 50% of the matched cases, NKp30 splice variant profiles were shifted from NKp30a/b to NKp30c and from NKp30c to NKp30a, indicating the flexible nature of NKp30 splice variant expression (Table 1; [37]).
NKp44 splice variant profiles
Among the NCR family, NKp44 is a unique receptor, as its expression on the cell membrane of NK cells requires stimulation [6, 7]. NCR2 gene is transcribed as six exons and three splice variants NKp44-1, NKp44-2, NKp44-3 (Fig. 2b, NCBI; Accession: NM_004828.3, NM_001199509.1, and NM_001199510.1, respectively). However, NKp44-1, but not NKp44-2 or NKp44-3, has an immunoreceptor tyrosine-based inhibition motif (ITIM)-like sequence, encoded by exon six, as part of its cytoplasmic tail. Alternative splicing events in NKp44-2 and NKp44-3 result in the expression of exon five, leading to an early stop codon on exon six and a shorter cytoplasmic tail without the ITIM (Figs. 1b, 2b). Proliferating cell nuclear antigen (PCNA) is an inhibitory ligand of NKp44 [38,39,40]. PCNA expression was associated with inhibition of NK cells in endometrial carcinoma [41]. The ITIM on the NKp44-1 intracellular domain was shown to be essential for NKp44-mediated inhibition of NK cell function when exposed to HeLa cells overexpressing PCNA (Table 2; [38]). The inhibitory nature of NKp44 was also observed in pDC and was linked to reduced secretion of IFNα and lysis function [11, 12]. Expressing the ITIM sequence of NKp44-1 fused to KIR3DL1 in NK92 cells, however, did not result in SHP-1, SHP-2, or SHIP recruitment, even when the ITIM tyrosine was phosphorylated by pervanadate stimulation [22]. Furthermore, the ITIM-containing NKp44 cytoplasmic domain in this chimeric receptor could not elicit inhibitory function following mAb engagement in redirected assays using the P815 model system [22]. These early results suggested that the ITIM-like sequence in NKp44-1 was incapable of inhibitory function using mAb-redirected assay (Table 2; [22]).
The effect of decidua-enriched cytokines on the pattern of NKp44 splice variants was also examined [34]. As with NKp30 splice variants, there is no specific mAb against each of the NKp44 isoforms and, therefore, the profiling of NKp44 splice variants is only possible by mRNA analysis. The expression of NKp44 splice variants was compared between pNK cells following overnight stimulation with IL-15 and dNK cells from the same donor, and these results were correlated with NK cell function based on NKp44 mAb ligation [34]. Both pNK and dNK expressed all NKp44 splice variants. However, pNK expressed higher levels of NKp44-2 and lower levels of NKp44-1 and NKp44-3, while dNK expressed similar levels of all NKp44 splice variants. dNK cells are known to express NKp44 protein and have a poor cytotoxic function. NKp44 mAb ligation of dNK did not result in CD107a degranulation, while NKp46 ligation did. However, co-ligation of NKp44 and NKp46 in dNK cells led to reduction in CD107a degranulation as compared to ligation of NKp46 alone, indicating that NKp44 has inhibitory function in these cells [34]. NKp44 mAb ligation did lead to the secretion of IFNγ, VEGF-A, or CCL3 by dNK cells. Stimulation of pNK cells with decidua-enriched cytokines (combination of IL-15, IL-18, and TGF-β) up-regulated the expression of both NKp44-1 and NKp44-3 after 6 days of culture and affected the ability of pNK cells to secrete TNFα, IFNγ, VEGF-A, and CCL3 (Table 2; [34]).
NKp44-mediated inhibition of NK cells following PCNA recognition requires the dominant expression of NKp44-1 relative to NKp44-2 and NKp44-3 [13]. Freshly isolated pNK cells or PBMC express basal levels of NKp44 splice variants that are characterized by similar levels of NKp44-1/NKp44-2/NKp44-3 mRNA, but with a very low-to-negative expression at the NKp44 protein level. Following 6 days of in vitro IL-2/IL-15 stimulation, purified pNK cells up-regulated the expression of NKp44-1, NKp44-2, and NKp44-3 with a dominant expression of NKp44-1 splice variant (NKp44-1 > 66% of NKp44 mRNA, i.e., NKp44-1 profile) [13]. On the other hand, in vitro stimulation of whole PBMC with IL-2/IL-15 leads to co-dominant expression of both NKp44-1 and NKp44-3 (NKp44-1 < 66% of NKp44 mRNA, i.e. NKp44-2/3 profile). KHYG1 cells also exhibit co-dominant expression of NKp44-1 and NKp44-3. In accordance, isolated human pNK cells were inhibited by PCNA overexpressing HeLa cells, while KHYG1 cells were not inhibited. The inhibitory effect of the NKp44-1 profile on pNK cells was blocked using an anti-NKp44 mAb, while overexpression of NKp44-1 but not NKp44-2 or NKp44-3 in NK-92 cells resulted in inhibition by PCNA overexpression (Table 2; [13]).
The differential role of NKp44 splice variant profiles in human pathology was first published from RNAseq data obtained from PBMC samples taken from newly diagnosed AML patients (The human Cancer Genome Atlas) [13]. The examination of RNAseq data obtained from PBMC can better reflect the immune status, as blood samples are more homogenous than solid tumors in their cell composition. Plotting all NKp46+/NKp44+ cases against NKp46+/NKP44− cases with day of death data revealed no difference in patient survival between the two groups. However, when dividing the NKp46+/NKp44+ cases according to their NKp44 splice variant profiles, a significant decrease in patient survival was observed in the group that had an NKp44 splice variant profile with solitary expression of NKp44-1 [13]. Examination of all available risk factors, NKp46 and NKp30 mRNA expression, and NKp30 splice variant expression did not influence the observation (Table 2; [13]).
Tumor cells are known to up-regulate PCNA expression in vivo [37, 41]. The NKp44-1-dominant expression profile was seen in 80% of NKp44 mRNA positive cases of tumor biopsies taken from cancers of the breast, lung, cervix/uterus, kidney, gastrointestinal (GI) tract organs, and GI tract accessory organs, and NKp44 mRNA incidence was increased in the tumor tissue as compared to matched normal tissue [37]. The NKp44-1 profile may facilitate inhibition of intratumoral NK cell function; however, this hypothesis still needs to be further examined in larger cohorts. The interaction of NKp44 splice variants with other reported NKp44 activating ligands is poorly defined. The combined effect of PCNA and NKp44 activating ligands also needs to be further addressed as NKp44-1 splice variant has also been reported to elicit NK cell activation [42]. Consistent with the tumor tissue, 80% of placental tissue samples from first trimester elective abortions also exhibit the NKp44-1-dominant profile (Table 2; [37]). Placental microenvironment is known to suppress the maternal immune system to protect the fetus from rejection and human primary trophoblasts cells express NKp44 ligands [8, 43]. On the other hand, 80% of placental samples from the first trimester spontaneous abortions have an NKp44-3-dominant profile. This shift in NKp44 splice variant profile may reflect changes in placental microenvironment factors, such as IL-15, IL-18, and TGF- β that are known to regulate the NCR2 gene. The direct effect of NKp44 splice variants on dNK in respect to the placental microenvironment and unique cell composition is still unclear and should be further explored.
NKp46 splice variant profiles
The NCR1 gene is transcribed as seven exons and five splice variants; NKp46a, NKp46b, NKp46c, NKp46d, and NKp46e (Figs. 1c, 2c, NCBI; Accession: NM_004829.6, NM_001145457.2, NM_001145458.2, NM_ 001242356.2, and NM_001242357.2 respectively). The variation in NKp46 splice variants is mainly in two forms; (1) deletion of exon 3, which encodes the extracellular Ig-like D1 domain (splice variants NKp46d and NKp46e), (2) deletion of exon 6 that encodes for 17 amino acids in the hinge domain (splice variants NKp46c and NKp46d). NKp46a and NKp46b encode the full-length receptor with variation of one amino acid (Fig. 1c). In contrast to NKp44 and NKp30 isoforms, NKp46 isoforms can be partially classified at the protein level with specific antibodies against D1 and D2 Ig-like domains. Long-term stimulation of pNK with IL2 revealed a small NK cell population that expresses NKp46 protein without D1 (NKp46D1−) [44]. The interaction of NKp46 with its ligands was shown to require the D2 but not the D1 domain [45, 46]. Following anti-NKp46/NKp30 mAb stimulation and assessment of CD107a degranulation, the NKp46D1− pNK population manifested higher activity as compared to other pNK cells [44]. Furthermore, the analysis of nasal wash samples with respiratory viral infection for NKp46D1−, NKp46D1+, and total NKp46 mRNA showed that most cases were positive for NKp46D1- mRNA. In contrast, the analysis of PBMC samples revealed that all samples were NKp46D1+ [44].
Innate immune checkpoints and new candidate(s) based on splice variants of NCRs
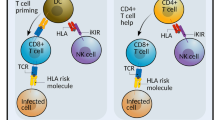
Activating and inhibitory receptors dynamically modulate the NK cell activation state, delicately balancing the state of activation vs. inhibition [47,48,49]. Immune surveillance via recognition of class I HLA molecules on the surface of normal or target cells by inhibitory killer-cell Ig-like receptors (KIR) and CD94/NKG2A provide a dominant early innate immune checkpoint [50]. The loss of class I HLA recognition favors activation, overcoming immune escape by the target cell and allowing signals from activating receptors (e.g., NCRs) to predominate [26, 38, 51]. Immune surveillance via pattern recognition provides a secondary late innate immune checkpoint and is characterized by engagement of NCRs that can recognize ligands on tumor and virus infected cells independently of HLA expression [52]. The pattern recognition by NCRs ultimately depends on how the pathogen or malignant transformation modulates the expression of endogenous proteins (e.g., B7-H6 or PCNA) or on direct recognition of pathogen-associated molecules (e.g., HA-NA or pp65) [38, 53,54,55]. Tumors can escape both immune checkpoints by either masking target cell surface class I HLA molecules or by taking advantage of the differential pattern recognition outcomes (i.e., suppression vs. activation) by specific NCRs isoforms [26, 38]. While the early innate immune checkpoint is often missed and, therefore, appears to be difficult to tackle, the later checkpoint provides more opportunities for targeting by either mAbs or small molecule drug-based therapy. These therapeutic approaches might potentially aim to interfere with a receptor–ligand interaction via targeting NCR itself or its suppressive ligand. When considering NCRs for checkpoint-based therapeutics, NKp44 and NKp30 do have inhibitory isoforms, but mAb-based treatment is challenging, since the structural differences between isoforms (inhibitory vs. activation) are within the transmembrane/cytoplasmic domains (Figs. 1, 2). Therefore, when considering NCR ligands, the membrane-associated PCNA (mediating inhibition through the NKp44-1 isoform) could be considered as a more appropriate target for mAb-based checkpoint therapeutics for cancer, as compared to B7–H6, which was reported to activate all isoforms of NKp30 (inhibitory and activating).
Concluding remarks
The data provided here show the importance of NCR splice variant profiles in a variety of human pathologies. The functional outcomes mediated through NKp30, NKp44, and NKp46 still need to be defined with respect to known and unknown NCR splice variants and other cell types. It is important to note that NCRs are expressed on a wide array of immune cells, including type 1, 2, and 3 innate lymphoid cells (ILCs), γδT and αβT cells, and NKT cells, which play important roles in the immune response toward a variety of bacteria and helminth infections, cancer immunity, and fat tissue metabolism. The characterization of NCR splice variants in NK cell function and tumor and placental microenvironment has opened a window that reveals novel molecular mechanisms by which the three NCRs regulate innate immune functions and identifies new targets to modulate these pathways in the clinic.
Abbreviations
- CCL:
-
Chemokine (C–C motif) ligand
- D1:
-
Domain-1
- D2:
-
Domain-2
- DAP12:
-
DNAX-activation protein 12
- GI:
-
Gastrointestinal
- GIST:
-
Gastrointestinal stromal tumor
- HA:
-
Hemagglutinin
- ILCs:
-
Innate lymphoid cells
- KIR:
-
Killer-cell immunoglobulin-like receptors
- LY94:
-
Lymphocyte antigen 94
- NA:
-
Neuraminidase
- NCRs:
-
Natural cytotoxicity receptors
- NCR1:
-
Natural cytotoxicity receptor 1
- NCR2:
-
Natural cytotoxicity receptor 2
- NCR3:
-
Natural cytotoxicity receptor 3
- NKG2A:
-
Natural killer G2A
- NKT:
-
Natural killer T
- PCNA:
-
Proliferating cell nuclear antigen
- pDCs:
-
Plasmacytoid dendritic cells
- pNK:
-
Peripheral blood NK
- pp65:
-
Cytomegalovirus pp65
- SHP:
-
Small heterodimer partner
- TGF-β:
-
Transforming growth factor beta
References
Kruse PH, Matta J, Ugolini S, Vivier E (2014) Natural cytotoxicity receptors and their ligands. Immunol Cell Biol 92(3):221–229
Pessino A, Sivori S, Bottino C et al (1998) Molecular cloning of NKp46: a novel member of the immunoglobulin superfamily involved in triggering of natural cytotoxicity. J Exp Med 188(5):953–960
Yokoyama WM, Kim S, French AR (2004) The dynamic life of natural killer cells. Annu Rev Immunol 22:405–429
Pende D, Parolini S, Pessino A et al (1999) Identification and molecular characterization of NKp30, a novel triggering receptor involved in natural cytotoxicity mediated by human natural killer cells. J Exp Med 190(10):1505–1516
Walzer T, Blery M, Chaix J et al (2007) Identification, activation, and selective in vivo ablation of mouse NK cells via NKp46. Proc Natl Acad Sci USA 104(9):3384–3389
Vitale M, Bottino C, Sivori S et al (1998) NKp44, a novel triggering surface molecule specifically expressed by activated natural killer cells, is involved in non-major histocompatibility complex-restricted tumor cell lysis. J Exp Med 187(12):2065–2072
Mattiola I, Pesant M, Tentorio PF et al (2015) Priming of human resting NK cells by autologous M1 macrophages via the engagement of IL-1beta, IFN-beta, and IL-15 pathways. J Immunol 195(6):2818–2828
Hanna J, Goldman-Wohl D, Hamani Y et al (2006) Decidual NK cells regulate key developmental processes at the human fetal-maternal interface. Nat Med 12(9):1065–1074
Tabiasco J, Rabot M, Aguerre-Girr M et al (2006) Human decidual NK cells: unique phenotype and functional properties—a review. Placenta 27(Suppl A):S34–S39
Spits H, Artis D, Colonna M et al (2013) Innate lymphoid cells—a proposal for uniform nomenclature. Nat Rev Immunol 13(2):145–149
Bonaccorsi I, Cantoni C, Carrega P et al (2010) The immune inhibitory receptor LAIR-1 is highly expressed by plasmacytoid dendritic cells and acts complementary with NKp44 to control IFNα production. PLoS One 5(11):e15080
Fuchs A, Cella M, Kondo T, Colonna M (2005) Paradoxic inhibition of human natural interferon-producing cells by the activating receptor NKp44. Blood 106(6):2076–2082
Shemesh A, Brusilovsky M, Hadad U et al (2016) Survival in acute myeloid leukemia is associated with NKp44 splice variants. Oncotarget 7(22):32933–32945
Platonova S, Cherfils-Vicini J, Damotte D et al (2011) Profound coordinated alterations of intratumoral NK cell phenotype and function in lung carcinoma. Cancer Res 71(16):5412–5422
Gur C, Porgador A, Elboim M et al (2010) The activating receptor NKp46 is essential for the development of type 1 diabetes. Nat Immunol 11(2):121–128
Waldhauer I, Steinle A (2008) NK cells and cancer immunosurveillance. Oncogene 27(45):5932–5943
Biassoni R, Cantoni C, Pende D et al (2001) Human natural killer cell receptors and co-receptors. Immunol Rev 181(1):203–214
Mandelboim O, Porgador A (2001) NKp46. Int J Biochem Cell Biol 33(12):1147–1150
Foster CE, Colonna M, Sun PD (2003) Crystal structure of the human natural killer (NK) cell activating receptor NKp46 reveals structural relationship to other leukocyte receptor complex immunoreceptors. J Biol Chem 278(46):46081–46086
Joyce MG, Tran P, Zhuravleva MA, Jaw J, Colonna M, Sun PD (2011) Crystal structure of human natural cytotoxicity receptor NKp30 and identification of its ligand binding site. Proc Natl Acad Sci USA 108(15):6223–6228
Cantoni C, Ponassi M, Biassoni R et al (2003) The three-dimensional structure of the human NK cell receptor NKp44, a triggering partner in natural cytotoxicity. Structure 11(6):725–734
Campbell KS, Yusa S, Kikuchi-Maki A, Catina TL (2004) NKp44 triggers NK cell activation through DAP12 association that is not influenced by a putative cytoplasmic inhibitory sequence. J Immunol 172(2):899–906
Hollyoake M, Campbell RD, Aguado B (2005) NKp30 (NCR3) is a pseudogene in 12 inbred and wild mouse strains, but an expressed gene in Mus caroli. Mol Biol Evol 22(8):1661–1672
De Maria A, Ugolotti E, Rutjens E et al (2009) NKp44 expression, phylogenesis and function in non-human primate NK cells. Int Immunol 21(3):245–255
Hong HS, Rajakumar PA, Billingsley JM, Reeves RK, Johnson RP (2013) No monkey business: why studying NK cells in non-human primates pays off. Front Immunol 4:32
Delahaye NF, Rusakiewicz S, Martins I et al (2011) Alternatively spliced NKp30 isoforms affect the prognosis of gastrointestinal stromal tumors. Nat Med 17(6):700–707
Prada N, Antoni G, Commo F et al (2013) Analysis of NKp30/NCR3 isoforms in untreated HIV-1-infected patients from the ANRS SEROCO cohort. Oncoimmunology 2(3):e23472
Mantovani S, Mele D, Oliviero B, Barbarini G, Varchetta S, Mondelli MU (2015) NKp30 isoforms in patients with chronic hepatitis C virus infection. Immunology 146(2):234–242
Messaoudene M, Fregni G, Enot D et al (2016) NKp30 isoforms and NKp46 transcripts in metastatic melanoma patients: unique NKp30 pattern in rare melanoma patients with favorable evolution. OncoImmunology 5(12):e1154251
Sivori S, Parolini S, Marcenaro E et al (2000) Involvement of natural cytotoxicity receptors in human natural killer cell-mediated lysis of neuroblastoma and glioblastoma cell lines. J Neuroimmunol 107(2):220–225
Semeraro M, Rusakiewicz S, Minard-Colin V et al (2015) Clinical impact of the NKp30/B7-H6 axis in high-risk neuroblastoma patients. Sci Transl Med 7(283):283ra55
Fend L, Rusakiewicz S, Adam J et al (2017) Prognostic impact of the expression of NCR1 and NCR3 NK cell receptors and PD-L1 on advanced non-small cell lung cancer. OncoImmunology 6(1):e1163456
Rusakiewicz S, Perier A, Semeraro M et al (2017) NKp30 isoforms and NKp30 ligands are predictive biomarkers of response to imatinib mesylate in metastatic GIST patients. OncoImmunology 6(1):e1137418
Siewiera J, Gouilly J, Hocine H et al (2015) Natural cytotoxicity receptor splice variants orchestrate the distinct functions of human natural killer cell subtypes. Nat Commun 6:10183
Shemesh A, Tirosh D, Sheiner E et al (2015) First trimester pregnancy loss and the expression of alternatively spliced NKp30 isoforms in maternal blood and placental tissue. Front Immunol 6:189
Levi I, Amsalem H, Nissan A et al (2015) Characterization of tumor infiltrating natural killer cell subset. Oncotarget 6(15):13835–13843
Shemesh A, Kugel A, Steiner N et al (2016) NKp44 and NKp30 splice variant profiles in decidua and tumor tissues: a comparative viewpoint. Oncotarget 7(43):70912–70923
Rosental B, Brusilovsky M, Hadad U et al (2011) Proliferating cell nuclear antigen is a novel inhibitory ligand for the natural cytotoxicity receptor NKp44. J Immunol 187(11):5693–5702
Horton NC, Mathew SO, Mathew PA (2013) Novel interaction between proliferating cell nuclear antigen and HLA I on the surface of tumor cells inhibits NK cell function through NKp44. PloS one 8(3):e59552
Horton NC, Mathew PA (2015) NKp44 and natural cytotoxicity receptors as damage-associated molecular pattern recognition receptors. Front Immunol 6:31
Garzetti GG, Ciavattini A, Goteri G et al (1994) Natural killer cell activity in stage I endometrial carcinoma: correlation with nuclear grading, myometrial invasion, and immunoreactivity of proliferating cell nuclear antigen. Gynecol Oncol 55(1):111–114
Rajagopalan S, Long EO (2013) Found: a cellular activating ligand for NKp44. Blood 122(17):2921–2922
Holtan SG, Creedon DJ, Haluska P, Markovic SN (2009) Cancer and pregnancy: parallels in growth, invasion, and immune modulation and implications for cancer therapeutic agents. Mayo Clin Proc 84(11):985–1000
Shemer-Avni Y, Kundu K, Shemesh A et al (2017) Expression of NKp46 splice variants in nasal lavage following respiratory viral infection: domain 1-negative isoforms predominate and manifest higher activity. Front Immunol 8:161
Hadad U, Thauland TJ, Martinez OM, Butte MJ, Porgador A, Krams SM (2015) NKp46 clusters at the immune synapse and regulates NK cell polarization. Front Immunol 6:495
Arnon TI, Achdout H, Lieberman N et al (2004) The mechanisms controlling the recognition of tumor- and virus-infected cells by NKp46. Blood 103(2):664–672
MacFarlane A, Campbell KS (2006) Signal transduction in natural killer cells. In: Compans R et al (eds) Immunobiology of natural killer cell receptors. Current topics in microbiology and immunology, vol 298. Springer, Berlin, pp 23–57.
Lanier LL (2008) Up on the tightrope: natural killer cell activation and inhibition. Nat Immunol 9(5):495–502
Vivier E, Tomasello E, Baratin M, Walzer T, Ugolini S (2008) Functions of natural killer cells. Nat Immunol 9(5):503–510
Campbell KS, Purdy AK (2011) Structure/function of human killer cell immunoglobulin-like receptors: lessons from polymorphisms, evolution, crystal structures and mutations. Immunology 132(3):315–325
Porgador A, Mandelboim O, Restifo NP, Strominger JL (1997) Natural killer cell lines kill autologous beta2-microglobulin-deficient melanoma cells: implications for cancer immunotherapy. Proc Natl Acad Sci USA 94(24):13140–13145
Bloushtain N, Qimron U, Bar-Ilan A et al (2004) Membrane-associated heparan sulfate proteoglycans are involved in the recognition of cellular targets by NKp30 and NKp46. J Immunol 173(4):2392–2401
Schlecker E, Fiegler N, Arnold A et al (2014) Metalloprotease-mediated tumor cell shedding of B7-H6, the ligand of the natural killer cell-activating receptor NKp30. Cancer Res 74(13):3429–3440
Mandelboim O, Lieberman N, Lev M et al (2001) Recognition of haemagglutinins on virus-infected cells by NKp46 activates lysis by human NK cells. Nature 409(6823):1055–1060
Arnon TI, Achdout H, Levi O et al (2005) Inhibition of the NKp30 activating receptor by pp65 of human cytomegalovirus. Nat Immunol 6(5):515–523
Author information
Authors and Affiliations
Contributions
AS, MB, KSC, and AP wrote the manuscript. AS, MB, KK, AO, KSC, and AP edited the manuscript.
Corresponding author
Ethics declarations
Funding
The splice variants studies from Angel Porgador laboratory described in this review were supported by the Israel Science Foundation Grant 1188/16 (Angel Porgador), the US/Israel Binational Science Foundation Grant 2015344 (Angel Porgador and Kerry S. Campbell), the Israeli Ministry of Science and Technology/German Cancer Research Center program Grant CA172 (Angel Porgador), and the Israeli Ministry of Science and Technology Grant 54180 (Angel Porgador).
Conflict of interest
The authors have declared that no conflict of interest exists.
Additional information
This paper is a Focussed Research Review based on a presentation given at the Fifth International Conference on Cancer Immunotherapy and Immunomonitoring (CITIM 2017), held in Prague, Czech Republic, 24th–27th April 2017. It is part of a series of Focussed Research Reviews and meeting report in Cancer Immunology, Immunotherapy.
Rights and permissions
About this article
Cite this article
Shemesh, A., Brusilovsky, M., Kundu, K. et al. Splice variants of human natural cytotoxicity receptors: novel innate immune checkpoints. Cancer Immunol Immunother 67, 1871–1883 (2018). https://doi.org/10.1007/s00262-017-2104-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00262-017-2104-x