Abstract
Melanoma reactive CTL were obtained by stimulating PBL from a melanoma patient in remission since 1994 following adjuvant TIL immunotherapy, with the autologous melanoma cell line. They were cloned by limiting dilution. One CTL clone recognized melanoma cell lines expressing tyrosinase and the B*4002 molecule, either spontaneously or upon transfection. We demonstrated that this clone recognizes the tyrosinase-derived nonapeptide 316-324 (ADVEFCLSL) and the overlapping decapeptide 315–324 (SADVEFCLSL). We derived two distinct additional specific CTL clones from this same patient that were also reactive against B*4002 melanoma cell lines, suggesting a relative diversity of this specific repertoire in this patient. Stimulating PBMC derived from four additional B*4002 melanoma patients with the tyrosinase 316–324 nonapeptide induced the growth of specific cells for two of the patients, demonstrating the immunogenicity of this new epitope. Our data show that this nonapeptide is a new tool that could be used to generate melanoma-specific T cells for adoptive immunotherapy or serve as a peptide vaccine for HLA-B*4002 melanoma patients.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
One of the main criteria to be respected for using a tumor epitope in cancer immunotherapy is its ability to stimulate a high-avidity T cell repertoire towards tumor cells. Several melanoma antigens and epitopes have already been identified, either after stimulation of melanoma patient PBMC with autologous cell lines (e.g. MAGE-A1/A1 [35], MAGE-A3/A1 [11], tyrosinase/B3501 [25], tyrosinase/B44 [5]), or directly from tumor infiltrating lymphocytes (e.g. Melan-A/A2 [17] or Melan-A/B35 [3]). However, the immunogenicity of all these potential targets will only be established if they work in immunotherapy for melanoma patients. Another possible way to discover tumor epitopes relevant for immunotherapy is to look for the presence of tumor-specific memory T cells in patients who have responded to immunotherapy and to analyze their specificities.
To identify such tumor epitopes, we looked for tumor-specific T cells in the blood of a melanoma patient (M88) who remained relapse-free for 12 years after TIL therapy [10, 21]. After complete lymph node excision, this stage III melanoma patient had received 6.7 × 109 TIL, among which 3 × 106 lymphocytes were reactive against the autologous M88 melanoma line [21, 27]. In a retrospective study, we analyzed the antigenic specificities of those TIL and found only a response against the NA88-A antigen restricted by the B*1302 allele [2, 24].
M88 CD8-sorted PBMC were stimulated with the melanoma cell line derived from the original tumor. These stimulated CD8+ lymphocytes exhibited high reactivity towards the autologous melanoma line. We report here the characterization of a tyrosinase epitope, presented in the HLA-B*4002 context, that is recognized by a CTL clone derived from the stimulated cells. Tyrosinase, an enzyme involved in melanin synthesis, is a melanocytic differentiation antigen and a number of class I epitopes derived from this antigen have been found to be presented in various HLA-A and B contexts [22].
Analysis of the reactivity of our melanoma-specific CTL clone towards COS cells transfected with fragments of the tyrosinase cDNA allowed us to narrow down the region containing the epitope to a 21-amino acid segment. Overlapping peptides within this region were then synthesized, and a 9-amino acid peptide, as well as an overlapping 10-amino acid peptide, were recognized by our CTL clone.
Materials and methods
Cell lines
Melanoma cells lines were established from fragments of metastatic tumors or tumor invaded lymph nodes. Mouse fibrosarcoma WEHI 164 clone 13, used for TNF production assays and COS-7 cells were obtained from T. Boon (Ludwig Institute for Cancer Research, Brussels, Belgium). COS-7 cells, WEHI 164 clone 13 and melanoma cells lines were cultured as described before [2]. The EBV cell line B*4002 “Adam” was obtained in our laboratory [6].
PBL stimulation with autologous melanoma cell line
PBL were isolated from M88 melanoma patient blood, collected on December 2003, approximately 10 years after TIL immunotherapy. This patient remained relapse-free after TIL therapy that occurred in February 1994. An autologous melanoma cell line was established from fragments of tumor invaded lymph node, which were also used to produce TIL for immunotherapy. The HLA genotype of M88 melanoma cell line was A*0201, A*0301, B*1302, B*4002, Cw*0602 and Cw*0202. 2 × 105 CD8-sorted PBL were then stimulated by 2 × 104 irradiated M88 melanoma cells in RPMI 1640 medium + 8% HS supplemented with 1,000 U/ml of IL-6 and 5 ng/ml IL-12 (Abcys), in 96-well plates. Irradiated melanoma cells were added again twice at 7-day intervals in a medium containing 10 U/ml IL-2 (Chiron, Amsterdam, the Netherlands) and 5 ng/ml IL-7 (Abcys). Seven days after the third stimulation, the specificity of stimulated culture was tested by TNF response against M88 cell line.
Cloning of T cells
Cells from polyclonal cultures containing specific T cells were cloned by limiting dilution as previously described [12]. Briefly, T cells were plated in U-bottom 96-well plates with irradiated feeder cells, at concentrations of 1, 0.6 or 0.3 T cells/well. Irradiated feeder cells consisted of 105 allogenic PBMC and 104 BLCL cells/well. Stimulatory medium consisted of RPMI 1640 + 8% HS containing 150 U/ml IL-2 and 1 μg/ml Phytohemagglutinin-L (PHA-L, Sigma). After 5 days, half of the volume of medium (75 μl/well) was replaced with culture medium containing 8% HS and 150 U/ml IL-2 without PHA-L. After 8 or 9 days, proliferating clones could be seen by microscopy and were maintained at a concentration of 106 T cells/ml. After 2 weeks, aliquots of each clone (3 × 104 cells) were tested for specificity using a TNF production assay. Specific clones were restimulated as described earlier. After 2 weeks, clones reached a resting state and were frozen in aliquots of 1.5 × 106 cells/ampoules in FCS containing 10% DMSO. In all experiments, clones were used at least 14 days after restimulation.
Synthetic peptides
Peptides were purchased from Eurogentec (Angers, France). Purity (>70%) was controlled by reversed-phase high-performance liquid chromatography. Peptides were lyophilized, dissolved in DMSO at 10 mg/ml and stored at −20°C.
HLA cDNAs
cDNAs coding for HLA-A*0201, A*0301, B*1302, Cw*0202 and Cw*0602 were obtained from T. Boon (LICR, Brussels, Belgium). The B*4002 coding cDNA has been cloned in our laboratory. cDNAs coding for cancer-germline and melanocytic differentiation antigens were provided by T. Boon. NA17-A NA88-A, NA-134-A and N-RAS (bearing 4 different mutations at position 61) cDNAs were cloned in our laboratory [13, 14, 23, 24]. Survivin, telomerase, Hsp70, Her-2 neu and EphA2 cDNAs were a gift from K. Kosmatopoulos (Unit 484 INSERM, Villejuif, France). MC1R cDNA was a gift from R. Kissling (Karolinska institute, Stockholm, Sweden).
Progressive deletions in tyrosinase cDNA
The plasmid was opened with Xba I and, after a protection of the 5′ protruding ends, the plasmid was digest by Xho I before digestion with exonuclease III. This treatment was performed with the Erase-a-base System (Promega, Madison, WI). After ligation, the plasmids were electroporated in TOP 10F′ Escherichia coli bacteria and selected with ampicillin (50 μg/ml). Clones were isolated, plasmid DNA was extracted from each clone and transfected into COS-7 cells together with the HLA-B*4002 cDNA.
Transient transfection of COS-7 cells and tumor cell lines
COS-7 cell transfection was performed by the DEAE-dextran-chloroquine method. Details of the procedure have been described previously [7, 8]. In brief, 16.5 × 103 COS-7 cells were co-transfected with 100 ng of plasmid coding for an HLA and 100 ng of plasmid coding for a melanoma-associated antigen. Transfected COS cells were then used to stimulate T cell populations. Tumor cells were transfected with 100 ng of HLA-B*4002 plasmid, with a lipofectamine reagent kit (Invitrogen), according to the manufacturer’s instructions.
RT-PCR for tyrosinase
Total RNA was extracted by the guanidinium–cesium chloride procedure. Reverse transcription was performed as previously described [2]. PCR amplification for tyrosinase was then performed on 50 ng of the cDNA with PCR buffer, MgCl2 1.5 mM (GIBCO-BRL), dNTP mix 0.8 mM, sense and antisense primers (5′GGATAGCGGATGCCTCTCAAAG3′ and 5′CCCAAGGAGCCATGACCAGAT3′) 1 μM, 0.1 U of Taq polymerase (GIBCO-BRL) in a final water volume of 25 μl. 25 PCR cycles were performed (1′ at 92°C, 1′ at 65°C and 1′ at 72°C). In total, 10 μl of PCR products were size fractionated on a 1% agarose gel. Expected length for tyrosinase cDNA was 383 bp.
Analysis of T cell responses by measurement of TNF release
Totally, 104 lymphocytes were stimulated in duplicate cultures by target cells : melanoma cells (3 × 104) pre-treated or not with IFN-γ (500 U/ml during 48 h), transfected COS-7 cells (48 h after transfection) or HLA-B*4002 B-EBV cell line, Adam [6], loaded with peptides. Culture supernatants were harvested 6 h later and tested for TNF content by a biological assay, as previously described [8]. mAb against HLA class I (clone W6.32), HLA-B/C (clone B1.23.2), HLA-A2 (clone BB7.2) added to cultures in some experiments, were produced in our laboratory from hybridomas obtained from the ATCC for W6.32 and BB7.2 antibodies and from F. Lemonier (Pasteur Institute, France) for B1.23.2 antibody.
Cytotoxicity assay
Target cells (Adam cell line or M88 melanoma line) were labeled with 100 μCi Na251CrO4 (Oris Industrie, Gif-sur-Yvette, France) for 1 h at 37°C before loading (or not) with synthetic peptides for 30 min. To enhance the cell surface expression of HLA molecules, M88 cells were pre-incubated (or not) for 48 h in medium containing 500 U/ml of IFN-γ (Tebu, Paris, France). Totally, 103 target cells were then mixed with 104 effectors T cells (in a final volume of 100 μl) and radioactivity was measured, 4 h later, with a β plate counter (EG&G Wallac, Evry, France).
Stimulation of B*4002 PBMC from melanoma patients with the tyrosinase-B*4002-specific peptide and detection of specific T cells
PBMC from M88 patient were obtained from a blood sample collected on July 2007, 13 years after TIL transfer. PBMC from four additional metastatic B*4002 melanoma patients were obtained from D. Schadendorf (patients MA324 and MA414) and from P. Coulie (Patients LUD01 and PAUMA). PBMC were seeded in 96-well plates (2 × 105 cells/well) in RPMI 1640 medium + 8% HS supplemented with 50 U/ml of IL-2 (Chiron) and 10 μM of the tyrosinase peptide 316–324 (Eurogentec, France). After a 14-day culture period, the specificity of stimulated microcultures was tested by intracellular TNF labeling, in response to the Adam cell line loaded with this tyrosinase peptide. In total, 105 stimulated PBL were incubated 5 h with 2 × 105 stimulator cells (peptide-pulsed Adam cells) in 200 μl of RPMI 1640 containing 10% FCS and 10 μg/ml brefeldin A (Sigma, St Louis MO, USA) [15]. Negative controls were performed with unpulsed Adam cells. Stimulated cells were then labeled with an anti-human CD8-FITC mAb (BD Pharmingen) and then fixed [15]. Lymphocytes were stained for 30 min at room temperature with the anti-human TNF-PE mAb (Pharmingen, San Diego, CA), at the concentration of 5 μg/ml in PBS containing 0.1% BSA and 0.1% saponin (Sigma). After staining, cells were resuspended in PBS and 104 events were analyzed on a FACScan flow cytometer using Cell Quest software (Beckton Dickinson, Grenoble, France).
Sequence analysis of TCR beta transcripts
RNA from 5 × 106 T cell clones was extracted with Trizol reagent (Invitrogen, Cergy Pontoise, France) according to the supplier’s instructions and dissolved in 15 μl of water. Reverse transcription, PCR amplifications and sequencing were performed as described [6]. We have followed throughout the article the TCR nomenclature from Rowen et al. [30].
Results
M88-stimulated PBMC recognize the M88 autologous cell line in a HLA-B/C restricted context
PBMC were derived from a patient who has remained relapse-free since 1994, after lymph node excision and adjuvant therapy with autologous TIL + IL-2 [10, 21]. CD8+ sorted T cells were stimulated for 3 weeks with the autologous irradiated melanoma cell line (A*0201, A*0301, B*1302, B*4002, Cw*0602, Cw*0202) and then tested for their reactivity against this line. As shown in Fig. 1, stimulated CD8+ PBMC recognized the autologous melanoma cell line in a class I-restricted manner. This recognition was not restricted by the HLA-A*0201 molecule, but was restricted by at least one of the four B/C alleles of the M88 cell line: recognition was dramatically reduced in the presence of a blocking anti HLA-B/C antibody.
TNF secretion by M88-stimulated CD8-sorted PBMC in response to the autologous M88 melanoma cell line. Stimulated PBMC were incubated for 6 h at 37°C with the M88 melanoma cell line at an E:T ratio of 1:3. As indicated, blocking antibodies directed against class I, A2 and B/C HLA were added at various concentrations during the stimulation. TNF-α secreted in the supernatant was measured by assessing its cytotoxicity for WEHI 164 clone 13
M88 T cell clones recognize a tyrosinase antigen in the HLA-B*4002 context
Stimulated CD8+ T cells were cloned by limiting dilution. Of the 50 T cell clones tested, 8 were highly reactive against the M88 cell line. Each of these T cell clones was then tested for reactivity against a panel of 46 antigens expressed in melanomas (Table 1) in the four HLA-B/C contexts corresponding to the M88 melanoma line. All eight T cell clones recognized the tyrosinase antigen in the HLA-B*4002 context. Four of the eight clones could be amplified, and their TCR Vβ chains were sequenced. The four T cell clones were identical as they expressed the same CDR3 on their Vβ7.6 TCR chains, and will be referred to hereafter as clone M88-74. Clone M88-74 gave a good TNF response in the presence of the autologous melanoma cell line and the B*4002/tyrosinase complex (Fig. 2a). It lysed the M88 autologous cell line poorly, though this cytotoxic response was greatly enhanced by culturing the M88 melanoma cell line for 48 h with IFN-γ. This is probably the result of increasing the expression of cell surface HLA class I molecules (Fig. 2b).
a TNF secretion by the M88-74 CTL clone in response to the autologous M88 melanoma cell line or to COS-7 cells transfected with cDNAs coding for the HLA-B*4002 molecule and the tyrosinase antigen. Fourty-eight hours after transfection, COS cells were co-cultured for 6 h at 37°C with the M88-74 T cell clone. Then TNF-α secreted in the supernatant was measured in a biological assay. b Lytic activity of the M88-74 T cell clone towards the M88 autologous melanoma line (circles) or K562 cells (squares). Effector and target cells were incubated at various E:C ratios. Before biological assay, the M88 cell line was either treated (black circles) or not (white circles) with 500 U/ml of IFN-γ
Identification of the tyrosinase cDNA region coding for the antigenic peptide
To identify the tyrosinase epitope recognized by the M88-74 T cell clone, multiple truncated tyrosinase variants were generated using the exonuclease method. The truncated tyrosinase cDNAs were then transfected into COS-7 cells along with the HLA-B*4002 cDNA. The reactivity of the M88-74 T cell clone towards COS-7 cells transfected either with the full-length cDNA, truncated cDNAs, or B*4002 alone was measured in a TNF release assay (Fig. 3). COS-7 cells transfected with one truncated cDNA (nucleotides 1-975) were recognized by the M88-74 T cell clone, whereas COS-7 cells transfected with a marginally shorter truncated cDNA (nucleotides 1–933) were not. This suggests that the region coding for the epitope is located between nucleotides 933 and 975 of the tyrosinase cDNA. This region codes for amino acids 304–325, a region that also contains two B*3501 restricted epitopes [3, 25].
a Identification of the cDNA regions coding for the B*4002 restricted tyrosinase epitope. Tyrosinase fragments were obtained by exonuclease digestion. Fragments recognized by the specific M88-74 CTL clone are indicated with a positive sign in the right column. The positions are indicated in base pairs. Fragments were co-transfected into COS-7 cells with the appropriate HLA cDNA. Fourty-eight hours after transfection, cells were incubated for 6 h at 37°C with the CTL clone and TNF secreted in the supernatant was measured by assessing its cytotoxicity for WEHI 164 clone 13. b TNF secretion by M88-74 CTL clone in response to COS-7 cells transfected with the B*4002 cDNA and the full-length cDNA coding for tyrosinase and with the tyrosinase fragments 1–933 and 1–975
Identification of antigenic tyrosinase peptides
Twelve overlapping decapeptides within the region 304–325 were synthesized, with a shift of a single amino acid from one peptide to the next. These peptides were screened by testing for their recognition by the M88–74 T cell clone when loaded onto a B*4002 B-EBV cell line. Figure 4 shows that the decapeptide 315–324 was recognized by our CTL clone. We then tested for the recognition of two nonapeptides derived from the decapeptide. We observed that deletion of the leucine at the C-terminal end (position 324) dramatically decreased the CTL clone response, whereas deletion of the serine at the N-terminus (position 315) enhanced the response of our CTL clone. The further deletion of the alanine at position 316 at the N-terminus of the nonapeptide 316–324, or the addition of a threonine at the C-terminus (position 325), dramatically reduced recognition by the CTL clone. In conclusion, the optimal peptide appeared to be the nonapeptide 316–324 (ADVEFCLSL), with a half-maximal lysis of 20 × 10−9 M.
Titration analysis of tyrosinase peptides recognized by the HLA-B*4002-restricted M88-74 CTL clone. Target cells were a B-EBV cell line (Adam) expressing the HLA-B*4002 molecule loaded with the indicated synthetic peptides at various concentrations. Lytic activity was measured, at an E:T ration of 10:1, by the classical 4-h 51Cr release assay at 37°C
Recognition of HLA-B*4002-positive melanoma cell lines expressing the tyrosinase antigen
To see if this B*4002 restricted tyrosinase epitope is naturally processed by melanoma cell lines, we tested for the recognition of melanoma cell lines expressing the tyrosinase antigen and the B*4002 molecule, either naturally or upon transient transfection with a B*4002 cDNA. The M44 and M88 B*4002-positive melanoma cell lines were spontaneously recognized by the tyrosinase-specific clone. Transfection of the B*4002 cDNA significantly enhanced the recognition of the M44 cell line, and induced recognition of cell lines that did not spontaneously expressed this HLA molecule (M199 and M134, Fig. 5). These results show that this new tyrosinase epitope can be processed spontaneously by several melanoma cell lines other than the autologous cell line.
TNF secretion by the M88-CTL clone in response to melanoma cell lines transfected (black bars) or not (white bars) with the B*4002 molecule. Tumor cells were transiently transfected with 100 ng of HLA-B*4002 plasmid, with a lipofectamine reagent kit (Invitrogen). The M88-74 CTL clone was then incubated for 6 h at 37°C with melanoma cell lines at an E:T ratio of 1:3
To determine whether melanoma cell line recognition by our CTL clone was strictly restricted by the B*4002 allele, we also tested the recognition of melanoma cell lines expressing tyrosinase and the closely related B*4001 allele. None of these melanoma cell lines was recognized by our CTL clone. This is consistent with the fact that COS-7 cells co-transfected with the tyrosinase cDNA and the B*4001 allele were not recognized by our CTL clone either (data not shown), showing that the recognition of this new epitope by the CTL clone was strictly restricted by the B*4002 allele.
Peptide stimulation of B*4002 patient PBMC with tyrosinase316–324 peptide
In order to address the question of the immunogenicity of the tyrosinase/B*4002 epitope, we cultured PBMC derived from the M88 patient (from a blood sample collected 13 years after TIL immunotherapy) and from four additional B*4002 melanoma patients with the tyrosinase 316–324 peptide for 14 days. Before stimulation, tyrosinase-specific lymphocytes in PBMC were almost undetectable (less than 0.05% of CD8+ lymphocytes). PBMC stimulation with the tyrosinase316–324 peptide induced the growth of specific CTL for three of the five patients tested (Fig. 6). The fraction of positive microcultures ranged from 5/96 (patients M88 and MA324) to 20/48 (patient MA414), with a percentage of specific CD8+ T cells up to 80% for patient MA414.
a % of TNF producing CD8+ T cells in individual microcultures in response to the Adam B*4002 cell line pulsed with 10 μM of the tyrosinase316–324 peptide. B*4002 melanoma patient-derived PBMC were previously cultured in 96-well plates (2 × 105 cells per well), in RPMI medium containing 10 μM of tyrosinase316–324 peptide and 50 U/ml of IL-2. After a 14 day-culture period, effector and target cells were incubated for 5 h, at a 1:2 ratio in the presence of Brefeldin A, stained with anti-TNF and anti-CD8 antibodies and analyzed by flow cytometry (104 T cells analyzed). Results show the percentages of TNF producing cells among CD8+ lymphocytes. b Examples of three positive wells from three different patients. Left panel: TNF response of CD8 T cells against unpulsed Adam cells, right panel: TNF response of CD8 T cells against peptide-pulsed Adam cells. The numbers between brackets correspond to the MFI of CD8 T cells producing TNF
Diversity of the specific repertoire in M88 patient PBMC
As stated earlier, stimulation of M88 PBMC with the tyrosinase/B*4002 peptide induced the growth of specific CTL in 5 of 96 microcultures (Fig. 6). The percentage of specific cells ranged from 1 to 11.6% among CD8+ T cells, as assessed by TNF production in response to peptide-loaded Adam cells. The microculture containing 11.6% of CD8+ specific cells was cloned and 8 tyrosinase-specific clones were obtained. Sequencing of the β chain of each of these T cell clones revealed the presence of two distinct CTL clones expressing Vβ7.9 and Vβ12 chains (Table 2). These clones are thus different from the M88.74 CTL clone. They were reactive against peptide pulsed Adam cells (Fig. 7a) and against the M88 autologous melanoma cell line, and to a lower extent, against the M44 melanoma cell line, especially when previously treated with IFN-γ (Fig. 7b).
a TNF produced by CTL clones in response to the Adam B*4002 cell line pulsed with 10 μM of the tyrosinase316–324 peptide. Black histograms, TNF produced in response to the unpulsed Adam cell line. Empty histograms, TNF produced in response to the peptide-pulsed Adam cell line. The numbers between brackets correspond to the MFI of CD8 T cells producing TNF. b TNF secretion by Tyrosinase-CTL clones in response to two melanoma cell lines pre-treated (black bars) or not (white bars) with IFN-γ. CTL clones were incubated for 6 h at 37°C with melanoma cell lines at an E:T ratio of 1:3
Discussion
We demonstrate here that a CTL response against the tyrosinase/B*4002 complex exists in the T cell repertoire of various B*4002 melanoma patients and that the derived CTL clones have enough avidity to react against B*4002 melanoma cell lines expressing tyrosinase.
Starting from PBMC isolated from a B*4002 patient who responded to TIL immunotherapy (M88), we isolated a CTL clone specific for this new tyrosinase epitope, following PBMC stimulation with the autologous melanoma cell line. This stimulation method allows us to define tumor epitopes spontaneously expressed by tumor cells. Interestingly, the two recently described class I tyrosinase epitopes (A26 and B38 restricted) were also identified starting from patient PBMC stimulated with the autologous melanoma line [22]. These data suggest that the response of autologous T cells to human melanoma is not systematically dominated by mutated neoantigens as stated in a previous report [22], in as far, as all the melanoma reactive CTL clones tested were specific for this shared melanoma antigen. Furthermore, in a second cloning experiment using M88-stimulated PBMC, we found another tyrosinase/B*4002-specific CTL clone and also a MAGE-A4/B*4002-specific CTL clone (data not shown), but we did not find any melanoma reactive CTL clones directed against private antigens.
The dominance of B*4002 restricted responses in CD8+ stimulated PBMC could be explained by a high expression of this HLA molecule on the surface of the M88 cell line. Indeed, this cell line has a relatively high surface expression of HLA B and C as compared with other melanoma cell lines (data not shown). This could favor the induction of HLA-B restricted responses. In support of this, we previously described a B*1302 restricted CTL response against the NA88-A antigen in tumor infiltrating lymphocytes from the M88 patient [4, 24]. Furthermore, we could obtain from M88 PBMC three distinct clonotypes specific for the B*4002/tyrosinase epitope and reactive against B*4002 melanoma cell lines. This also suggests a relative diversity in the T cell repertoire directed against this epitope in this particular patient.
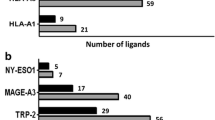
Tyrosinase is a melanocytic differentiation antigen expressed in about 90% of melanoma tumors [1]. Several tyrosinase peptides that are recognized by CD8+ or CD4+ T cells have already been identified (Fig. 8). They are presented by HLA class I molecules A1 [18, 19], A2 [29, 33, 37], A24 [16] A26 [22], B35 [3, 25], B38 [22] and B44 [5] and class II molecules DR4 [34] and DR15 [20]. The B*4002-restricted tyrosinase epitope described here bears a leucine at the C-terminus. Such a leucine is a consensus binding anchor residue for B*4002 [28]. Various viral epitopes presented in this HLA context have a glutamic acid in position 2 [26, 31, 32, 36], residue also proposed to be an anchor for B*4002. This was not the case for our new B*4002 epitope (ADVEFCLSL), suggesting that the aspartic acid in position 2 can also be an anchor residue for this HLA allele.
Finally, in order to assess the immunogenicity of this new epitope for potential use in immunotherapy, we performed peptide stimulations on PBMC deriving from four distinct HLA-B*4002 melanoma patients. We observed the expansion of tyrosinase-specific T cells in 2/4 patients, showing that such cells are present in several B*4002 patients. This suggests the epitope is potentially interesting for immunotherapy for B*4002 patients. However, the HLA-B*4002 allele (B61) is rather unfrequent in the caucasian population (3%) [9], and this will limit its application for immunotherapy. However, the new tyrosinase epitope shares its five first amino acids with the five last amino acids of two previously described B*3501 restricted epitopes (Fig. 8) [3, 25]. The existence of a panel of tyrosinase epitopes in the same region suggests that this region is fully accessible to the proteasome, and thus could represent an interesting region to target in multi-epitopic peptide-based vaccine protocols.
In conclusion, these results suggest that the characterization of long-term persistent anti-tumor responses in melanoma patients could be an excellent way to define relevant targets for future immunotherapy protocols, without any previous selection of a particular HLA context.
References
Barrow C, Browning J, MacGregor D, Davis ID, Sturrock S, Jungbluth AA, Cebon J (2006) Tumor antigen expression in melanoma varies according to antigen and stage. Clin Cancer Res 12:764–771
Benlalam H, Labarriere N, Linard B, Derre L, Diez E, Pandolfino MC, Bonneville M, Jotereau F (2001) Comprehensive analysis of the frequency of recognition of melanoma-associated antigen (MAA) by CD8 melanoma infiltrating lymphocytes (TIL): implications for immunotherapy. Eur J Immunol 31:2007–2015
Benlalam H, Linard B, Guilloux Y, Moreau-Aubry A, Derre L, Diez E, Dreno B, Jotereau F, Labarriere N (2003) Identification of five new HLA-B*3501-restricted epitopes derived from common melanoma-associated antigens, spontaneously recognized by tumor-infiltrating lymphocytes. J Immunol 171:6283–6289
Benlalam H, Vignard V, Khammari A, Bonnin A, Godet Y, Pandolfino MC, Jotereau F, Dreno B, Labarriere N (2007) Infusion of Melan-A/Mart-1 specific tumor-infiltrating lymphocytes enhanced relapse-free survival of melanoma patients. Cancer Immunol Immunother 56:515–526
Brichard VG, Herman J, Van Pel A, Wildmann C, Gaugler B, Wolfel T, Boon T, Lethe B (1996) A tyrosinase nonapeptide presented by HLA-B44 is recognized on a human melanoma by autologous cytolytic T lymphocytes. Eur J Immunol 26:224–230
Couedel C, Bodinier M, Peyrat MA, Bonneville M, Davodeau F, Lang F (1999) Selection and long-term persistence of reactive CTL clones during an EBV chronic response are determined by avidity, CD8 variable contribution compensating for differences in TCR affinities. J Immunol 162:6351–6358
Coulie PG, Brichard V, Van Pel A, Wolfel T, Schneider J, Traversari C, Mattei S, De Plaen E, Lurquin C, Szikora JP, Renauld JC, Boon T (1994) A new gene coding for a differentiation antigen recognized by autologous cytolytic T lymphocytes on HLA-A2 melanomas. J Exp Med 180:35–42
De Plaen E, Lurquin C, Lethe B, van der Bruggen P, Brichard V, Renauld JC, Coulie P, Van Pel A, Boon T (1997) Identification of genes coding for tumor antigens recognized by cytolytic T lymphocytes. Methods 12:125–142
Domena JD, Johnston-Dow L, Parham P (1992) The B*4002 allele encodes the B61 antigen: B40* is identical to B61. Tissue Antigens 40:254–256
Dreno B, Nguyen JM, Khammari A, Pandolfino MC, Tessier MH, Bercegeay S, Cassidanius A, Lemarre P, Billaudel S, Labarriere N, Jotereau F (2002) Randomized trial of adoptive transfer of melanoma tumor-infiltrating lymphocytes as adjuvant therapy for stage III melanoma. Cancer Immunol Immunother 51:539–546
Gaugler B, Van den Eynde B, van der Bruggen P, Romero P, Gaforio JJ, De Plaen E, Lethe B, Brasseur F, Boon T (1994) Human gene MAGE-3 codes for an antigen recognized on a melanoma by autologous cytolytic T lymphocytes. J Exp Med 179:921–930
Gervois N, Labarriere N, Le Guiner S, Pandolfino MC, Fonteneau JF, Guilloux Y, Diez E, Dreno B, Jotereau F (2000) High avidity melanoma-reactive cytotoxic T lymphocytes are efficiently induced from peripheral blood lymphocytes on stimulation by peptide-pulsed melanoma cells. Clin Cancer Res 6:1459–1467
Godefroy E, Moreau-Aubry A, Diez E, Dreno B, Jotereau F, Guilloux Y (2005) Alpha v beta3-dependent cross-presentation of matrix metalloproteinase-2 by melanoma cells gives rise to a new tumor antigen. J Exp Med 202:61–72
Guilloux Y, Lucas S, Brichard VG, Van Pel A, Viret C, De Plaen E, Brasseur F, Lethe B, Jotereau F, Boon T (1996) A peptide recognized by human cytolytic T lymphocytes on HLA-A2 melanomas is encoded by an intron sequence of the N-acetylglucosaminyltransferase V gene. J Exp Med 183:1173–1183
Jung T, Schauer U, Heusser C, Neumann C, Rieger C (1993) Detection of intracellular cytokines by flow cytometry. J Immunol Methods 159:197–207
Kang X, Kawakami Y, el-Gamil M, Wang R, Sakaguchi K, Yannelli JR, Appella E, Rosenberg SA, Robbins PF (1995) Identification of a tyrosinase epitope recognized by HLA-A24-restricted, tumor-infiltrating lymphocytes. J Immunol 155:1343–1348
Kawakami Y, Eliyahu S, Sakaguchi K, Robbins PF, Rivoltini L, Yannelli JR, Appella E, Rosenberg SA (1994) Identification of the immunodominant peptides of the MART-1 human melanoma antigen recognized by the majority of HLA-A2-restricted tumor infiltrating lymphocytes. J Exp Med 180:347–352
Kawakami Y, Robbins PF, Wang X, Tupesis JP, Parkhurst MR, Kang X, Sakaguchi K, Appella E, Rosenberg SA (1998) Identification of new melanoma epitopes on melanosomal proteins recognized by tumor infiltrating T lymphocytes restricted by HLA-A1, -A2, and -A3 alleles. J Immunol 161:6985–6992
Kittlesen DJ, Thompson LW, Gulden PH, Skipper JC, Colella TA, Shabanowitz J, Hunt DF, Engelhard VH, Slingluff CL Jr (1998) Human melanoma patients recognize an HLA-A1-restricted CTL epitope from tyrosinase containing two cysteine residues: implications for tumor vaccine development. J Immunol 160:2099–2106
Kobayashi H, Kokubo T, Takahashi M, Sato K, Miyokawa N, Kimura S, Kinouchi R, Katagiri M (1998) Tyrosinase epitope recognized by an HLA-DR-restricted T-cell line from a Vogt-Koyanagi-Harada disease patient. Immunogenetics 47:398–403
Labarriere N, Pandolfino MC, Gervois N, Khammari A, Tessier MH, Dreno B, Jotereau F (2002) Therapeutic efficacy of melanoma-reactive TIL injected in stage III melanoma patients. Cancer Immunol Immunother 51:532–538
Lennerz V, Fatho M, Gentilini C, Frye RA, Lifke A, Ferel D, Wolfel C, Huber C, Wolfel T (2005) The response of autologous T cells to a human melanoma is dominated by mutated neoantigens. Proc Natl Acad Sci USA 102:16013–16018
Linard B, Bezieau S, Benlalam H, Labarriere N, Guilloux Y, Diez E, Jotereau F (2002) A ras-mutated peptide targeted by CTL infiltrating a human melanoma lesion. J Immunol 168:4802–4808
Moreau-Aubry A, Le Guiner S, Labarriere N, Gesnel MC, Jotereau F, Breathnach R (2000) A processed pseudogene codes for a new antigen recognized by a CD8(+) T cell clone on melanoma. J Exp Med 191:1617–1624
Morel S, Ooms A, Van Pel A, Wolfel T, Brichard VG, van der Bruggen P, Van den Eynde BJ, Degiovanni G (1999) A tyrosinase peptide presented by HLA-B35 is recognized on a human melanoma by autologous cytotoxic T lymphocytes. Int J Cancer 83:755–759
Oseroff C, Kos F, Bui HH, Peters B, Pasquetto V, Glenn J, Palmore T, Sidney J, Tscharke DC, Bennink JR, Southwood S, Grey HM, Yewdell JW, Sette A (2005) HLA class I-restricted responses to vaccinia recognize a broad array of proteins mainly involved in virulence and viral gene regulation. Proc Natl Acad Sci USA 102:13980–13985
Pandolfino MC, Labarriere N, Tessier MH, Cassidanius A, Bercegeay S, Lemarre P, Dehaut F, Dreno B, Jotereau F (2001) High-scale expansion of melanoma-reactive TIL by a polyclonal stimulus: predictability and relation with disease advancement. Cancer Immunol Immunother 50:134–140
Rammensee HG, Friede T, Stevanoviic S (1995) MHC ligands and peptide motifs: first listing. Immunogenetics 41:178–228
Riley JP, Rosenberg SA, Parkhurst MR (2001) Identification of a new shared HLA-A2.1 restricted epitope from the melanoma antigen tyrosinase. J Immunother 24:212–220
Rowen L, Koop BF, Hood L (1996) The complete 685-kilobase DNA sequence of the human beta T cell receptor locus. Science 272:1755–1762
Scotet E, David-Ameline J, Peyrat MA, Moreau-Aubry A, Pinczon D, Lim A, Even J, Semana G, Berthelot JM, Breathnach R, Bonneville M, Houssaint E (1996) T cell response to Epstein–Barr virus transactivators in chronic rheumatoid arthritis. J Exp Med 184:1791–1800
Sidney J, Southwood S, Pasquetto V, Sette A (2003) Simultaneous prediction of binding capacity for multiple molecules of the HLA B44 supertype. J Immunol 171:5964–5974
Skipper JC, Hendrickson RC, Gulden PH, Brichard V, Van Pel A, Chen Y, Shabanowitz J, Wolfel T, Slingluff CL Jr, Boon T, Hunt DF, Engelhard VH (1996) An HLA-A2-restricted tyrosinase antigen on melanoma cells results from posttranslational modification and suggests a novel pathway for processing of membrane proteins. J Exp Med 183:527–534
Topalian SL, Gonzales MI, Parkhurst M, Li YF, Southwood S, Sette A, Rosenberg SA, Robbins PF (1996) Melanoma-specific CD4 + T cells recognize nonmutated HLA-DR-restricted tyrosinase epitopes. J Exp Med 183:1965–1971
Traversari C, van der Bruggen P, Luescher IF, Lurquin C, Chomez P, Van Pel A, De Plaen E, Amar-Costesec A, Boon T (1992) A nonapeptide encoded by human gene MAGE-1 is recognized on HLA-A1 by cytolytic T lymphocytes directed against tumor antigen MZ2-E. J Exp Med 176:1453–1457
Trivedi D, Williams RY, O’Reilly RJ, Koehne G (2005) Generation of CMV-specific T lymphocytes using protein-spanning pools of pp65-derived overlapping pentadecapeptides for adoptive immunotherapy. Blood 105:2793–2801
Wolfel T, Van Pel A, Brichard V, Schneider J, Seliger B, Meyer zum Buschenfelde KH, Boon T (1994) Two tyrosinase nonapeptides recognized on HLA-A2 melanomas by autologous cytolytic T lymphocytes. Eur J Immunol 24:759–764
Acknowledgments
This work was supported by grants from the “Ligue Nationale contre le Cancer and Comite 44”, the “INSERM” and by a grant from ENACT network number 503306. We acknowledge Profs. F. Lang and R. Breathnach for carefully reading the manuscript and Prof. P. Coulie for sending us melanoma patient PBMC.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Godet, Y., Bonnin, A., Guilloux, Y. et al. A new tyrosinase epitope recognized in the HLA-B*4002 context by CTL from melanoma patients. Cancer Immunol Immunother 58, 271–280 (2009). https://doi.org/10.1007/s00262-008-0551-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00262-008-0551-0