Abstract
Purposes
To evaluate the capability of PET/CT images for differentiating the histologic subtypes of non-small cell lung cancer (NSCLC) and to identify the optimal model from radiomics-based machine learning/deep learning algorithms.
Methods
In this study, 867 patients with adenocarcinoma (ADC) and 552 patients with squamous cell carcinoma (SCC) were retrospectively analysed. A stratified random sample of 283 patients (20%) was used as the testing set (173 ADC and 110 SCC); the remaining data were used as the training set. A total of 688 features were extracted from each outlined tumour region. Ten feature selection techniques, ten machine learning (ML) models and the VGG16 deep learning (DL) algorithm were evaluated to construct an optimal classification model for the differential diagnosis of ADC and SCC. Tenfold cross-validation and grid search technique were employed to evaluate and optimize the model hyperparameters on the training dataset. The area under the receiver operating characteristic curve (AUROC), accuracy, precision, sensitivity and specificity was used to evaluate the performance of the models on the test dataset.
Results
Fifty top-ranked subset features were selected by each feature selection technique for classification. The linear discriminant analysis (LDA) (AUROC, 0.863; accuracy, 0.794) and support vector machine (SVM) (AUROC, 0.863; accuracy, 0.792) classifiers, both of which coupled with the ℓ2,1NR feature selection method, achieved optimal performance. The random forest (RF) classifier (AUROC, 0.824; accuracy, 0.775) and ℓ2,1NR feature selection method (AUROC, 0.815; accuracy, 0.764) showed excellent average performance among the classifiers and feature selection methods employed in our study, respectively. Furthermore, the VGG16 DL algorithm (AUROC, 0.903; accuracy, 0.841) outperformed all conventional machine learning methods in combination with radiomics.
Conclusion
Employing radiomic machine learning/deep learning algorithms could help radiologists to differentiate the histologic subtypes of NSCLC via PET/CT images.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Lung cancer is one of the most common malignant tumours that endangers human health. In China and worldwide, the incidence and mortality of lung cancer are at the top of the list, and the trend is increasing annually [1]. Lung cancer represents a highly heterogeneous malignant epithelial tumour with distinct pathological features and clinical behaviour [2]. According to the histological size of cancer cells under a microscope, lung cancer can be divided into small cell lung cancer (SCLC) and NSCLC. The latter, which accounts for approximately 85% of lung cancer cases, includes ADC (~ 50%), SCC (~ 40%) and large cell lung cancer [3, 4]. The accurate staging and pathologic grading of lung cancer has important significance in the determination of a rational treatment regimen. For instance, Scagliotti et al. found that compared with docetaxel, pemetrexed significantly prolonged the overall survival and progression-free survival of ADC but had the opposite effect on SCC [5]. Hence, it is important to accurately distinguish between the two subtypes of NSCLC prior to initiating treatment.
Although experienced physicians can often diagnose the type of lung cancer based on clinical presentation and radiographic appearance, NSCLC is sometimes poorly differentiated and is distinguishable only by immunohistochemical staining and molecular testing. SCC is distinguished from ADC in the clinic by immunostaining for cytokeratin 5 and cytokeratin 6 and/or the transcription factors SRY-box 2 (SOX2) and p63 [3, 4, 6]. CT-guided transthoracic needle aspiration is typically the first-line method for peripheral lesions, and obtaining an adequate tissue sample is imperative for optimizing the diagnosis and treatment plan [7]. For small or peripherally located lung cancers, the current needle biopsy procedures sample only a small amount of tissue and have low accuracy. In some cases, CT-guided needle biopsy cannot be performed or is not suitable. Moreover, for deeply located lesions or lesions close to airways or blood vessels, needle biopsy is challenging. In patients with unfavourable situations, biopsy would not be recommended. Moreover, the tumour is often heterogeneous, which may affect the biopsy results. A non-invasive method for pathological classification prior to biopsy or surgery has not yet been developed.
Positron emission tomography and computed tomography (PET/CT) with the use of the 18F-fluorodeoxyglucose (18F-FDG) marker is an essential imaging modality for lung cancer [8, 9], and the majority of patients undergo 18F-FDG PET/CT before treatment initiation. However, radiologists have difficulty distinguishing ADC from SCC based on PET/CT images, and interobserver agreement is usually low. Recent studies have indicated that radiomic features can provide additional useful information based on PET/CT images, reflecting the potential of biological heterogeneity [10,11,12]. There is increasing interest in radiomics, which involves the conversion of medical images into mineable high-dimensional quantitative data. The use of these data to predict treatment responses and patient outcomes has been reported across a range of primary tumours [13, 14]. Combined with machine learning techniques, radiomic features extracted from FDG PET/CT performed better in predicting the progression-free survival (PFS) prognosis of anal squamous cell carcinoma (ASCC) than did conventional staging parameters [15], in addition to performing well in identifying bone marrow involvement (BMI) in patients with suspected relapsed acute leukaemia (AL) [16]. To the best of our knowledge, only one published study has evaluated FDG PET/CT radiomics in NSCLC to discriminate between ADC and SCC. The study applied a linear discriminant analysis (LDA) classifier and three feature selection algorithms [17]. The linear separability of LDA was 0.90; however, as the authors stated, it was a preliminary study that enrolled only 30 patients, and the feature extraction, feature selection and classification methods were simple. A larger-scale comprehensive study is necessary to explore the value of PET/CT imaging for the discrimination of NSCLC subtypes.
In this study, we investigated 10 feature selection methods, 10 ML classifiers and a DL algorithm (VGG16) for differentiating the histological subtypes of NSCLC [18]. These methods were chosen because of their popularity in the literature. The aim of this study was to evaluate whether FDG PET/CT images are powerful for differentiating ADC and SCC and to further search for the optimal model among numerous radiomics-based ML approaches and the DL algorithm. This work serves as a promising diagnostic tool for informing treatment decisions and fostering personalized therapy for patients with lung cancer.
Materials and methods
Patients
This retrospective, single-centre study included two cohorts of patients undergoing pulmonary PET/CT examination between January 2018 and August 2019 at the Department of Nuclear Medicine, Peking University Cancer Hospital. The inclusion criteria were as follows: (1) available pretreatment PET/CT images and (2) available definite pathological diagnosis of ADC or SCC. There were 1419 cases enrolled in the study, with 867 in the ADC cohort and 552 in the SCC cohort. The patients were first split into a training set and a testing set in an 8:2 ratio, and the positive-negative sample ratios in these sets were approximately the same as that in the complete dataset. Then, the training set was used to fit and tune models via tenfold cross-validation, and the testing set was used to evaluate the predictive and generalization ability of the models. The simple statistics of the training and testing set are summarized in Table 1.
PET/CT acquisition
The PET/CT system we used is an integrated PET and CT machine, a Philips Gemini TF 16 (Dutch Philips Corporation). Before the examination, each patient fasted for more than 6 h, and the blood glucose level was determined by fingertip blood sampling < 10 mmol/L. After 18F-FDG was intravenously injected according to the patient’s body weight (3.0~3.7 MBq/kg), the patient rested calmly. After 60 min, the body was scanned from the cranial to upper thigh regions. Emission collection was conducted from the end of the bed, scanning lasted 1.5 min per bed and 9 to 10 beds per patient. The ordered-subset expectation maximization (OSEM) iterative method was used for PET image reconstruction. The configuration parameters of the transmission scan (CT) were a tube voltage of 120 kV, a tube current of 100 mA and a scan layer thickness of 3 mm. Attenuation correction of the PET images was performed by means of CT data.
Radiomic features
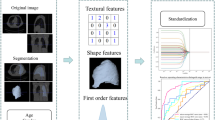
The region of interest (ROI) was segmented from the whole PET/CT image semiautomatically using the region growth method, which was performed by a radiologist with 5 years’ working experience and conducted with MATLAB 2017a (MathWorks, Natick, MA, USA). Level-3 subband of two-dimensional discrete wavelet transform (2D DWT) was conducted on the ROI to obtain eight subband images, and feature extraction was then performed on the subbands. A schematic of the wavelet transform is shown in Supplementary Fig. 1. The acquired imaging features comprised the first-order intensity statistical features and texture features, including the first-order statistics (18 features), grey-level co-occurrence matrix (GLCM, 22 features), grey-level run-length matrix (GLRLM, 16 features), grey-level size-zone matrix (GLSZM, 16 features) and grey-level dependence matrix (GLDM, 14 features) features. A total of 688 features were obtained from each ROI. Feature extraction was performed by a comprehensive open-source platform called PyRadiomics [19], which enables the processing and extraction of radiomic features from medical image data and is implemented in Python. The scoring/selection criteria of the feature selection process are available from the online documentation of PyRadiomics (http://pyradiomics.readthedocs.io/). An overview of the study workflow is illustrated in Fig. 1.
Feature selection methods
The main purposes of feature selection methods are to simplify the model, decrease the computational costs, avoid the curse of dimensionality and enhance the generalization ability of the model [20]. We considered ten feature selection methods that are widely used in the literature: Laplacian score (LS), ReliefF (ReF), spectral feature selection (SPEC), ℓ2,1-norm regularization (ℓ2,1NR), efficient and robust feature selection (RFS), multi-cluster feature selection (MCFS), chi-square score (CSS), Fisher score based on statistics (FS), t score (TS) and Gini index (GINI). The first three methods are feature selection methods based on similarity that assess feature importance in terms of the ability to preserve data similarity. The next three methods are based on sparse learning and employ regularization terms to reduce the weights of unimportant features in the model. The last four methods are statistical based methods that rely on various statistical measures to assess feature importance [21]. A feature selection repository of Python named “scikit feature”, which was released by Li et al. [21], was implemented. The web page of the repository is available at http://featureselection.asu.edu/.
Classification algorithms
Classification, a supervised learning task in which function is inferred based on labelled training data [22], is one of the most widely studied areas of machine learning. This study investigated 10 popular classifiers: AdaBoost (AdaB), bagging (BAG), decision tree (DT), naive Bayes (NB), K-nearest neighbours (KNN), logistic regression (LR), multilayer perceptron (MLP), linear discriminant analysis (LDA), random forest (RF) and support vector machine (SVM). Tenfold cross-validation was conducted on the training dataset to evaluate the performance of the models and to identify the optimal hyperparameters, in combination with grid search, after manually setting the bounds and discretization. The AUROC of the estimators, which is suitable for unbalanced classification, was used to evaluate the parameter settings.
In this study, we also considered an end-to-end deep learning approach and compared its classification efficiency with that of the traditional machine learning methods mentioned above. The selected deep learning model was VGG16, which adopts a transfer learning strategy and data argument technique. The pretraining weights were derived from training on the ImageNet dataset. The parameter configurations of each ML algorithm, as well as the data argumentation and transfer learning details of the DL algorithm, are provided in the Supplementary Materials.
Statistical analysis
Statistical descriptions of the demographic characteristics of the training and testing datasets are presented as the mean and standard deviation (SD) or percentage, and the statistical analysis of differences between two datasets was performed using the chi-square test and Student’s t test. The performance of the radiomics ML classifiers and the VGG16 DL algorithm was compared in terms of the AUROC, accuracy, precision, sensitivity (i.e., recall) and specificity. The 95% confidence interval (95% CI) of the AUROC was also calculated based on a binomial exact test [23]. The method of Delong et al. was adopted to conduct pairwise comparisons of the ROC curves [24]. Accuracy was defined as the ratio of the number of samples correctly classified by the classifier to the total number of samples in the testing dataset. Sensitivity was defined as the number of correct positive results that occurred among all ADC samples available during the test. Specificity was defined as the number of correct positive results that occurred among all SCC samples available during the test. Statistical analyses were performed using the “scikit-learn”, “sciPy” and “math” packages in the Python programming language. The 95% CIs of the AUROC were obtained by means of MedCalc statistical software (version 19.0.7, Ostend, Belgium). P values < 0.05 were deemed to be statistically significant.
Results
Demographic and clinical characteristics of the patients
A total of 1419 samples were collected in this study, and the patients had an average age of 65.20 ± 9.59 years, 473 (33.40%) were female, 338 (23.82%) had a history of smoking, and 300 (21.14%) had metastasis. A stratified random sampling method was conducted to extract 20% (283 samples) of the total sample as the testing set to evaluate the model performance. The remaining 80% were used for training the models and were divided into 10 subsets during cross-validation. The details of the demographic and clinical characteristics of the training and testing cohorts are shown in Table 1.
Performance of radiomics machine learning algorithm
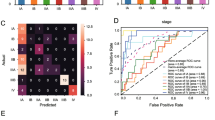
We extracted 688 quantitative features from each of the segmented tumour regions and then applied ten feature selection methods to rank the features in the training set. In the pilot study, we used the 10, 20, 30… 100 top-ranked features obtained by each feature selection method one by one to fit 10 machine learning classifiers. Almost all models achieved the highest AUROC when using the top 50 features. Therefore, we selected the top 50 features of each selection method for our current study. The heatmap in Fig. 2 depicts the AUROC and accuracy on the testing dataset with the optimal hyperparameter configuration. The results for the 30, 40, 60 and 70 top-ranked features are reported in Supplementary Fig. 2, 3, 4, 5 in the Supplementary Material. The white numerals in the grid correspond to the top ten best-performing models. The combined LDA (classifier) and ℓ2,1NR (feature selection method) model achieved the best classification performance, with an AUROC of 0.863 and an accuracy of 0.794. The second-ranked model was the combination of SVM (classifier) and ℓ2,1NR (feature selection method), with an AUROC of 0.863 and an accuracy of 0.792. The mean AUROC and accuracy of all 10 feature selection methods are calculated as the representative AUROC and accuracy for each classifier. Similarly, for each feature selection method, the mean AUROC and accuracy of 10 classifiers are used as the representative AUROC and accuracy. These representative AUROC and accuracy values for the feature selection and classification methods are given in Tables 2 and 3, respectively. The FR classifier and ℓ2,1NR feature selection method showed optimal performance. Figure 3 illustrated the change in AUROC values of the classifiers (LDA, SVM and RF) when using different numbers of top-ranked features selected by ℓ2,1NR on the testing dataset.
AUROC value of the top-three combined models vs. number of top-ranked features selected via ℓ2,1NR. The dashed curves represent the AUROC of the model on the testing dataset. The relative importance weights of the first 100 features are shown by the stem-and-leaf diagram in the lower half of the figure, corresponding to the right Y-axis
The average absolute values of the correlation coefficients (CCAA) between the top 50 features selected by each feature selection method are listed in Table 3. A lower CCAA denotes less redundant information is included in the selected features. The 50 features selected by the ℓ2,1NR feature selection method have the lowest correlation among features. The matrix diagram of Fig. 4 explicitly illustrates the correlation between each of the 50 features selected by ℓ2,1NR and three other feature selection methods (ReF, RFS and GINI).
Performance of the deep learning algorithm
We adopted a deep convolution neural network (DCNN) algorithm, VGG16, to train the classification model on the same training dataset. To improve the performance of the model and accelerate the convergence speed of training, pre-initialization using weights from the same network trained to classify objects in the ImageNet dataset [25], as well as data argumentation on the training dataset, was executed. As expected, the VGG16 deep learning model achieved excellent performance (AUROC, 0.903; accuracy, 0.841) on the testing dataset and outperformed the top-ranked model, namely, the combination of LDA classifier and ℓ2,1NR feature selection method. The detailed network iteration process is illustrated in Fig. 5.
Performance comparison of top four models
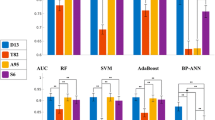
Table 4 presents a comprehensive performance comparison of the optimal three combinations of classifiers and feature selection methods and the VGG16 model on the testing dataset. Pairwise comparisons of the ROC curves were conducted using the method proposed by Delong et al. [24], and no statistically significant differences were observed. The results are shown in Table 4 and Fig. 6.
Discussion
Cancer management has entered the era of precision medicine, which relies on validated biomarkers to classify patients with respect to their probable disease risk, prognosis and/or response to the treatment. Therefore, early and accurate subtype diagnosis of lung cancer is particularly important. PET/CT scanning, a well-established hybrid-functional imaging technique, enables non-invasive tumour evaluation for grading, staging and measuring the response to treatment of certain cancers; however, its value in the differential diagnosis of ADC and SCC is limited for radiologists to interpret the images in a routine manner.
In this study, we used machine learning/deep learning algorithms to discover the value of PET/CT images by means of radiomics for the differential diagnosis of ADC and SCC. The results showed that the LDA (AUROC, 0.863; accuracy, 0.794) and SVM (AUROC, 0.863; accuracy, 0.792) classifiers, both combined with the ℓ2,1NR feature selection method, achieved optimal performance compared with other combinations. Furthermore, the VGG16 DL algorithm (AUROC, 0.903; accuracy, 0.841) outperformed all conventional machine learning algorithms with radiomics. To the best of our knowledge, this is the first study to report the potential of PET/CT images with the application of a panel of machine learning/deep learning algorithms for the identification of ADC and SCC.
Radiomics, a young and emerging discipline that bridges the gap between medical imaging and personalized medicine [26, 27], attempts to explore the value of medical images in disease diagnosis, grading and prognosis prediction using medical image analysis technology and machine learning algorithms in artificial intelligence. However, due to the numerous available feature selection methods and ML algorithms [21, 28], the optimal method to use for specific medical images or specific target tasks remains unclear. To determine which feature selection and machine learning algorithms are suitable for the given medical image data, the performance of various feature selection and machine learning algorithms in medical image classification has been studied in recent years [29,30,31,32]. For example, Parmar et al. [33] investigated 14 feature selection methods and 12 classification methods for predicting overall survival of patients with lung cancer, with 440 radiomic features extracted from three-dimensional CT images. The Wilcoxon feature selection approach and its variants showed a higher prediction accuracy than that of the other methods, and the naive Bayes classifier outperformed other classifiers and achieved the highest AUROC (0.72). Zhang B et al. [30] evaluated the performance of 6 feature selection methods and 9 classification methods for the radiomics-based prediction of local failure and distant failure in advanced nasopharyngeal carcinoma. They extracted 970 radiomic features from T2-weighted and contrast-enhanced T1-weighted MRI images of each patient and observed that the combination of the RF classifier and RF feature selection methods yielded the highest prognostic performance.
In the current study, we found that the LDA and SVM classifiers coupled with the ℓ2,1NR feature selection method showed the best performance on our dataset. Unlike the other feature selection methods considered in our study, ℓ2,1NR, RFS and MCFS are embedded methods that embed feature selection into a typical learning algorithm (such as logistic regression). Such methods take into account the correlations between two features. Hence, they can handle redundant feature during the selection phase. By contrast, the other 7 feature selection methods are filter methods. One disadvantage of these methods is that they typically analyse features individually and hence fail to address feature redundancy. The abovementioned research is in agreement with the results of our study. Our findings are consistent with those of Qian [31], who discovered that SVM combined with the least absolute shrinkage and selection operator (LASSO) yielded the highest prediction efficacy in differentiating glioblastoma from solitary brain metastases. Clearly, both LASSO and ℓ2,1NR are sparse-based feature selection methods. According to the literature, RFS is more suitable for multi-label tasks [34, 35], and MCFS is used for unsupervised feature selection [36], which may be the reason why their performance is inferior to that of ℓ2,1NR in this study. The feature analysis also indicated that though radiomics is conducive to obtaining high-throughput features from images, which is accompanied by abundant redundant information. SVM is a robust, powerful and effective machine learning classifier that has been predominately used in the field of radiomics [37, 38]. The results of this study indicate that SVM with a linear kernel is superior to the radial basis function and other kernels. Meanwhile, LDA also performed well in the study. SVM and LDA have an identical function class – the linear decision boundary. A possible explanation for the good performance might be that the data of our study are linearly separable. Another important finding was that the random forest classifier showed good discrimination performance and was the least sensitive to the feature selection methods. These results are in line with those of the previous studies [29, 30, 33]. The random forest algorithm, proposed by L. Breiman [39], has been extremely successful as a general purpose classification method and is easily adaptable to various ad hoc learning tasks. However, it has been observed to have an overfitting problem [39], which was also observed in the current study.
Compared with similar published studies, we found that the optimal ML classifier and feature selection method are inconsistent, which may be due to various reasons, including the differentiation of image modalities, feature extraction algorithms, number of selected features, target task and cohort size. For instance, Zhang Y et al. [32] compared the predictive performance of different combinations of classifiers and feature reduction methods for three clinical outcomes based on the same radiomic features extracted from the same CT image dataset and found that the best model for recurrence is the RF classifier combined with near zero variance (NZV) feature selection; for death, the best model is the NB classifier combined with zero variance (ZV) feature selection; and for recurrence-free survival (RFS), the best model is the mixture discriminant analysis (MDA) classifier without a feature selection method.
In addition to the traditional machine learning algorithms, we also assessed a state-of-the-art DL algorithm. DL algorithms, especially convolution neural networks (CNN), have become the most popular algorithms in computer vision, which is widely used in medical image recognition, target detection, image segmentation and other fields. CNN algorithms are implemented by means of convolution and pooling strategies and are able to simultaneously perform feature construction, feature selection and prediction modelling, essentially performing an end-to-end analysis from inputting raw images to prediction. As such, they are very powerful and labour-saving learning algorithms compared with radiomics. The VGG16 CNN utilized in the current study yielded the best performance among all models. However, as with any other tool, DL algorithms have strengths and limitations. Generally, the DL model needs millions of parameters for training; that is, a large amount of data is needed to train an ideal model. However, medical images are often difficult to collect in large quantities. In addition, a complete theory to explain how the hidden layers that lie between the inputs and outputs is not yet available. The lack of transparency makes it difficult to monitor when model prediction may fail or require troubleshooting [40]. We used dropout layers and data augmentation techniques in training the VGG16 CNN model, which is advantageous for improving the performance of the models and avoiding overfitting, especially for small datasets.
We collected PET/CT fusion images as the image modality in this study. Compared with other modalities, such as CT and MRI, PET has an inherent defect of a low signal-to-noise ratio and resolution and is considered unsuitable for texture feature research. However, tumours are heterogeneous entities at all scales (macroscopic, physiological, microscopic, genetic) [41]. CT and MRI reflect mainly the anatomical structure of tumours, while PET/CT can be used to explore intratumour heterogeneity in both the anatomical and functional dimensions. Texture features calculated from PET modalities have exclusive advantages in reflecting metabolic heterogeneity, which is a recognized characteristic of malignant tumours, presumably linked to basal metabolism, cell necrosis and hypoxia [42]. The results of a prospective study in 54 patients with head and neck cancer demonstrated that some PET texture features could be linked to signalling pathway alterations associated with cell proliferation and apoptosis [43]. In our study, the texture features of PET/CT demonstrated excellent performance in identifying the histological subtypes of NSCLC, which were roughly the same as that of CT images [44].
Our study has several limitations. First, the sample size was small compared with that required for the machine learning methods we used, especially the deep learning algorithm, which may prevent the possibility of establishing better predictive models and is prone to model instability. However, this problem is prevalent in the research on machine learning for medical images [45]. In addition, the images in this study came from a single PET/CT scanner, and a previous study showed that differences in the image acquisition parameters or reconstruction among PET/CT devices affect the extracted texture feature values [46]. Furthermore, we need to collect samples from multiple centres for model training to enhance the robustness and generalization ability of the models. Third, our study used only PET/CT fused images. We may obtain better results if we extract texture features from PET and CT images separately and build models with the merged texture features.
In conclusion, machine learning/deep learning algorithms can be used to differentiate the histological subtypes of NSCLC, namely, ADC and SCC, based on PET/CT images. This work serves as a promising diagnostic tool for informing treatment decisions and fostering personalized therapy for patients with lung cancer in a non-invasive manner.
References
Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–32.
Chen Z, Fillmore CM, Hammerman PS, Kim CF, Wong K-K. Non-small-cell lung cancers: a heterogeneous set of diseases. Nat Rev Cancer. 2014;14(8):535–46.
Davidson MR, Gazdar AF, Clarke BE. The pivotal role of pathology in the management of lung cancer. J Thorac Dis. 2013;5(Suppl 5):S463.
Langer CJ, Besse B, Gualberto A, Brambilla E, Soria J-C. The evolving role of histology in the management of advanced non–small-cell lung cancer. J Clin Oncol. 2010;28(36):5311–20.
Scagliotti G, Hanna N, Fossella F, Sugarman K, Blatter J, Peterson P, et al. The differential efficacy of pemetrexed according to NSCLC histology: a review of two phase III studies. Oncologist. 2009;14(3):253–63.
Lu Y, Futtner C, Rock JR, Xu X, Whitworth W, Hogan BL, et al. Evidence that SOX2 overexpression is oncogenic in the lung. PLoS One. 2010;5(6):e11022.
Rivera MP, Mehta AC, Wahidi MM. Establishing the diagnosis of lung cancer: diagnosis and management of lung cancer: American College of Chest Physicians evidence-based clinical practice guidelines. Chest. 2013;143(5):e142S–e65S.
Takeuchi S, Khiewvan B, Fox PS, Swisher SG, Rohren EM, Bassett RL, et al. Impact of initial PET/CT staging in terms of clinical stage, management plan, and prognosis in 592 patients with non-small-cell lung cancer. Eur J Nucl Med Mol Imaging. 2014;41(5):906–14.
Liu B, Dong J, Xu C, Zuo C, Le J, Guan Y, et al. Accuracy of 18F-FDG PET/CT for lymph node staging in non-small-cell lung cancers. Chin Med J. 2009;122(15):1749–54.
Kirienko M, Cozzi L, Antunovic L, Lozza L, Fogliata A, Voulaz E, et al. Prediction of disease-free survival by the PET/CT radiomic signature in non-small cell lung cancer patients undergoing surgery. Eur J Nucl Med Mol Imaging. 2018;45(2):207–17.
Lovinfosse P, Polus M, Van Daele D, Martinive P, Daenen F, Hatt M, et al. FDG PET/CT radiomics for predicting the outcome of locally advanced rectal cancer. Eur J Nucl Med Mol Imaging. 2018;45(3):365–75.
Lucia F, Visvikis D, Desseroit M-C, Miranda O, Malhaire J-P, Robin P, et al. Prediction of outcome using pretreatment 18 F-FDG PET/CT and MRI radiomics in locally advanced cervical cancer treated with chemoradiotherapy. Eur J Nucl Med Mol Imaging. 2018;45(5):768–86.
Philippe L, Emmanuel RV, Ralph L, Sara C, Stiphout RGPM, Patrick G, et al. Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer. 2007;43(8):441–6.
Larue RT, Defraene G, De Ruysscher D, Lambin P, Van Elmpt W. Quantitative radiomics studies for tissue characterization: a review of technology and methodological procedures. Brit J Radiol. 2017;90(1070):20160665.
Brown P, Zhong J, Frood R, Currie S, Gilbert A, Appelt A, et al. Prediction of outcome in anal squamous cell carcinoma using radiomic feature analysis of pre-treatment FDG PET-CT. Eur J Nucl Med Mol Imaging. 2019;46(13):2790–9.
Li H, Xu C, Xin B, Zheng C, Zhao Y, Hao K, et al. 18F-FDG PET/CT Radiomic analysis with machine learning for identifying bone marrow involvement in the patients with suspected relapsed acute leukemia. Theranostics. 2019;9(16):4730.
Ha S, Choi H, Cheon GJ, Kang KW, Chung J-K, Kim EE, et al. Autoclustering of non-small cell lung carcinoma subtypes on 18 F-FDG PET using texture analysis: a preliminary result. Nucl Med Mol Imaging. 2010;48(4):278–86.
Simonyan K, Zisserman A. Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:14091556. 2014.
Van Griethuysen JJ, Fedorov A, Parmar C, Hosny A, Aucoin N, Narayan V, et al. Computational radiomics system to decode the radiographic phenotype. Cancer Res. 2017;77(21):e104–e7.
Motoda H, Liu H. Feature selection, extraction and construction. Communication of IICM (Institute of Information and Computing Machinery, Taiwan). 2002;5(67–72):2.
Li J, Cheng K, Wang S, Morstatter F, Trevino RP, Tang J, et al. Feature selection: a data perspective. ACM Computing Surveys (CSUR). 2018;50(6):94.
Mohri M, Rostamizadeh A, Talwalkar A. Foundations of machine learning. 1st ed. Cambridge: MIT Press; 2012.
Cortes C, Mohri M. Confidence intervals for the area under the ROC curve. Adv Neural Inf Process Syst. 2005;17:305–12.
Giovannoni SJ, Delong EF, Olsen GJ, Pace NR. Phylogenetic group-specific oligodeoxynucleotide probes for identification of single microbial cells. J Bacteriol. 1988;170(2):720.
Russakovsky O, Deng J, Su H, Krause J, Satheesh S, Ma S, et al. ImageNet large scale visual recognition challenge. Int J Comput Vis. 2015;115(3):211–52.
Gillies RJ, Kinahan PE, Hricak H. Radiomics: images are more than pictures. They Are Data Radiology. 2015;278(2):151169.
Lambin P, Leijenaar RT, Deist TM, Peerlings J, De Jong EE, Van Timmeren J, et al. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol. 2017;14(12):749.
Fernandez-Delgado M, Cernadas E, Barro S, Amorim D. Do we need hundreds of classifiers to solve real world classification problems? J Mach Learn Res. 2014;15(1):3133–81.
Wu W, Parmar C, Grossmann P, Quackenbush J, Lambin P, Bussink J, et al. Exploratory study to identify radiomics classifiers for lung cancer histology. Front Oncol. 2016;6:71.
Zhang B, He X, Ouyang F, Gu D, Dong Y, Zhang L, et al. Radiomic machine-learning classifiers for prognostic biomarkers of advanced nasopharyngeal carcinoma. Cancer Lett. 2017;403:21–7.
Qian Z, Li Y, Wang Y, Li L, Li R, Wang K, et al. Differentiation of glioblastoma from solitary brain metastases using radiomic machine-learning classifiers. Cancer Lett. 2019;451:128–35.
Zhang Y, Oikonomou A, Wong A, Haider MA, Khalvati F. Radiomics-based prognosis analysis for non-small cell lung cancer. Sci Rep. 2017;7:46349.
Parmar C, Grossmann P, Bussink J, Lambin P, Aerts HJWL. Machine learning methods for quantitative radiomic biomarkers. Sci Rep. 2015;5(1):13087.
Chang X, Nie F, Yang Y, Huang H. A convex formulation for semi-supervised multi-label feature selection. AAAI 2014. 2014;2:1171–7.
Jian L, Li J, Shu K, Liu H. Multi-label informed feature selection. IJCAI'16. 2016;1627–33.
Cai D, Zhang C, He X. Unsupervised feature selection for multi-cluster data. KDD'10. 2010;333–42.
Wang F, Zhang B, Wu X, Liu L, Fang J, Chen Q, et al. Radiomic nomogram improves preoperative T category accuracy in locally advanced laryngeal carcinoma. Front Oncol. 2019;9:1064.
Wu S, Jiao Y, Zhang Y, Ren X, Li P, Yu Q, et al. Imaging-based individualized response prediction of carbon ion radiotherapy for prostate cancer patients. Cancer Manag Res. 2019;11:9121.
Breiman L. Random forests. Mach Learn. 2001;45(1):5–32.
Hosny A, Parmar C, Quackenbush J, Schwartz LH, Aerts HJ. Artificial intelligence in radiology. Nat Rev Cancer. 2018;18(8):500.
Marco G, Rowan AJ, Stuart H, James L, David E, Eva G, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366(10):883–92.
Cheng NM, Fang YHD, Yen TC. The promise and limits of PET texture analysis. Ann Nucl Med. 2013;27(9):867–9.
Tixier F, Hatt M, Le Rest CC, Simon B, Key S, Corcos L, et al. Signaling pathways alteration involved in head and neck cancer can be identified through textural features analysis in 18F-FDG PET images: a prospective study. J Nucl Med. 2015;56(supplement 3):449.
Zhu X, Dong D, Chen Z, Fang M, Zhang L, Song J, et al. Radiomic signature as a diagnostic factor for histologic subtype classification of non-small cell lung cancer. Eur Radiol. 2018;28(7):2772–8.
Obermeyer Z, Emanuel EJ. Predicting the future—big data, machine learning, and clinical medicine. N Engl J Med. 2016;375(13):1216.
Galavis PE, Hollensen C, Jallow N, Paliwal B, Jeraj R. Variability of textural features in FDG PET images due to different acquisition modes and reconstruction parameters. Acta Oncol. 2010;49(7):1012–6.
Funding
This work was supported by funds from the National Natural Science Foundation of China (No. 81773542 and No. 81703318) and the Key Projects of Science and Technology Plan from Beijing Municipal Education Commission (No. KZ201810025031).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Ethical approval
This study was approved by the Institutional Review Board of Capital Medical University and is in line with the Declaration of Helsinki and its later amendments.
Informed consent
The need for informed consent was waived by the Ethics Committee of Peking University Cancer Hospital due to the retrospective design of the present study.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the Topical Collection on Advanced Image Analyses (Radiomics and Artificial Intelligence)
Electronic supplementary material
ESM 1
(PDF 1055 kb)
Rights and permissions
About this article
Cite this article
Han, Y., Ma, Y., Wu, Z. et al. Histologic subtype classification of non-small cell lung cancer using PET/CT images. Eur J Nucl Med Mol Imaging 48, 350–360 (2021). https://doi.org/10.1007/s00259-020-04771-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00259-020-04771-5