Abstract
Despite the long-term interest in solventogenic clostridia-based ABE (acetone-butanol-ethanol) fermentation, clostridial butanol tolerance and its underlying mechanism remain poorly understood, which is a major obstacle hindering further improvements of this important fermentative process. In this study, a two-component system (TCS), BtrK/BtrR, was identified and demonstrated to positively regulate butanol tolerance and ABE solvent formation in Clostridium acetobutylicum, a representative species of solventogenic clostridia. The transcriptomic analysis results showed that BtrK/BtrR has a pleiotropic regulatory function, affecting a large number of crucial genes and metabolic pathways. Of the differentially expressed genes, btrTM, encoding a putative ABC-type transporter (named BtrTM), was shown to be under the direct control of BtrR, the response regulator of the BtrK/BtrR TCS. Furthermore, BtrTM was shown to contribute to more butanol tolerance (46.5% increase) by overexpression, revealing a novel regulatory mechanism consisting of the BtrK/BtrR TCS and the BtrTM transporter in C. acetobutylicum. Based on these findings, we achieved faster growth and solvent production of C. acetobutylicum by overexpressing BtrK/BtrR or its direct target BtrTM, although no significant improvement in the final butanol titer and yield. These results further confirm the importance of BtrK/BtrR and BtrTM in this organism. Also, of significance, a specific number of btrR-btrT-btrM-btrK-like gene clusters were identified in other Clostridium species, including the pathogens Clostridium perfringens and Clostridium botulinum, indicating a broad role for this regulatory module in the class Clostridia.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Solventogenic clostridia-based ABE (acetone-butanol-ethanol) solvent production is an important industrial fermentative process used worldwide (Darkwah et al. 2018; Lee et al. 2008), which still attracts great interest due to its potential economic competitiveness to the petrochemical production of acetone and butanol. Among the ABE solvents, butanol (i.e., n-butanol or 1-butanol) is regarded as a promising alternative fuel with better characteristics than ethanol (Schiel-Bengelsdorf et al. 2013). However, a major issue obstructing the high-level production of ABE solvents by clostridia is their poor ability to tolerate high butanol concentration, which has received a great deal of research interest (Patakova et al. 2018).
To realize more butanol tolerance of solventogenic clostridia, several strategies have been used, e.g., the overexpression of stress and heat shock–encoding genes and small RNAs (Jones et al. 2016; Liao et al. 2017; Mann et al. 2012; Ventura et al. 2013), repetitive evolutionary domestication and genome shuffling (Li et al. 2016; Liu et al. 2013), and medium optimization (Wu et al. 2016). Despite the progresses made using these approaches, how solventogenic clostridia sense butanol and then activate adaptive mechanisms to promote survival and maintain growth remains poorly understood, hindering the ability to efficiently modify strains for high butanol production.
Two-component systems (TCSs), which are normally composed of a membrane-bound histidine kinase and a corresponding response regulator, are the predominant signal transduction and regulatory systems used by bacteria to monitor, respond, and adapt to different environmental changes (Bekker et al. 2006; Skerker et al. 2005). In Bacillus subtilis, a representative Gram-positive bacterium, a large number of TCSs have been identified and shown to be involved in various physiological activities, e.g., the production of secondary metabolites (Martin 2004), cell division (Fukuchi et al. 2000), and adaptation to environmental stress (Darmon et al. 2002). However, for solventogenic clostridia, a large group of Gram-positive bacteria, few reports have described TCSs that are associated with crucial biological processes. Furthermore, only a single orphan histidine kinase (encoded by CAC3319) was observed to be associated with butanol tolerance in Clostridiumacetobutylicum through a comparative genomic analysis of C. acetobutylicum ATCC 55025 and its mutant strain JB200, but the mechanism through which this orphan kinase promotes butanol tolerance is unclear (Xu et al. 2015).
To date, numerous comparative transcriptomics and genomic analyses have been performed to elucidate butanol tolerance mechanisms in C. acetobutylicum (Alsaker et al. 2010; Borden and Papoutsakis 2007; Janssen et al. 2012; Schwarz et al. 2012; Tomas et al. 2004; Xu et al. 2017). In one of these studies, the CAC0863 gene, encoding a putative histidine kinase, was found to be significantly upregulated in the presence of added butanol (Schwarz et al. 2012). Interestingly, the transcription of this gene was also shown to be significantly upregulated with a transient butanol pulse in another study (Janssen et al. 2012). These findings indicate an involvement of the CAC0863-encoded kinase in the cellular response of C. acetobutylicum to butanol stress. Furthermore, this also raised a number of interesting questions, such as whether this kinase has a cognate response regulator protein to constitute a complete TCS, and if so, which genes or metabolic pathways are under the control of this TCS. The elucidation of these questions may uncover a novel regulatory mechanism used by C. acetobutylicum to cope with the stress from high butanol concentrations.
In this study, we performed a detailed investigation regarding the abovementioned issues. Through in vitro phosphorylation assays, the cognate response regulator of the CAC0863-encoded kinase was identified, demonstrating a paired two-component system (named BtrK/BtrR) in C. acetobutylicum ATCC 824. Subsequent biochemical and physiological experiments showed that BtrK/BtrR can directly regulate its adjacent putative ABC-type transporter BtrTM to exert improved growth under butanol stress. The overexpression of either BtrK/BtrR or BtrTM led to much faster growth and solvent formation in C. acetobutylicum ATCC 824. Furthermore, btrR-btrT-btrM-btrK-like gene clusters were identified in a number of other Clostridium species, including the pathogens Clostridium perfringens and Clostridium botulinum, indicating a broad role of this functional module in clostridial stress responses.
Materials and methods
Media and growth conditions
Escherichia coli strain Top10 was used as a host strain for gene cloning and plasmid construction. E. coli strains ER2275 and Rosetta (DE3) were used for plasmid methylation and protein purification, respectively. These E. coli strains were grown in Luria–Bertani (LB) medium or agar plate by adding appropriate quantity of antibiotics (100 μg/ml ampicillin, 50 μg/ml kanamycin, 25 μg/ml chloramphenicol, and 100 μg/ml spectinomycin) when needed.
Methylated plasmids were transferred into C. acetobutylicum ATCC 824 by electroporation. All the strains derived from C. acetobutylicum ATCC 824 was firstly inoculated into CGM medium (Wiesenborn et al. 1988) for inoculum preparation, and then transferred into P2 medium (Baer et al. 1987) for solvent production. Erythromycin (10 μg/ml) and thiamphenicol (8 μg/ml) were added to the P2 medium when needed.
Bacterial strains and plasmids
The plasmids and strains used in this study are listed in Table S3. All the C. acetobutylicum strains used are derivatives of C. acetobutylicum ATCC 824.
To purify the BtrK and BtrR protein, the btrK (CAC0863) and btrR (CAC0860) gene were PCR-amplified from the genome of C. acetobutylicum ATCC 824 by using the primers btrK-ex-for/btrK-ex-rev and btrR-ex-for/btrR-ex-rev, respectively. The resulting DNA fragment of btrK and btrR were digested with BamHI/XhoI and SacI/XhoI, respectively, and then ligated to pET28a and pQ8, yielding the plasmid pET28a-BtrK and pQ8-BtrR, respectively. These two plasmids were transformed into the E. coli Rosetta (DE3) strain for protein expression. The following purification method was the same as previously reported (Ren et al. 2012). Here, the 20 amino acid transmembrane domain at the 5′-end, which may adversely impact the purification of BtrK from the E. coli strain, was deleted in the purified His-BtrK.
The plasmid pIMP1-Pthl was constructed as previously reported (Xiao et al. 2011). To obtain the plasmid for btrK and btrR overexpression, the btrK and btrR gene were PCR-amplified from the genome of C. acetobutylicum ATCC 824 using the primers btrK-for/btrK-rev and btrR-for/btrR-rev, respectively. The resulting DNA fragments were digested and then inserted into the plasmid pIMP1-Pthl and pIMP1-Pptb (Xue et al. 2016), yielding the plasmid pIMP1-Pthl-btrK and pIMP1-Pptb-btrR, respectively.
The plasmid pIMP1-Pthl-btrR-Pthl-btrK was constructed as follows: the DNA fragments of btrR, Pthl, and btrK were PCR-amplified from the genome of C. acetobutylicum ATCC 824 using the primers listed in Table S4. Next, a large DNA fragment that contains btrR, Pthl, and btrK was obtained by overlapping PCR using the primers btrR-for-1 and btrK-rev-1. This DNA fragment was digested with BamHI/SmaI, and then inserted into the plasmid pIMP1-Pthl, yielding the plasmid pIMP1-Pthl-btrR-Pthl-btrK.
The plasmid pIMP1-Pptb-btrTM for btrTM overexpression was constructed as follows: the DNA fragment of btrTM was PCR-amplified from the genome using the primers listed in Table S2. The resulting DNA fragment was digested with BamHI/SmaI, and then inserted into the plasmid pIMP1-Pptb that was digested with the same restriction enzymes, yielding the plasmid pIMP1-Pptb-btrTM.
Inactivation of btrK and btrR was achieved by using the TargeTron method reported previously (Shao et al. 2007).
In vitro phosphorylation assay
Autophosphorylation assays were performed as follows: the reactions were performed in 20 μl kinase buffer (50 mM Tris-HCl (pH 8.0), 0.5 mM EDTA, 10 mM MgCl2,10 mM MnCl2, 50 μM ATP, 10% glycerol), in which 5 μM His-BtrK protein and 1 μl [γ-32P] ATP were included. The reactions were carried out at 30 °C for 0, 25, and 50 min, and then quenched by addition of 5 μl 5× SDS-PAGE loading buffer. Next, all the reactions were heated to 56 °C for 10 min. The phosphorylated proteins were resolved by 12% SDS-PAGE, and the gel was exposed to a phosphor screen (Starion FLA-9000 Scanner, Fujifilm, Japan) for 2 h at the room temperature.
Fermentations of Clostridium acetobutylicum strains
Inoculum preparation and fermentations of C. acetobutylicum ATCC 824 and its derivatives were carried out as previously reported (Zhang et al. 2018). In the fermentations, d-glucose (70 g/l) was used as the sole carbon source. Samples were removed from the medium at different time points and then stored at − 20 °C.
Analytical methods
OD600 was measured by spectrophotometer (DU730, Beckman Coulter). The concentration of acetone, butanol, and ethanol was determined as described previously (Ren et al. 2010). Isobutyl alcohol and isobutyric acid were used as the internal standards for solvents quantification.
Butanol tolerance assays
The wild-type and engineered strains of C. acetobutylicum ATCC 824 were inoculated into 5 ml liquid CGM medium and cultured anaerobically at 37 °C overnight. Then, 5 ml of inoculum was transferred into 50 ml CGM medium and grown for another 4 h until OD600 reached 0.8~1.0. Next, 2 ml of the grown cells were inoculated into 28 ml liquid CGM medium that contained no or 1% (vol./vol.) butanol for cultivation. The cell growth (OD600) was determined after 48-h fermentation. The relative butanol tolerance was calculated as the following equation: OD600 (with butanol addition)/OD600 (no butanol).
Here, the CGM medium (Wiesenborn et al. 1988) rather than P2 medium (Baer et al. 1987) was used for butanol tolerance assays because the former, as a nutrient-rich growth medium for C. acetobutylicum, is more suitable to observe cellular growth changes under butanol stress.
Microarray analysis and real-time qRT-PCR
The engineered C. acetobutylicum strain 824-btrK/R and the control strain (824-C) were grown in P2 medium (500 ml) using d-glucose as the sole carbon source. Erythromycin (10 μg/ml) was added into the medium when needed. Samples were taken at 24 and 48 h. The microarray analysis was performed as previously reported (Ren et al. 2012), in which the Agilent custom 60-mer oligonucleotide microarrays were used with a 60-mer oligonucleotide probe for each gene. Single-color microarray assays were carried out by Shanghai Biochip Co., Ltd. (Shanghai, China). The microarray data have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE138466. Genes that exhibited over 2-fold transcriptional change in the 824-btrK/R versus 824-C strain were considered to be significantly affected by the overexpression of btrK and btrR.
For real-time qRT-PCR analysis, total RNAs were extracted from the 824-btrK/R and 824-C strain using TRIzol (Invitrogen, Carlsbad, CA) and then purified using RNeasy™ cleanup kit (Qiagen, Inc., Valencia, CA). Next, RNAs were reverse transcribed to cDNA using the PrimeScript RT reagent kit (TaKaRa, cat. no. RR047A). qRT-PCR experiments were performed by using Bio-Rad iQ5 real-time PCR detection system (Bio-Rad, Palo Alto, USA). The reaction conditions were listed as follows: 95 °C for 2 min, followed by 40 cycles of 95 °C for 15 s, 55 °C for 20 s, and lastly 72 °C for 20 s. The CAC2679 gene (encoding a pullulanase) was used as the internal control according to the previous report (Alsaker et al. 2005).
Electrophoretic mobility shift assay
The Cy5-labeled probe (0.04 pmol) used for electrophoretic mobility shift assay (EMSA) was generated by a two-step PCR amplification as previously reported (Ren et al. 2012). Next, the probe was pre-incubated with the BtrR protein in a buffer containing 20 mM Tris-HCl (pH 7.9), 40 ng/ml bovine serum albumin (BSA), 5% glycerol, 10 mM MgCl2, 40 mM KCl, 0.25 mM DTT, and 50 ng/μl fish sperm DNA (Wu et al. 2015). The reaction mixture was incubated at 25 °C for 20 min and then loaded onto a 5% polyacrylamide gel. The gel was run in 0.5× TBE buffer at 120 V for 70 min in an ice bath. Finally, the gel was visualized by using a FLA-9000 Phosphorimager (Fujifilm, Japan).
Results
BtrR and BtrK constitute a paired two-component system in C. acetobutylicum
As previously mentioned, the results of the comparative transcriptomic analysis showed that the expression of CAC0863, encoding a histidine kinase BtrK, was significantly upregulated when C. acetobutylicum was exposed to an increased butanol concentration in the medium (Schwarz et al. 2012). It has been known that butanol is able to affect the membrane composition and fluidity of C. acetobutylicum (Baer et al. 1987; Vollherbst-Schneck et al. 1984; Wang et al. 2016). In addition, histidine kinases of TCSs often act as the sensors of membrane fluidity, and their transmembrane domains are essential for sensing membrane fluidity and controlling the changes between the kinase and phosphatase conformation (Albanesi et al. 2004; Fernandez et al. 2019; Hunger et al. 2004). Therefore, we speculated that BtrK, as a sensor histidine kinase, may be associated with the response and adaptation of C. acetobutylicum to butanol stress.
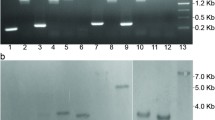
To characterize the role of BtrK, we first investigated whether this histidine kinase has a cognate response regulator. While scrutinizing the chromosomal regions adjacent to CAC0863, we noticed that the upstream CAC0860 gene is annotated as a putative response regulator–encoding gene (Fig. 1a). Given that histidine kinases (HKs) and response regulators (RRs) constituting TCSs are often closely located on the chromosome, we speculated that CAC0860 and CAC0863 constitute a paired TCS. To test this hypothesis, an in vitro phosphorylation assay was performed using the purified His6-labeled BtrK and MBP-labeled BtrR (Fig. 1b). Importantly, to ensure the release of BtrK from the plasma membrane for purification, BtrK was expressed without its transmembrane domain (20 amino acids at the 5′ end) (Fig. S1). As expected, the results of the phosphorylation assay showed that BtrK can autophosphorylate itself in the presence of ATP (Fig. 1c), and subsequently phosphorylate BtrR (Fig. 1c). These data demonstrate that BtrK and BtrR form a paired TCS in C. acetobutylicum. Interestingly, the homologs of BtrK and BrtR were found to be widespread in clostridia, indicating their broad roles in the Clostridium genus. This will be elaborated in the subsequent section.
Transphosphorylation between BtrK and BtrR. a Locations of btrR and btrK in the genome of C. acetobutylicum. CAC0859: anti-SigV factor; CAC0860: two-component response regulator; CAC0861: multidrug ABC transporter ATPase; CAC0862: transmembrane protein; CAC0863: histidine kinase; CAC0864: histidine kinase-like ATPase; CAC0865: two-component response regulator. b Purification of His6-BtrK and MBP-BtrR. MK, molecular mass markers. c Autophosphorylation of BtrK and phosphoryl transfer between BtrK and BtrR. K: BtrK; R: BtrR
BtrK/BtrR regulates the butanol tolerance and fermentation performance of C. acetobutylicum
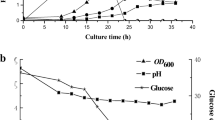
We subsequently investigated whether BtrK/BtrR plays a role in butanol tolerance in C. acetobutylicum. Since BtrK expression was upregulated in the presence of butanol stress, we simultaneously overexpressed btrK and btrR under the control of a constitutive promoter in wild-type C. acetobutylicum (Fig. 2a). As shown in Fig. 2b, the btrK/btrR-overexpression strain (824-btrK/R) reached a much higher biomass (OD600) and butanol tolerance than the control strain (824-C) with 1% (vol./vol.) butanol added to the medium, suggesting a positive impact of the BtrK/BtrR TCS on the growth of C. acetobutylicum under butanol stress. Additionally, btrK and btrR overexpression resulted in much faster growth and solvent synthesis of the 824-btrK/R strain compared with the 824-C strain, although their biomasses and ABE solvent titers were similar at the end of the fermentation process (Fig. 2c). These findings further suggest that BtrK/BtrR plays an important role in C. acetobutylicum.
Influence of the overexpression of both btrR and btrK on butanol tolerance, growth, and solvent production in C. acetobutylicum. a Map of the plasmid overexpressing btrR and btrK. Pthl, thl promoter. b Changes in butanol tolerance in C. acetobutylicum after overexpressing btrR and btrK. 824-C: the C. acetobutylicum strain carrying the control plasmid; 824-btrK/R: the C. acetobutylicum strain carrying the btrR/btrK-overexpressing plasmid. Cell growth (OD600) was measured after 24 h and 49 h of fermentation using CGM medium. A 1% (vol./vol.) of butanol was added into the medium when needed. The butanol tolerance was calculated as the following equation: OD600 (with butanol addition)/OD600 (no butanol). The data are presented as the means ± standard deviations calculated from triplicate independent experiments (*p < 0.05; ***p < 0.001; t test). c Fermentation profiles of the btrR/btrK-overexpressing strain (824-btrK/R) and the control strain (824-C). The data are presented as the means ± standard deviations calculated from triplicate independent experiments (*p < 0.05;**p < 0.01; ***p < 0.001; t test)
We also inactivated btrR and btrK separately (Fig. 3a) to examine the resulting phenotypic changes in C. acetobutylicum. As shown in Fig. 3 b and c, inactivation of either btrK (824m-btrK-P, the btrK-disrupted strain 824m-btrK carrying an empty plasmid pIMP1-Pthl) or btrR (824m-btrR-P, the btrR-disrupted strain 824m-btrR carrying an empty plasmid pIMP1-Pptb) resulted in markedly impaired growth and solvent production in C. acetobutylicum. These phenotypic changes could be completely or largely restored through the plasmid-based expression of btrK or btrR in these mutants (824m-btrK-C and 824m-btrR-C).
Influence of btrR or btrK disruption on the cellular performance of C. acetobutylicum. a PCR screening of the desired mutants with btrR or btrK disruption. 824m-btrR: the btrR-disrupted strain; 824m-btrK: the btrK-disrupted strain. b Fermentation profiles of the btrK-disrupted mutant and its genetic complementation. 824-Pthl: the C. acetobutylicum strain carrying an empty plasmid pIMP1-Pthl; 824m-btrK-P: the btrK-disrupted strain (824m-btrK) carrying an empty plasmid pIMP1-Pthl; 824m-btrK-C: the btrK-disrupted strain (824m-btrK) carrying the pIMP1-Pthl-btrK plasmid for btrK expression. c Fermentation profiles of the btrR-disrupted mutant and its genetic complementation. 824-Pptb: the C. acetobutylicum strain carrying an empty plasmid pIMP1-Pptb; 824m-btrR-P: the btrR-disrupted strain (824m-btrR) carrying an empty plasmid pIMP1-Pptb; 824m-btrR-C: the btrR-disrupted strain (824m-btrR) carrying the plasmid pIMP1-Pptb-btrR for btrR expression. The means ± standard deviations between the control strain and btrK or btrR-disrupted strain were calculated from two independent experiments (*p < 0.05; **p < 0.01; ***p < 0.001; t test)
Identification of putative genes under the regulation of BtrK/BtrR
To identify the putative genes under the regulation of the response regulator BtrR, we performed a comparative transcriptomic analysis of the abovementioned 824-btrK/R and 824-C strains. As shown in Fig. 4a, samples for RNA extraction and microarray analysis were collected after 24 and 48 h of fermentation. From this analysis, 691 and 424 genes exhibited significantly altered transcription (fold change ≥ 2.0) after the overexpression of btrK/btrR at 24 and 48 h, respectively (Fig. 4b and Tables S1 and S2). Among these genes, 153 genes showed significantly different expression at both 24 and 48 h (Fig. 4b). These findings indicate a pleiotropic regulatory role for BtrR in C. acetobutylicum.
Overview of the genes affected by the co-overexpression of btrR and btrK. a Growth curve of the BtrK/BtrR-overexpressing (824-btrK/R) strain and the control strain (824-C). Samples were taken at 24 and 48 h for microarray assays. Data are means ± standard deviations calculated from two independent experiments. b The genes that showed altered expression (≥ 2-fold) after BtrK/BtrR overexpression. c Functional categories of the differentially expressed genes (fold change ≥ 2.0) after BtrK/BtrR overexpression. PTM, post-translational modification
All of the differentially expressed genes at 24 and 48 h, except those with unknown functions, can be generally grouped into 15 and 16 subsets, respectively, according to their predicted functions (Fig. 4c). Many genes that are known or potentially associated with important physiological characteristics of C. acetobutylicum showed significantly altered expression, e.g., those with functions in sporulation (CAC0686, CAC0859, CAC1276, CAC1689, CAC1694, CAC1713, CAC2780, CAC2898, CAC2908, CAC3205, CAC3244, and CAC3731) and solvent synthesis (CAP0035, CAP0059, CAP0078, and CAC3484). It should be noted that in C. acetobutylicum, some sporulation-related genes have been previously shown to have the potential relationship to butanol stress (Alsaker et al. 2004; Borden and Papoutsakis 2007). Besides, we compared the expressional changes (> 5-fold) of several solvent synthesis–related genes in this study with previously reported microarray data and found that their changes are not totally consistent. For example, the CAP0035 gene, encoding a aldehyde/alcohol dehydrogenase (AdhE1) that are responsible for alcohol production in C. acetobutylicum, was previously shown to be upregulated under butanol stress (Alsaker et al. 2004; Janssen et al. 2012), but was greatly downregulated (11-fold downregulation, 24 h; 14-fold downregulation, 48 h) in this study; another gene, CAP0078 (encoding acetyl-CoA acetyltransferase), also known to be associated with solvent synthesis in C. acetobutylicum, was significantly downregulated (20-fold downregulation, 24 h) after btrK/btrR overexpression in this study, whereas no obvious changes under butanol stress in the previous report (Janssen et al. 2012). All these findings suggest a complex regulatory mechanism of C. acetobutylicum in response to butanol stress.
An ABC-type transporter under the direct control of BtrR has a role in butanol tolerance in C. acetobutylicum
Among the genes with significantly altered expression after btrK/btrR overexpression, we noticed two genes (CAC0861 and CAC0862), encoding a putative ABC-type transporter (named BtrTM in this study) that contains a transmembrane protein and an ATPase component, located between btrR and btrK in the chromosome (Fig. 1a). A real-time qRT-PCR analysis showed a 1.52- and 2.23-fold increase in the transcription of btrTM at 24 and 48 h, respectively, after the overexpression of btrK/btrR (Fig. 5a). To further verify whether the btrTM genes are under the direct control of the response regulator BtrR, electrophoretic mobility shift assays (EMSAs) were performed using the purified BtrR protein and a DNA fragment spanning the entire noncoding region upstream of btrTM (PbtrTM). As shown in Fig. 5b, a substantial DNA band shift was observed for the PbtrTM fragment after adding the BtrR protein, which could be suppressed by the addition of an unlabeled DNA competitor but not a nonspecific competitor. These data demonstrate that BtrR can bind to the noncoding region upstream of btrTM, suggesting that BtrR directly controls the transcription of btrTM.
Direct regulation of BtrR on the ABC-type transporter BtrTM (CAC0861-0862) and identification of the BtrR-binding site. a The difference of btrTM expression between the 824-btrK/R (BtrK/BtrR overexpression) and 824-C strain (control). The data are presented as the means ± standard deviations calculated from two independent experiments (*p < 0.05; **p < 0.01; t test). b EMSAs for verifying the in vitro binding of the BtrR protein and the promoter region (PbtrTM) of the btrTM genes using 0 to 0.4 μM BtrR. The final concentration of Cy5-labeled PbtrTM was 0.04 pM. The DNA fragment of CAC1790 (120 μM) was used as a nonspecific competitor. Spec. comp., specific competitor. Nonspec. comp., nonspecific competitor. c Truncation of P-I and EMSAs for investigating the in vitro BtrR-binding to P-II, P-III, and P-IV. The region containing the potential binding site (green box) was indicated by the double arrow line (red). d Detection and verification of the BtrR-binding site in the truncated fragment from P-II by EMSAs, in which CCTTAATGTACATATAAG was mutated to GGGGGGGGGACATATAAG
Next, to identify the potential BtrR-binding sites within the noncoding region upstream of btrTM (P-I) (Fig. 5c), this region was gradually truncated, yielding the fragments P-II, P-III, and P-IV as probes for EMSAs. As shown in Fig. 5c, an obviously shifted band was observed in assays with P-I and P-II, but this band was eliminated when P-III and P-IV were used, thereby suggesting that a BtrR-binding site resides in the truncated region from P-II to P-III. Since this truncated fragment is only 40 bp long (indicated by the red double arrow line) (Fig. 5c), we visually scanned it and fortunately found an 18-nt imperfect palindromic sequence (CCTTAATGTACATATAAG). To further examine whether this sequence is a BtrR-binding site, it was mutated, resulting in a derived P-II-mu fragment that was used as a DNA probe for EMSAs (Fig. 5d). As expected, the mutation completely abolished the binding between BtrR and the DNA probe (Fig. 5d), thereby verifying the role of this 18-nt sequence.
Based on the above findings, we next investigated whether the ABC-type transporter BtrTM has a role in butanol tolerance in C. acetobutylicum. Because the btrTM genes showed higher expression level in the 824-btrK/R strain compared with the control strain 824-C (Fig. 5a), we overexpressed these two genes simultaneously in the wild-type C. acetobutylicum to evaluate any changes in butanol tolerance. As expected, compared with the 824-P strain (the C. acetobutylicum strain carrying the plasmid pIMP1-Pptb), the 824-btrTM strain (the C. acetobutylicum strain carrying the plasmid pIMP1-Pptb-btrTM for btrTM overexpression) showed increased growth and butanol tolerance (46.5% increase) in the presence of butanol stress (1%, vol./vol.) (Fig. 6a), consisting with the results from the btrK/btrR-overexpression analysis (Fig. 2b). Furthermore, when cultivated in medium without added butanol, the 824-btrTM strain also grew and produced solvents much faster than the 824-P strain (Fig. 6b).
Influence of btrTM overexpression on butanol tolerance, growth, and solvent production in C. acetobutylicum. a Changes in the butanol tolerance of C. acetobutylicum after overexpressing btrT and btrM. 824-P: the C. acetobutylicum strain carrying the plasmid pIMP1-Pptb; 824-btrTM: the C. acetobutylicum strain carrying the plasmid pIMP1-Pptb-btrTM for btrTM overexpression. The cell growth (OD600) was measured after 48 h of fermentation using CGM medium. A 1% (vol./vol.) of butanol was added into the medium when needed. The butanol tolerance was calculated as the following equation: OD600 (with butanol addition)/OD600 (no butanol). The data are presented as the means ± standard deviations calculated from two independent experiments (*p < 0.05; ***p < 0.001; t test). b The fermentation profiles of the 824-P and 824-btrTM strains. The data are presented as the means ± standard deviations calculated from triplicate independent experiments (*p < 0.05; **p < 0.01; ***p < 0.001; t test)
In summary, these data, together with results regarding the influence of BtrK/BtrR on butanol tolerance and cellular performance (Figs. 2 and 3), suggest that BtrK/BtrR and their direct regulation of the ABC transporter–encoding genes btrTM form a functional module that plays a role in the response and tolerance of C. acetobutylicum to butanol stress.
btrR-btrT-btrM-btrK-like gene clusters are present in other Clostridium species
TCSs and ABC transporters are often located in closely associated bacterial chromosomes and form a functional module to respond to different stimuli (Dintner et al. 2011). Therefore, we assessed whether btrR-btrT-btrM-btrK-like gene clusters are also present in other Clostridium species. Based on the amino acid sequences of BtrK/BtrR and BtrTM, we searched the genomes of many other representative Clostridium species using the BLASTp, and interestingly, a number of homologs with high amino acid sequence identity and similar gene arrangements as the btrR-btrT-btrM-btrK gene cluster were identified (Fig. 7). Notably, this gene cluster occurred in two pathogenic Clostridium species (C. perfringens and C. botulinum), indicating a broad role of this TCS-based regulatory module for stress response and adaptation in clostridia.
Discussion
In this study, a functional module consisting of a two-component system (BtrK/BtrR) and a two-gene ABC-type transporter (BtrTM) was identified and shown to play an important role in butanol tolerance of the solventogenic bacterium C. acetobutylicum. The comparative transcriptomics analysis revealed that BtrK/BtrR has a pleiotropic regulatory function, affecting numerous genes and metabolic pathways. Furthermore, we revealed that BtrK/BtrR directly and positively regulates the expression of btrTM, ABC-type transporter-encoding genes, which consequently led to improved butanol tolerance, growth, and solvent synthesis in C. acetobutylicum.
To date, multiple methods by which bacteria counter butanol stress have been uncovered, among which efflux pumps, including ABC-type transporters, play important roles (Patakova et al. 2018). ABC-type transporters are a group of membrane proteins that mediate diverse ATP-driven transport processes (Davidson et al. 2008; Locher 2016). These transporters are highly dynamic membrane proteins capable of extruding numerous substances from the cytosol (Rees et al. 2009), and thereby associated with many crucial cellular processes (Hofmann et al. 2019; Robey et al. 2018; Trowitzsch and Tampe 2018). Among Clostridium species, a few examples of ABC transporters associated with phenolic compound tolerance and multidrug resistance have been reported in industrial C. beijerinckii and pathogenic C. difficile (Liu et al. 2018; Ngernsombat et al. 2017), respectively, although a native butanol tolerance-related ABC transporter remains unobserved. In this study, the comparative transcriptomic analysis revealed various ABC transporters potentially related to butanol tolerance, in which the BtrTM-coding genes (CAC0861-0862) showed a significant upregulation upon butanol stress. The BtrTM proteins represent an ABC-type 2 transporter, consisting of a membrane protein and an ATP-binding protein (Filippova et al. 2014), which should belong to the multidrug resistance (MDR) transporter systems based on the common characteristics (Martinez et al. 2009). Since MDR transporter systems have been known to act as solvent-extruding pumps (Martinez et al. 2009), a possible hypothesis for the function of BtrTM is that it helps in alcohol (ethanol and butanol) excretion from C. acetobutylicum cells. Actually, we performed an investigation of the role of BtrTM in transporting substrates by searching for homologs in other bacteria; however, no definitive annotations were uncovered, despite the identification of some homologs with high amino acid sequence identity to BtrTM in the NCBI database. This result indicates that BtrTM and its homologs may belong to a new category of ABC transporters with functions that remain to be elucidated.
The expression of the btrR-btrT-btrM-btrK cluster (CAC0860–CAC0863) have been found to be significantly upregulated in the presence of butanol stress in the previous studies (Schwarz et al. 2012; Janssen et al. 2012). Here, a further screening revealed that the btrR-btrT-btrM-btrK-like gene clusters, encoding a TCS and an ABC-type transporter, are present in multiple Clostridium species. Interestingly, some TCSs have been observed to be chromosomally located adjacent to ABC-type transporters in bacteria, and these TCSs contribute to cellular resistance to different stresses (e.g., antibodies and bacitracin) by regulating the expression of the adjacent transporters (Dintner et al. 2011; Meehl et al. 2007). Therefore, it seems that such a TCS-transporter module on the chromosome may enable bacteria to initiate response and adaptation mechanisms as quickly as possible upon sensing external signals. In some cases, bacteria adopt coevolved TCSs and ABC transporters to respond to environmental stresses. For example, the B. subtilis protein BceS, a histidine kinase without an extracellular signal-sensing domain, can activate its cognate response regulator BceR with the assistance of the adjacently encoded ABC transporter BceAB. In turn, the phosphorylated BceR can activate the expression of BceAB, allowing B. subtilis cells to tolerate bacitracin (Dintner et al. 2011). Unlike the above example, a distinct feature of BtrK in the “btrR-btrT-btrM-btrK” module is that it contains a large extracellular domain (Fig. S1), indicating that BtrK is capable of independently sensing external signals.
In this study, the induction of more butanol tolerance in C. acetobutylicum, either by overexpressing BtrK/BtrR or BtrTM, was observed, although no significant increase was observed in the final concentration of butanol and the other two assayed solvents (Figs. 2 and 6). This finding further suggests that high butanol tolerance is not in total accord with high butanol production in C. acetobutylicum, a phenomenon that has been observed in some previous studies (Jones et al. 2016; Mann et al. 2012). Based on the current understanding of butanol tolerance, it seems that butanol synthesis and butanol tolerance are two characteristics that are not tightly linked. However, the generation of a strain with high butanol tolerance represents a good chassis for further improvements to hopefully generate strains with high butanol production. Besides, considering that the metabolic energy for substrate transport by ABC-type transporters is derived from ATP binding and hydrolysis, the overexpression of BtrTM is very likely to cause extra energy consumption and metabolic burden in the C. acetobutylicum cells, which may partly explain why there were no increases in biomass and butanol titer despite the enhanced butanol tolerance after overexpressing BtrTM (Fig. 6b).
In addition, it should be noted that the TCS BtrK/BtrR has a pleiotropic regulatory role in C. acetobutylicum, according to the results of the comparative transcriptomic analysis. A large number of genes exhibited significant changes in transcription after the overexpression of BtrK/BtrR. Therefore, it is highly possible that BtrR has other direct targets besides the ABC transporter BtrTM that have roles in butanol tolerance. To confirm this hypothesis, high-throughput screening techniques, e.g., ChIP-Seq, are necessary.
In summary, in this study, we identified a butanol tolerance-related module that is composed of a two-component pleiotropic regulatory system (BtrK/BtrR) and a novel two-gene ABC-type transporter (BtrTM) in the representative solventogenic C. acetobutylicum. The positive regulation of BtrK/BtrR on BtrTM provides an explanation for the improved growth of C. acetobutylicum under butanol stress after the overexpression of BtrK/BtrR. To the best of our knowledge, BtrK/BtrR is the first reported paired TCS that exerts a pleiotropic regulatory function and is responsible for butanol tolerance in C. acetobutylicum, suggesting a previously uncharacterized role of TCSs in this important anaerobic bacterium. Moreover, btrR-btrT-btrM-btrK-like homologs are present in many other Clostridium species, including pathogens, indicating a broad role of this module in stress response and adaptation in clostridia.
References
Albanesi D, Mansilla MC, de Mendoza D (2004) The membrane fluidity sensor DesK of Bacillus subtilis controls the signal decay of its cognate response regulator. J Bacteriol 186:2655–2663
Alsaker KV, Spitzer TR, Papoutsakis ET (2004) Transcriptional analysis of spo0A overexpression in Clostridium acetobutylicum and its effect on the cell’s response to butanol stress. J Bacteriol 186:1959–1971
Alsaker KV, Paredes CJ, Papoutsakis ET (2005) Design, optimization and validation of genomic DNA microarrays for examining the Clostridium acetobutylicum transcriptome. Biotechnol Bioprocess Eng 10:432–443
Alsaker KV, Paredes C, Papoutsakis ET (2010) Metabolite stress and tolerance in the production of biofuels and chemicals: gene-expression-based systems analysis of butanol, butyrate, and acetate stresses in the anaerobe Clostridium acetobutylicum. Biotechnol Bioeng 105:1131–1147
Baer SH, Blaschek HP, Smith TL (1987) Effect of butanol challenge and temperature on lipid composition and membrane fluidity of butanol-tolerant Clostridium acetobutylicum. Appl Environ Microbiol 53(12):2854–2861
Bekker M, Teixeira de Mattos MJ, Hellingwerf KJ (2006) The role of two-component regulation systems in the physiology of the bacterial cell. Sci Prog 89(Pt 3–4):213–242
Borden JR, Papoutsakis ET (2007) Dynamics of genomic-library enrichment and identification of solvent tolerance genes for Clostridium acetobutylicum. Appl Environ Microbiol 73:3061–3068
Darkwah K, Nokes SE, Seay JR, Knutson BL (2018) Mechanistic simulation of batch acetone-butanol-ethanol (ABE) fermentation with in situ gas stripping using Aspen Plus (TM). Bioprocess Biosyst Eng 41(9):1283–1294
Darmon E, Noone D, Masson A, Bron S, Kuipers OP, Devine KM, van Dijl JM (2002) A novel class of heat and secretion stress-responsive genes is controlled by the autoregulated CssRS two-component system of Bacillus subtilis. J Bacteriol 184(20):5661–5671
Davidson AL, Dassa E, Orelle C, Chen J (2008) Structure, function, and evolution of bacterial ATP-binding cassette systems. Microbiol Mol Biol Rev 72:317–364
Dintner S, Staron A, Berchtold E, Petri T, Mascher T, Gebhard S (2011) Coevolution of ABC transporters and two-component regulatory systems as resistance modules against antimicrobial peptides in Firmicutes Bacteria. J Bacteriol 193(15):3851–3862
Fernandez P, Porrini L, Albanesi D, Abriata LA, Dal Peraro M, de Mendoza D, Mansilla MC (2019) Transmembrane prolines mediate signal sensing and decoding in Bacillus subtilis DesK histidine kinase. mBio 10(6):e02564-19
Filippova EV, Tkaczuk KL, Chruszcz M, Xu X, Savchenko A, Edwards A, Minor W (2014) Structural characterization of the putative ABC-type 2 transporter from Thermotoga maritima MSB8. J Struct Funct Genom 15(4):215–222
Fukuchi K, Kasahara Y, Asai K, Kobayashi K, Moriya S, Ogasawara N (2000) The essential two-component regulatory system encoded by yycF and yycG modulates expression of the ftsAZ operon in Bacillus subtilis. Microbiology 146:1573–1583
Hofmann S, Januliene D, Mehdipour AR, Thomas C, Stefan E, Bruchert S, Kuhn BT, Geertsma ER, Hummer G, Tampe R, Moeller A (2019) Conformation space of a heterodimeric ABC exporter under turnover conditions. Nature 571:580–583
Hunger K, Beckering CL, Marahiel MA (2004) Genetic evidence for the temperature-sensing ability of the membrane domain of the Bacillus subtilis histidine kinase DesK. FEMS Microbiol Lett 230:41–46
Janssen H, Grimmler C, Ehrenreich A, Bahl H, Fischer RJ (2012) A transcriptional study of acidogenic chemostat cells of Clostridium acetobutylicum—solvent stress caused by a transient n-butanol pulse. J Biotechnol 161:354–365
Jones AJ, Venkataramanan KP, Papoutsakis T (2016) Overexpression of two stress-responsive, small, non-coding RNAs, 6S and tmRNA, imparts butanol tolerance in Clostridium acetobutylicum. FEMS Microbiol Lett 363(8):fnw063
Lee SY, Park JH, Jang SH, Nielsen LK, Kim J, Jung KS (2008) Fermentative butanol production by Clostridia. Biotechnol Bioeng 101(2):209–228
Li SB, Qian Y, Liang ZW, Guo Y, Zhao MM, Pang ZW (2016) Enhanced butanol production from cassava with Clostridium acetobutylicum by genome shuffling. World J Microbiol Biotechnol 32:53
Liao ZP, Zhang YN, Luo S, Suo YK, Zhang SZ, Wang JF (2017) Improving cellular robustness and butanol titers of Clostridium acetobutylicum ATCC 824 by introducing heat shock proteins from an extremophilic bacterium. J Biotechnol 252:1–10
Liu XB, Gu QY, Yu XB (2013) Repetitive domestication to enhance butanol tolerance and production in Clostridium acetobutylicum through artificial simulation of bio-evolution. Bioresour Technol 130:638–643
Liu J, Lin QL, Chai XY, Luo YC, Guo T (2018) Enhanced phenolic compounds tolerance response of Clostridium beijerinckii NCIMB 8052 by inactivation of Cbei_3304. Microb Cell Factories 17(1):35
Locher KP (2016) Mechanistic diversity in ATP-binding cassette (ABC) transporters. Nat Struct Mol Biol 23:487–493
Mann MS, Dragovic Z, Schirrmacher G, Lutke-Eversloh T (2012) Over-expression of stress protein-encoding genes helps Clostridium acetobutylicum to rapidly adapt to butanol stress. Biotechnol Lett 34(9):1643–1649
Martin JF (2004) Phosphate control of the biosynthesis of antibiotics and other secondary metabolites is mediated by the PhoR-PhoP system: an unfinished story. J Bacteriol 186(16):5197–5201
Martinez JL, Sanchez MB, Martinez-Solano L, Hernandez A, Garmendia L, Fajardo A, Alvarez-Ortega C (2009) Functional role of bacterial multidrug efflux pumps in microbial natural ecosystems. FEMS Microbiol Rev 33:430–449
Meehl M, Herbert S, Gotz F, Cheung A (2007) Interaction of the GraRS two-component system with the VraFG ABC transporter to support vancomycin-intermediate resistance in Staphylococcus aureus. Antimicrob Agents Chemother 51(8):2679–2689
Ngernsombat C, Sreesai S, Harnvoravongchai P, Chankhamhaengdecha S, Janvilisri T (2017) CD2068 potentially mediates multidrug efflux in Clostridium difficile. Sci Rep 7(1):9982
Patakova P, Kolek J, Sedlar K, Koscova P, Branska B, Kupkova K, Paulova L, Provaznik I (2018) Comparative analysis of high butanol tolerance and production in clostridia. Biotechnol Adv 36(3):721–738
Rees DC, Johnson E, Lewinson O (2009) ABC transporters: the power to change. Nat Rev Mol Cell Biol 10:218–227
Ren C, Gu Y, Hu SY, Wu Y, Wang P, Yang YL, Yang C, Yang S, Jiang WH (2010) Identification and inactivation of pleiotropic regulator CcpA to eliminate glucose repression of xylose utilization in Clostridium acetobutylicum. Metab Eng 12(5):446–454
Ren C, Gu Y, Wu Y, Zhang WW, Yang C, Yang S, Jiang WH (2012) Pleiotropic functions of catabolite control protein CcpA in Butanol-producing Clostridium acetobutylicum. BMC Genomics 13:349
Robey RW, Pluchino KM, Hall MD, Fojo AT, Bates SE, Gottesman MM (2018) Revisiting the role of ABC transporters in multidrug-resistant cancer. Nat Rev Cancer 18:452–464
Schiel-Bengelsdorf B, Montoya J, Linder S, Durre P (2013) Butanol fermentation. Environ Technol 34(13–16):1691–1710
Schwarz KM, Kuit W, Grimmler C, Ehrenreich A, Kengen SW (2012) A transcriptional study of acidogenic chemostat cells of Clostridium acetobutylicum—cellular behavior in adaptation to n-butanol. J Biotechnol 161(3):366–377
Shao L, Hu SY, Yang Y, Gu Y, Chen J, Yang YL, Jiang WH, Yang S (2007) Targeted gene disruption by use of a group II intron (targetron) vector in Clostridium acetobutylicum. Cell Res 17(11):963–965
Skerker JM, Prasol MS, Perchuk BS, Biondi EG, Laub MT (2005) Two-component signal transduction pathways regulating growth and cell cycle progression in a bacterium: a system-level analysis. PLoS Biol 3(10):e334
Tomas CA, Beamish J, Papoutsakis ET (2004) Transcriptional analysis of butanol stress and tolerance in Clostridium acetobutylicum. J Bacteriol 186:2006–2018
Trowitzsch S, Tampe R (2018) ABC transporters in dynamic macromolecular assemblies. J Mol Biol 430:4481–4495
Ventura JR, Hu H, Jahng D (2013) Enhanced butanol production in Clostridium acetobutylicum ATCC 824 by double overexpression of 6-phosphofructokinase and pyruvate kinase genes. Appl Microbiol Biotechnol 97(16):7505–7516
Vollherbst-Schneck K, Sands JA, Montenecourt BS (1984) Effect of butanol on lipid composition and fluidity of Clostridium acetobutylicum ATCC 824. Appl Environ Microbiol 47:193–194
Wang YF, Tian J, Ji ZH, Song MY, Li H (2016) Intracellular metabolic changes of Clostridium acetobutylicum and promotion to butanol tolerance during biobutanol fermentation. Int J Biochem Cell Biol 78:297–306
Wiesenborn DP, Rudolph FB, Papoutsakis ET (1988) Thiolase from Clostridium acetobutylicum ATCC 824 and its role in the synthesis of acids and solvents. Appl Environ Microbiol 54(11):2717–2722
Wu Y, Yang YP, Ren C, Yang C, Yang S, Gu Y, Jiang WH (2015) Molecular modulation of pleiotropic regulator CcpA for glucose and xylose coutilization by solvent-producing Clostridium acetobutylicum. Metab Eng 28:169–179
Wu YD, Xue C, Chen LJ, Yuan WJ, Bai FW (2016) Synergistic effect of calcium and zinc on glucose/xylose utilization and butanol tolerance of Clostridium acetobutylicum. FEMS Microbiol Lett 363(5):fnw023
Xiao H, Gu Y, Ning YY, Yang YL, Mitchell WJ, Jiang WH, Yang S (2011) Confirmation and elimination of xylose metabolism bottlenecks in glucose phosphoenolpyruvate-dependent phosphotransferase system-deficient Clostridium acetobutylicum for simultaneous utilization of glucose, xylose, and arabinose. Appl Environ Microbiol 77(22):7886–7895
Xu MM, Zhao JB, Yu L, Tang IC, Xue C, Yang ST (2015) Engineering Clostridium acetobutylicum with a histidine kinase knockout for enhanced n-butanol tolerance and production. Appl Microbiol Biotechnol 99(2):1011–1022
Xu MM, Zhao JB, Yu L, Yang ST (2017) Comparative genomic analysis of Clostridium acetobutylicum for understanding the mutations contributing to enhanced butanol tolerance and production. J Biotechnol 263:36–44
Xue Q, Yang YP, Chen J, Chen L, Yang S, Jiang WH, Gu Y (2016) Roles of three AbrBs in regulating two-phase Clostridium acetobutylicum fermentation. Appl Microbiol Biotechnol 100(21):9081–9089
Zhang L, Liu YQ, Yang YP, Jiang WH, Gu Y (2018) A novel dual-cre motif enables two-way autoregulation of CcpA in Clostridium acetobutylicum. Appl Environ Microbiol 84(8):e00114-18
Funding
This work was supported by the National Natural Science Foundation of China (31630003 and 31921006), and Open Funding Project of the State Key Laboratory of Bioreactor Engineering.
Author information
Authors and Affiliations
Contributions
W. J. and Y. G. designed the research; Y. Y., N. L., and L. Z. performed the research; H. W. analyzed the data; and Y.Y., W. J., and Y. G. wrote the paper.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest. This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(PDF 328 kb)
Rights and permissions
About this article
Cite this article
Yang, Y., Lang, N., Zhang, L. et al. A novel regulatory pathway consisting of a two-component system and an ABC-type transporter contributes to butanol tolerance in Clostridium acetobutylicum. Appl Microbiol Biotechnol 104, 5011–5023 (2020). https://doi.org/10.1007/s00253-020-10555-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-020-10555-6