Abstract
Genetic transfer of T-cell receptor (TCR) chains provides a means of transferring tumor antigen specificity onto an alternate T-cell population. To determine which tumor reactive TCRs are best suitable for such adoptive transfer, careful evaluation of the resulting TCR modified populations need to be performed. We have previously cloned, and expressed TCRs from melanoma, EBV, HCV, and HPV reactive T-cell clones and found that several routine indicators of T-cell function do not always predict the relative strength of a TCR. Using a combination of tetramer binding assays and antigen recognition assays, we identified TCRs that fall into three classes. One class of TCR did not bind tetramers yet resulted in cells with high avidity for antigen. A second TCR class bound tetramer but did not secrete cytokines in response to antigen. Finally, the third class of TCRs bound tetramer and reacted to antigen as would be expected. We conclude that tetramer binding is not always a good indicator of the function of a cloned TCR or the avidity of a TCR gene modified T cell. Furthermore, our data indicate that the use of tetramer binding alone to identify antigen reactive TCRs may result in the exclusion of TCRs that may be highly reactive or cross reactive to the relevant tumor antigen.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Although considered to be highly specific, the binding between any given T-cell receptor (TCR) and MHC-peptide complex is characterized by a low affinity and fast off rates [7, 31]. It is argued that during T-cell recognition, these low affinity and fast off rates are necessary to enable serial contacts of each TCR molecule with multiple peptide ligands [32]. This unfavorable interaction, however, initially limited the ability to directly stain T cells by fluorescent labeled monomer MHC-peptide molecules [1]. Therefore, the concept of tetramerization of MHC class I molecules can be seen as a major innovation in cellular immunology [1]. The low affinity of the single MHC unit is thus compensated for by the higher affinity gained by cooperative binding [20]. The development of MHC-peptide tetramers has introduced the possibility of identifying and isolating T lymphocytes carrying an antigen-specific receptor, independently of their effector function and has proven to be a powerful diagnostic tool in human clinical studies for monitoring CD8+ T-cell responses in viral and tumor immunity [17, 18] and to probe TCR–MHC interactions [15].
Selection events during T-cell development favor a TCR repertoire that excludes high affinity TCRs [14, 30]. Therefore, cytotoxic T lymphocytes (CTLs) cloned from patient blood as a source of TCRs can be expected to contain only naturally selected, low-affinity TCRs, particularly in the case of a self-antigen. These TCRs may function efficiently in the context of the T cell, however, the inherent low affinity of αβ pairs may limit their utility when the TCR is transferred to alternate effectors. Furthermore, several groups have reported that following viral infection, the number of anti-viral T cells evaluated by tetramers largely exceeds the anti-viral T cells that have cytolytic activity or secrete cytokines [2, 11, 12, 36]. This led to the notion that not all tetramer positive cells initiate an effective immune response. Furthermore, it raises the possibility that tetramer-negative cells that are still cross-reactive to tumor antigens exist [9]. These are critical issues to be considered when choosing TCRs that may be appropriate for adoptive transfer.
The rationale of adoptive T-cell therapy is based on the attempt to circumvent a cancer patient’s impaired cellular immune response by isolating the anergic, potentially tumor reactive T cells from the tolerizing host environment and activating them ex vivo. Following the expansion of tumor reactive T cells in vitro, large numbers of T cells can be adoptively transferred to the patient. Unfortunately, in the case of most cancer patients, adoptive immunotherapy is unlikely to be successful due to the exceedingly low levels of circulating antigen-specific T cells. Furthermore, immunotherapy for solid tumors is much more difficult due to the fact that specific tumor-associated antigens (TAA) have been defined in only a minority of tumors. For other cancers, several TAAs have been identified and can be used as targets for immunotherapy. Therefore, adoptive transfer strategies based on TCRs specific for epitopes from these and other poorly immunogenic tumors could overcome the difficulty of isolating and propagating tumor specific T cells from advanced cancer patients. It has been shown that the retroviral transfer of genes encoding tumor specific TCRs into peripheral blood lymphocytes (PBLs) could enhance antigen-specific immunity by increasing the frequency of tumor-specific T cells [4, 6, 16, 29].
To better understand the properties of a TCR that are important for adoptive transfer studies, we have cloned and transferred TCRs from tumor and virus reactive T-cell clones. Here we report the analysis of three HLA-A2 restricted TCRs, which recognize MART-127–35 (TIL 5 TCR), tyrosinase368–376 (TIL 1383I TCR), and HPV16 E711–20 (D4 TCR) [5, 25, 35]. Using retroviral vectors encoding the α and β chains of these TCRs, we transferred them into alternate T cells. Based on the pattern of tetramer staining and effector function, we found that tetramer staining did not always correlate with TCR function. Therefore, tetramer binding alone cannot be used to predict a TCR with adequate signaling properties for TCR gene transfer.
Materials and methods
Cells
Cell lines were purchased from American Type Culture Collection (Manassas, VA, USA) unless otherwise noted. Jurkat cells are a CD8 negative human T-cell line, SupT1 cells are a CD4/CD8 double positive human T-cell line and T2 cells are a human HLA-A*0201 B/T lymphoma. All of these cells were maintained in human complete medium (hCM), which consisted of RPMI 1640 (Mediatech, Herndon, VA, USA) supplemented with 10% FCS (Invitrogen Life Technologies, Gaithersburg, MD, USA) and 100 U/ml penicillin (Mediatech), 100 μg/ml streptomycin (Mediatech), and 2.92 mg/ml l-glutamine (Mediatech). 293 GP retroviral producer cells were maintained in DMEM (Mediatech) supplemented as stated earleir. The D4 T-cell clone was isolated by tetramer sorting from a cervical cancer patient at the University of Wales, Cardiff, UK [35]. The original TIL5 and TIL 1383I lines were established from surgical specimens obtained from melanoma patients undergoing immunotherapy in the Surgical Branch, National Cancer Institute. T-cell clones were grown in X-Vivo 15 (BioWhittaker, Walkersville, MD, USA) medium supplemented with 10% heat inactivated human AB serum (Valley Biomedical, Winchester, VA, USA) 100 U/ml penicillin (Invitrogen), 100 μg/ml streptomycin (Invitrogen), 2.92 mg/ml l-glutamine (Invitrogen), and 6,000 IU/ml recombinant human IL-2 (Chiron). Since the parental TIL5 clone is no longer available for analysis, another MART-127–35 reactive clone was used as a positive control for HLA-A*0201 MART-127–35 tetramer staining.
Reagents and antibodies
The HLA-A*0201 binding peptides, HPV16 E711–20 (YMLDLQPETT), MART-1 27–35 (AAGIGILTV), and tyrosinase368–376 (YMDGTMSQV) were synthesized at the University of Chicago by Dr. Stephen Meredith and purified by reverse phase HPLC. Purity was assessed by analytical HPLC and was determined to be >99%. Anti-human CD4-FITC, anti-human CD8-FITC, anti-human CD3-PE, anti-human CD3-FITC, anti-human HLA-A2-FITC, and anti-human αβ TCR-PE were purchased from BD Biosciences (San Diego, CA, USA). HLA-A*0201 tetramers labeled with phycoerythrin and containing the MART-127–35, tyrosinase368–376 , and HPV16 E711–20 peptide were obtained from Beckman Coulter (Fullerton, CA, USA).
TCRα/β chain identification, cloning and analysis
The identification and cloning of TCR α and β chains from the TIL5 and TIL 1383I have been described by Clay et al. and Roszkowski et al. [4, 26]. The TCR from the CTL clone D4 was identified by RT-PCR using a panel of TCR α chain V region (AV) and TCR β-chain V region (BV) subfamily specific primers [4]. Total RNA was isolated from 5×106 T cells using RNAeasy kits (Qiagen, Valencia, CA, USA) according to the manufacturer’s instructions. First-strand c-DNA was prepared from 1 μg of total RNA using Superscript II reverse transcriptase (Invitrogen) and oligo (dT)12–18 (Invitrogen). Ten nanograms of cDNA was PCR amplified in a 50 μl reaction consisting of one time PCR buffer (Invitrogen), 1.5 mM MgCl2 (Invitrogen), 200 nM dNTP (Invitrogen), 400 nM TCR AV or BV subfamily specific forward primer (Invitrogen), 400 nM TCR α-chain constant region specific or β-chain constant region specific reverse primer, and 2.5 U Taq DNA polymerase (Continental Lab Products, San Diego, CA, USA). PCR amplification was performed using a MJ Research thermocycler (Watertown, MA, USA) under the following conditions: 5 min at 92°C (one cycle) followed by 30 s at 92°C, 30 s at 58°C, and 1 min at 72°C (35 cycles), followed by 5 min at 72°C (one cycle). The resulting PCR products were separated on 1% agarose gels containing ethidium bromide (Continental Lab Products) and were visualized under UV light. The presence of a band of the appropriate size in a reaction indicated the presence of that TCR AV or BV subfamily in the T-cell clones. For identification of the D4 TCR α and β chains, 5′RACE (Rapid Amplification of C-tailed Ends) was performed according to the manufacturer’s instructions (Invitrogen) using RNA isolated from HLA-A*0201 HPV16 E711–20 tetramer sorted cells. Once the identity of the α and β chain was confirmed, forward cloning primers were designed from the genomic sequences of the α and β genes, which contained elements of the 5′ untranslated region, the ATG start codon, and SalI and XhoI sites, respectively for subsequent cloning. Reverse cloning primers were designed from the genomic sequence of the human α constant and β constant regions, which contained the termination codon and SalI and XhoI sites, respectively, for subsequent subcloning. The products of each PCR reaction were ligated into the pCR 2.1 TA cloning vector (Invitrogen) and transformed into Escherichia coli DH5α competent cells (Life Technologies, MA, USA). Bacterial clones were screened by PCR using the specific α and β primers to identify clones containing inserts of the predicted size. DNA was isolated from those clones and their inserts were sequenced by cycle sequencing using BigDye Terminator Cycle Sequencing kits (Applied Biosystems, Foster City CA, USA) and analyzed on an ABI Prism 310 Genetic Analyzer (Applied Biosystems) to ensure there were no PCR errors in the sequence.
RNA was isolated from transduced Jurkat cells using the Qiagen RNeasy Mini Kit (Qiagen) for total RNA isolation. RT-PCR was performed on freshly isolated RNA using the SuperScript First-Strand Synthesis System for RT-PCR (Invitrogen) according to the manufacturer’s instructions. The primers used to clone the three full-length TCR α and β chains were used to amplify the cDNA. Untransduced Jurkat cells were used as a positive control and constant region primers was also used.
Retroviral vector construction
The SAMEN CMV/SRα retroviral vector was designed specifically for introducing TCR genes into alternate T cells. The 5′ long terminal repeat (LTR) in the SAMEN SRα backbone was replaced with a hybrid LTR consisting of the human CMV enhancer and promoter fused with the Moloney murine leukemia virus 5′LTR. This modification permits production of retroviral supernatants by transiently transfecting 293 GP cells [26]. Other key elements of the SAMEN CMV/SRα vector include an internal SRα promoter to permit the expression of multiple genes, unique SalI and XhoI restriction sites for ease of inserting TCR chains, and an internal ribosome entry site/neor cassette for G418 selection. A rapid ligation strategy was used to subclone the D4 TCR α and β chain genes into SAMEN CMV SRα. TCR β-chain genes were excised from pCR2.1 with XhoI and ligated into the SalI restriction site in SAMEN CMV/SRα using a mixture of T4 DNA ligase and and SalI restriction endonuclease. The resulting SalI/XhoI hybrid sites are resistant to digestion by SalI and XhoI. Ligation reactions were re-digested with SalI, resulting in linearization of plasmids not containing the TCR β-chain insert, allowing for enrichment of recombinant clones. The ligation reactions were cloned into E. coli DH5α competent cells (Invitrogen), and bacterial clones were screened by PCR using primers that flanked the cloning sites in SAMEN CMV/SRα. The DNA sequence of clones containing inserts was determined using BigDye Terminator Cycle Sequencing kits and analyzed using an ABI Prism 310 Genetic Analyzer (Perkin Elmer/ABI, Foster City, CA, USA) to ensure that TCR gene inserts were in the proper orientation. This method was repeated to insert the TCR α-chain into the retroviral vector by ligating a SalI fragment containing the α-chain into the XhoI site of SAMEN CMV/SRα to create the D4 TCR retroviruses.
Generation of retroviral supernatants
100 mm tissue culture plates were coated with 0.02% type B Bovine skin gelatin (Sigma, MO, USA) in HBSS for 15 min at room temperature. 293 GP cells were seeded onto coated plates at sufficient density to provide 60% confluency after 24 h (approximately 3×106 cells). Monolayers were rinsed three times with PBS and transiently co-transfected using Lipofectamine and PLUS reagents (Life Technologies), 3 μg of the retroviral plasmid containing the TCR genes, and 3 μg of vesicular stomatitis virus envelope gene in 6.0 ml of serum-free DMEM. Following a 3-h incubation, 10 ml of DMEM with 20% FCS was added to the plates, and cells were incubated at 37°C. Medium was discarded after 24 h and replaced with 10 ml RPMI 1640 medium supplemented with 10% FCS. Retroviral supernatants were collected after 24 h, replaced with medium, and collected again after an additional 24 h. Retroviral supernatants were either used immediately or frozen at −70°C for later use.
Retroviral transduction
Fresh retroviral supernatants were supplemented with 8 μg/ml polybrene (Sigma) and filter sterilized. Jurkat and SupT1 cells were suspended at 1×106 cells/ml in retroviral supernatant. An amount of 1 ml of cells/supernatant was added to each well of 24-well tissue culture plate, and the plates were centrifuged at 1,000g for 90 min at 32°C. Plates were returned to the incubator and after 4 h, 1 ml of fresh RPMI was added to each well. Transduced cells were incubated overnight, and this procedure was repeated the next day with fresh supernatant as described earlier.
Antigen recognition assay
Retrovirally transduced Jurkat cells were tested for reactivity to tumor antigens in cytokine release assays. T2 cells pre-incubated for 2 h with 10 μg/ml peptides were washed two times with PBS and then added to effector cells at a 1:1 ratio in a total volume of 200 μl of RPMI medium supplemented with 10% FBS per well of a 96-well, U-bottom tissue culture plate. The actual numbers were 2×105 cell per well. Co-cultures were incubated at 37°C in a humidified CO2 incubator for 24 h. Supernatants were harvested, and the amount of human IL-2 (R&D Systems, Minneapolis, MN, USA) or TNF-α (Pierce, Rockford, IL, USA) released by Jurkat cells was measured by ELISA.
Results
Retroviral transduction of alternate T cells
We have previously shown that the TIL 5, TIL 1383I, and D4 clones all recognize their appropriate HLA-A2 restricted epitopes (Table 1) [5, 25, 35]. Furthermore, we have reported that transferring the TIL5, TIL1383I as well as other TCR receptors to Jurkat cells and PBL-derived T cells, results in antigen reactivity against their corresponding peptide epitopes and tumor cells [4, 5, 24, 26]. However, we have observed that not all transferred TCRs retain similar reactivity for their peptide antigen as their parental clone when transferred to alternate cells. We therefore, set out to further analyze which TCRs will be the best candidate for adoptive transfer studies in patients by assessing their abilities to bind tetramer and to retain the antigen reactivity seen in the parental clone.
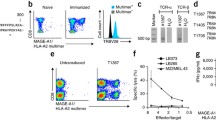
To do this, SupT1 and Jurkat T cell lines were transduced to express the TIL 5, TIL1383I, and D4 TCRs. SupT1 is a CD4/CD8 double positive T-cell line that does not express a native TCR and thus, due to the lack of competing TCR genes, is easily transduced [23]. Since SupT1 cells do not contain a native TCR, they also do not express the CD3 complex. Therefore, TCR expression can be measured by the upregulation of the TCRαβ chains as well as the CD3 complex. TCR transduced cells were stained with anti-CD3, and anti-TCRαβ and were determined to be approximately 99% TCR positive (Fig. 1). To determine if the transferred TCR could bind their peptide/MHC tetramers, the transduced SupT1 cells were stained with their appropriate tetramers and negative controls. The binding of each of the tetramers used in these studies was confirmed using T-cell clones or TIL cultures to ensure that they had been properly assembled (data not shown). While both the D4 and TIL 1383I TCR transduced cells stained with tetramers, the TIL5 TCR transduced cells did not stain with the HLA-A*0201 MART-127–35 tetramer (Fig. 2). Furthermore, although almost 100% of the TIL 1383I and D4 TCR transduced SupT1 cells were TCRαβ and CD3 positive, only 50% were tetramer positive. This is possibly due to β homodimers on the surface of transduced cells or lack of TCR clusters required for optimal tetramer staining. Therefore, two of these three TCRs are capable of binding peptide/MHC tetramers.
Cell surface TCR expression of TCR transduced SupT1 cells. Untransduced SupT1 cells (empty) and SupT1 cells transduced with the TIL5, TIL 1383I, or D4 TCR (shaded) were stained with anti-CD3 FITC (a) or anti-TCRαβ (b) and fluorescence was quantified by flow cytometry. Each histogram represents the relative log fluorescence of 104 viable cells
Peptide specific tetramer staining of TCR transduced SupT1 cells. Untransduced SupT1 cells and SupT1 cells transduced with the TIL5, TIL 1383I, or D4 TCR were stained with their corresponding PE-tetramers or negative control tetramers and fluorescence was quantified by flow cytometry. The MART-1 tetramer was used as a negative control for TIL1383I and D4 and the HPV1611–20 tetramer was used as a negative control for TIL5. Each plot represents the relative log fluorescence of 104 viable cells
Antigen recognition by TCR transduced Jurkat cells
SupT1 cells have proven to be useful for confirming the expression of our transduced TCR. However, SupT1 cells do not signal upon antigen stimulation. In order to determine if there was a correlation between the ability of a TCR to bind tetramer and its ability to mediate antigen recognition, we transduced Jurkat cells with each of our TCRs. Jurkat cells are a human T lymphoma cell line that does not express CD8 but does contain a native TCR and secretes IL-2 and TNFα in response to antigen recognition.
Transferring our TAA reactive TCRs to Jurkat cells should allow for recognition of their appropriate targets. Therefore, we co-cultured TIL 5, TIL 1383I, and D4 TCR transduced Jurkats with T2 cells alone or T2 cells loaded with either MART-127–35, Tyrosinase368–376, or HPV16 E711–20. Responder and stimulator cell co-cultures were incubated overnight and the next day supernatants were removed and the amount of IL-2 or TNFα released was measured by ELISA. Jurkat cells transduced with the TIL5 and TIL 1383I TCR secreted both TNFα and IL-2 in response to their relevant peptides while the D4 TCR transduced Jurkat cells did not respond to the HPV16 E711–20 peptide loaded T2 cells (Fig. 3). These results were inconsistent with the tetramer binding data. Surprisingly, the D4 TCR retroviral construct was capable of binding to tetramers when transferred to SupT1 but did not signal against peptide antigen, while the TIL5 transduced cells were unable to bind tetramer but were strongly reactive against peptide loaded cells. Furthermore, the TIL5 TCR proved to be a very high avidity TCR by recognizing nanomolar quantities of peptide (Fig. 4). Therefore, the relative avidity of each TCR gene-modified cells was not proportional to the TCRs ability to bind tetramers.
Relative sensitivity of TIL5, TIL 1383I, and D4 TCR transduced Jurkat cells for peptide loaded T2 cells. The antigen reactivity of transduced cells was measured in TNF-α and IL-2 release assays. The amount of cytokine released was measured by ELISA. Each column represents the average of triplicate wells and is a representative of three replicate experiments. PMA and ionomycin were used as positive controls and showed that all TCR transduced cells released cytokines upon non-specific stimulation (data not shown)
Relative avidity of TCR transduced Jurkat cells for peptide loaded stimulators. The relative avidity of TIL5, TIL 1383I, and D4 TCR transduced Jurkat cells was measured in IL-2 release assays using T2 cells loaded with four different concentrations of peptides: MART-127–35, Tyrosinase368–376, and HPV16 E711–20. The amount of IL-2 released was then measured by ELISA. Data points represent IL-2 release against the transduced cells’ relevant peptide. There was no IL-2 release against irrelevant control peptides. Each point represents the average of triplicate wells and is representative of three independent experiments
Since Jurkat cells have a native TCR, we cannot stain the cells with TCRαβ or CD3 to measure the expression of the transferred TCRs. Furthermore, since the Jurkat cells are CD8 negative, we cannot efficiently measure the transferred TCR expression with tetramers [8]. Therefore, we performed RT-PCR to ensure that both α and β chain transgenes were expressed in the transduced Jurkat cells. RNA was isolated from untransduced Jurkat cells and from the Jurkats transduced with the D4 TCR, TIL5 TCR, or TIL 1383I TCR retroviral vectors. RT-PCR was performed using TCR α and β chain specific primers and constant region primers as positive controls. Results show that the transduced Jurkat cells expressed the transferred TCR α and β chain and that these chains were absent in untransduced Jurkats cells (Fig. 5).
Gel Electrophoresis profiles of RT-PCR products amplified with TCR α- and β-chain-specific primers. RNA was isolated from untransduced Jurkats and Jurkats transduced with either the TIL5, TIL1383I, or D4 TCRs. cDNA was made with and without reverse transcriptase and PCR was performed using Vα and Vβ cloning primers specific for the TCR as well as TCR constant region primers as positive controls
Discussion
It has become clear that the use of tetramers to selectively sort high-affinity tumor-specific CTLs from the peripheral blood of tumor patients has opened new strategies for the therapy of cancers. However, our data suggest that not all antigen-reactive TCRs will efficiently bind to tetramer and that this technology may result in the loss of potentially reactive T cells in patient PBLs. Furthermore, not all TCRs that bind tetramer are able to signal against their peptide antigen when transferred into new T cells and are, therefore, not suitable for adoptive transfer.
In these studies, we have utilized an alternative strategy to target tumor antigens by genetically modifying T cells to produce anti-tumor reactivity. Genes encoding for TCR α and β chains were cloned from tumor reactive CTL clones and transferred into human T cells. These transduced T cells should then redirect their specificity toward the antigen recognized by the transferred TCR and should display the same or similar avidity and affinity for peptide antigen as its parental clone. We have effectively transferred the TCRs into T-cell lines, as confirmed by cell surface staining for α and β genes and the CD3 complex. The D4 TCR and TIL1383I TCR transduced SupT1 cells also stained with HLA-A*0201 restricted tetramers. However, the TIL5 TCR transduced cells did not bind tetramer despite the clear indication of a TCR on the transduced cells surface. We would expect that since the TIL5 TCR did not bind tetramer on the SupT1 cells, it would not recognize its peptide antigen when transferred into Jurkat cells. However, this was not the case. The TIL 5 receptors proved to be highly reactive against the MART-1 peptide in cytokine ELISAs and had an extremely high avidity for its peptide as well. In direct contrast, the D4 TCR, which repeatedly bound tetramer when transferred to SupT 1 cells, never signaled against its peptide antigen.
It has been suggested that tumor cell recognition by a T cell is a property of the TCR and not the cell itself [26]. A TCR with high affinity for its ligand is predicted to increase the sensitivity or avidity of the T cell, thus requiring lower levels of antigen for efficient target cell recognition [26]. Due to its inability to recognize its target, we have postulated that the D4 TCR has a low affinity for the HPV16 E711–20 antigen. Thus, transduced T cells that received the D4 TCR had a very low avidity for the HPV16 E711–20 antigen and did not secrete cytokines against it. While the D4 parental clone was able to recognize HPV16 E711–20 antigen, it may be difficult to use its low affinity TCR to engineer T cells with similar avidity as the naturally occurring parental T-cell clone. Therefore, the affinity of the D4 TCR limited the practical utility of the TCR-transduced cells. There is also a possibility that the D4 TCR that was transferred into human T cells did not completely preserve the peptide fine specificities of its parental TCR. The D4 TCR genes, in particular their CDR3 regions may not have folded properly when they were synthesized in the endoplasmic reticulum. In this scenario, the TCR could still bind to its tetramer but due to misfolding was not able to signal. Although this is a possibility with every TCR transduction, a higher affinity TCR would require fewer correctly assembled TCRs for antigen recognition. There is also the possibility that the exogenous TCR genes combined with the endogenous TCR of the transduced T cell [27, 28]. This is an important issue that needs to be addressed before the clinical use of any TCR gene transduced T cells because of the potential of autoimmune reactions against the TCR αβ heterodimers comprising exogenous and endogenous TCR chains [16]. There is a possibility that the α and β chains of the D4 TCR are less fit to compete with the endogenous TCR of PBLs and Jurkat cells and thus significant numbers are not transported to the cell surface to recognize antigens. Analyzing the D4 TCR has shed light on the fact that some TCRs, though reactive on the parental clone, may not signal when transferred to alternate T cells.
The lack of tetramer binding by the highly reactive TIL5 TCR is a different situation entirely. Although CTLs specifically recognize 8–10 amino acid peptides and MHC class I molecular complexes, this interaction is notably cross-reactive or degenerate [13, 19, 21, 33]. Not only have single T-cell clones been shown to recognize and respond to a large number of different peptide/MHC complexes but numerous different T-cell clones are also responsive to a single peptide/MHC complex [34]. Furthermore, studies have shown that only a few anchor residue amino acids of a given peptide are required in the TCR-peptide/MHC interaction [3, 10, 22]. Therefore, what is necessary for tetramer binding may be different from the minimal homology required for an activation signal. CTL activity to the MART-127–35 epitope is frequently observed in TIL and unlike other melanoma reactive T cells can be readily isolated from PBL of melanoma patients. It has been suggested that CTL responses to MART-127–35 might be augmented in part by T-cell encounters with peptides that show sequence similarity to MART-127–35. By searching a protein database for peptides with homology to MART-127–35 and testing their reactivity, it has been shown that indeed, epitope mimicry may play a role in modulating the CTL response to MART-1 [19]. Therefore, it is possible that the TIL5 clone was derived from exposure to one of many pathogens containing similar epitopes, such that it cross-reacts to the MART-127–35 peptide but would not bind tetramer. Unfortunately, the TIL5 parental clone is no longer available for studies and although it was previously shown to be highly reactive against MART-127–35, it is not known whether this clone bound MART-127–35 tetramers. It has been shown that low affinity/low avidity correlates with high cross-reactivity [34]. Therefore, the TIL5 clone may have just been highly cross-reactive against the MART-1 antigen and not necessarily specific for it. Regardless of this fact, due to its high reactivity and avidity toward the MART-127–35 peptide this receptor is an excellent candidate for adoptive transfer and tetramer studies alone would have neglected to identify it.
Although tetramers have proven to be an excellent tool to isolate antigen reactive T cells from patients and to monitor T-cell reactivity in vitro, we find that they are not always accurate in predicting TCR avidity for antigen. Due to the possibility of cross-reactive T cells, which may not bind tetramer but still may be excellent targets for immunotherapy, specific antigen reactivity should also be assessed.
Abbreviations
- TCR:
-
T-cell receptor
- TIL:
-
Tumor-infiltrating lymphocytes
- TAA:
-
Tumor-associated antigen
- AV/BV, TCR:
-
α/β chain variable regions
References
Altman JD, Moss PA, Goulder PJ, Barouch DH, McHeyzer-Williams MG, Bell JI, McMichael AJ, Davis MM (1996) Phenotypic analysis of antigen-specific T lymphocytes. Science 274:94–96
Appay V, Nixon DF, Donahoe SM, Gillespie GM, Dong T, King A, Ogg GS, Spiegel HM, Conlon C, Spina CA, Havlir DV, Richman DD, Waters A, Easterbrook P, McMichael AJ, Rowland-Jones SL (2000) HIV-specific CD8(+) T cells produce antiviral cytokines but are impaired in cytolytic function. J Exp Med 192:63–75
Bjorkman PJ, Saper MA, Samraoui B, Bennett WS, Strominger JL, Wiley DC (2005) Structure of the human class I histocompatibility antigen, HLA-A2. J Immunol 174:6–19
Clay TM, Custer MC, Sachs J, Hwu P, Rosenberg SA, Nishimura MI (1999) Efficient transfer of a tumor antigen-reactive TCR to human peripheral blood lymphocytes confers anti-tumor reactivity. J Immunol 163:507–513
Cole DJ, Weil DP, Shilyansky J, Custer M, Kawakami Y, Rosenberg SA, Nishimura MI (1995) Characterization of the functional specificity of a cloned T-cell receptor heterodimer recognizing the MART-1 melanoma antigen. Cancer Res 55:748–752
Cooper LJ, Kalos M, Lewinsohn DA, Riddell SR, Greenberg PD (2000) Transfer of specificity for human immunodeficiency virus type 1 into primary human T lymphocytes by introduction of T-cell receptor genes. J Virol 74:8207–8212
Davis MM, Boniface JJ, Reich Z, Lyons D, Hampl J, Arden B, Chien Y (1998) Ligand recognition by alpha beta T-cell receptors. Annu Rev Immunol 16:523–544
Denkberg G, Cohen CJ, Reiter Y (2001) Critical role for CD8 in binding of MHC tetramers to TCR: CD8 antibodies block specific binding of human tumor-specific MHC-peptide tetramers to TCR. J Immunol 167:270–276
Denkberg G, Klechevsky E, Reiter Y (2002) Modification of a tumor-derived peptide at an HLA-A2 anchor residue can alter the conformation of the MHC-peptide complex: probing with TCR-like recombinant antibodies. J Immunol 169:4399–4407
Fremont DH, Matsumura M, Stura EA, Peterson PA, Wilson IA (1992) Crystal structures of two viral peptides in complex with murine MHC class I H-2Kb. Science 257:919–927
Gallimore A, Glithero A, Godkin A, Tissot AC, Pluckthun A, Elliott T, Hengartner H, Zinkernagel R (1998) Induction and exhaustion of lymphocytic choriomeningitis virus-specific cytotoxic T lymphocytes visualized using soluble tetrameric major histocompatibility complex class I-peptide complexes. J Exp Med 187:1383–1393
Gray CM, Lawrence J, Schapiro JM, Altman JD, Winters MA, Crompton M, Loi M, Kundu SK, Davis MM, Merigan TC (1999) Frequency of class I HLA-restricted anti-HIV CD8+ T cells in individuals receiving highly active antiretroviral therapy (HAART). J Immunol 162:1780–1788
Hemmer B, Fleckenstein BT, Vergelli M, Jung G, McFarland H, Martin R, Wiesmuller KH (1997) Identification of high potency microbial and self ligands for a human autoreactive class II-restricted T cell clone. J Exp Med 185:1651–1659
Holler PD, Chlewicki LK, Kranz DM (2003) TCRs with high affinity for foreign pMHC show self-reactivity. Nat Immunol 4:55–62
Holmberg K, Mariathasan S, Ohteki T, Ohashi PS, Gascoigne NR (2003) TCR binding kinetics measured with MHC class I tetramers reveal a positive selecting peptide with relatively high affinity for TCR. J Immunol 171:2427–2434
Kessels HW, Wolkers MC, van der Burg S, van der Valk MA, Schumacher TN (2001) Immunotherapy through TCR gene transfer. Nat Immunol 2:957–961
Kuroda MJ, Schmitz JE, Barouch DH, Craiu A, Allen TM, Sette A, Watkins DI, Forman MA, Letvin NL (1998) Analysis of Gag-specific cytotoxic T lymphocytes in simian immunodeficiency virus-infected rhesus monkeys by cell staining with a tetrameric major histocompatibility complex class I-peptide complex. J Exp Med 187:1373–1381
Kuroda MJ, Schmitz JE, Lekutis C, Nickerson CE, Lifton MA, Franchini G, Harouse JM, Cheng-Mayer C, Letvin NL (2000) Human immunodeficiency virus type 1 envelope epitope-specific CD4(+) T lymphocytes in simian/human immunodeficiency virus-infected and vaccinated rhesus monkeys detected using a peptide-major histocompatibility complex class II tetramer. J Virol 74:8751–8756
Loftus DJ, Castelli C, Clay TM, Squarcina P, Marincola FM, Nishimura MI, Parmiani G, Appella E, Rivoltini L (1996) Identification of epitope mimics recognized by CTL reactive to the melanoma/melanocyte-derived peptide MART-1 (27–35). J Exp Med 184:647–657
Mallone R, Nepom GT (2004) MHC Class II tetramers and the pursuit of antigen-specific T cells: define, deviate, delete. Clin Immunol 110:232–242
Mason D (1998) A very high level of crossreactivity is an essential feature of the T-cell receptor. Immunol Today 19:395–404
Matsumura M, Fremont DH, Peterson PA, Wilson IA (1992) Emerging principles for the recognition of peptide antigens by MHC class I molecules. Science 257:927–934
Morgan RA, Dudley ME, Yu YY, Zheng Z, Robbins PF, Theoret MR, Wunderlich JR, Hughes MS, Restifo NP, Rosenberg SA (2003) High efficiency TCR gene transfer into primary human lymphocytes affords avid recognition of melanoma tumor antigen glycoprotein 100 and does not alter the recognition of autologous melanoma antigens. J Immunol 171:3287–3295
Orentas RJ, Roskopf SJ, Nolan GP, Nishimura MI (2001) Retroviral transduction of a T-cell receptor specific for an Epstein-Barr virus-encoded peptide. Clin Immunol 98:220–228
Rosenberg SA, Aebersold P, Cornetta K, Kasid A, Morgan RA, Moen R, Karson EM, Lotze MT, Yang JC, Topalian SL et al (1990) Gene transfer into humans—immunotherapy of patients with advanced melanoma, using tumor-infiltrating lymphocytes modified by retroviral gene transduction. N Engl J Med 323:570–578
Roszkowski JJ, Yu DC, Rubinstein MP, McKee MD, Cole DJ, Nishimura MI (2003) CD8-independent tumor cell recognition is a property of the T-cell receptor and not the T cell. J Immunol 170:2582–2589
Sadelain M, Riviere I, Brentjens R (2003) Targeting tumours with genetically enhanced T lymphocytes. Nat Rev Cancer 3:35–45
Schumacher TN (2002) T-cell-receptor gene therapy. Nat Rev Immunol 2:512–519
Stanislawski T, Voss RH, Lotz C, Sadovnikova E, Willemsen RA, Kuball J, Ruppert T, Bolhuis RL, Melief CJ, Huber C, Stauss HJ, Theobald M (2001) Circumventing tolerance to a human MDM2-derived tumor antigen by TCR gene transfer. Nat Immunol 2:962–970
Starr TK, Jameson SC, Hogquist KA (2003) Positive and negative selection of T cells. Annu Rev Immunol 21:139–176
Sykulev Y, Brunmark A, Jackson M, Cohen RJ, Peterson PA, Eisen HN (1994) Kinetics and affinity of reactions between an antigen-specific T-cell receptor and peptide-MHC complexes. Immunity 1:15–22
Valitutti S, Muller S, Cella M, Padovan E, Lanzavecchia A (1995) Serial triggering of many T-cell receptors by a few peptide-MHC complexes. Nature 375:148–151
Wilson DB, Pinilla C, Wilson DH, Schroder K, Boggiano C, Judkowski V, Kaye J, Hemmer B, Martin R, Houghten RA (1999) Immunogenicity. I. Use of peptide libraries to identify epitopes that activate clonotypic CD4+ T cells and induce T-cell responses to native peptide ligands. J Immunol 163:6424–6434
Wilson DB, Wilson DH, Schroder K, Pinilla C, Blondelle S, Houghten RA, Garcia KC (2004) Specificity and degeneracy of T cells. Mol Immunol 40:1047–1055
Youde SJ, Dunbar PR, Evans EM, Fiander AN, Borysiewicz LK, Cerundolo V, Man S (2000) Use of fluorogenic histocompatibility leukocyte antigen-A*0201/HPV 16 E7 peptide complexes to isolate rare human cytotoxic T-lymphocyte-recognizing endogenous human papillomavirus antigens. Cancer Res 60:365–371
Zajac AJ, Blattman JN, Murali-Krishna K, Sourdive DJ, Suresh M, Altman JD, Ahmed R (1998) Viral immune evasion due to persistence of activated T cells without effector function. J Exp Med 188:2205–2213
Acknowlegments
This work was supported by generous contributions from the National Institutes of Health (CA90873, CA100240, CA97296 and CA74397). W.M. Kast holds the Walter A. Richter Cancer Research Chair. C. Yee is a Damon Runyon-Lilly Clinical Investigator supported (in part) by the Damon Runyon Cancer Research Foundation.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Lyons, G.E., Roszkowski, J.J., Man, S. et al. T-cell receptor tetramer binding or the lack there of does not necessitate antigen reactivity in T-cell receptor transduced T cells. Cancer Immunol Immunother 55, 1142–1150 (2006). https://doi.org/10.1007/s00262-005-0103-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00262-005-0103-9