Abstract
Chemotaxis towards root exudates and subsequent biofilm formation are very important for root colonization and for providing the beneficial functions of plant growth-promoting rhizobacteria (PGPRs). In this study, in comparison with other root-secreted compounds, d-galactose in the root exudates of cucumber was found to be a strong chemoattractant at the concentration of 1 μM for Bacillus velezensis SQR9. Chemotaxis assays with methyl-accepting chemotaxis proteins (MCPs) deletion strains demonstrated that McpA was solely responsible for chemotaxis towards d-galactose. Interestingly, d-galactose significantly enhanced the biofilm formation of SQR9 in an McpA-dependent manner. Further experiment showed that d-galactose also enhanced root colonization by SQR9. In addition, the secretion of d-galactose by cucumber roots could be induced by inoculation with SQR9, indicating that d-galactose may be an important signal in the interaction between plant and SQR9. These findings suggested that the root-secreted d-galactose was a signal, the secretion of which was induced by the beneficial bacteria, and which in turn induced colonization of the bacteria.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Rhizosphere colonization is important for plant growth-promoting rhizobacteria (PGPRs) to provide their beneficial functions to the plant host (Philippot et al. 2013). The root colonization of PGPR is an integrated process comprising chemotaxis from the rhizosphere to the root surface and the subsequent biofilm formation on the root surface (Belas 2014). A class of low molecular weight compounds in the plant root exudates induce the chemotactic movement of PGPR to the root surface and the biofilm formation of PGPR (de Weert et al. 2002; Yaryura et al. 2008; Haichar et al. 2014).
The composition of root exudates varies depending on the plant species and the microbes in the environment. For instance, the low molecular weight organic acids in the root exudates of tomato, cucumber, and Arabidopsis differ in both composition and abundance (Lugtenberg et al. 1999; de Weert et al. 2002; Badri et al. 2013). Arabidopsis roots secreted more d-malic acid upon infection by Pseudomonas syringae pv tomato (Rudrappa et al. 2008). In many cases, the change of root exudation is an active response of plants to the complex rhizosphere and plays an important role in regulating the behavior of rhizobacteria, such as the chemotaxis towards the root surface (Yaryura et al. 2008).
Rhizospheric Bacillus could employ methyl-accepting chemotaxis proteins (MCPs) to sense the root-secreted chemoattractants. Bacillus uses a chemosensory pathway involving the CheA histidine kinase and the CheY response regulator to transmit signals from the receptors to the flagellar motors (Hess et al. 1988; Rao et al. 2008). After ligand binding to the bacterial chemoreceptor, CheY is rapidly phosphorylated in response to the change of the chemoeffector levels and subsequently change forms of bacterial movement (Rao et al. 2008; Glekas et al. 2012).
Typical chemoreceptors comprise a ligand-binding domain (LBD) at the N-terminus, which is frequently located in the extracytoplasmic space and is responsible for binding extracellular compounds (Milburn et al. 1991), two transmembrane domains at the two end of the LBD, a HAMP (histidine kinase, adenyl cyclase, methyl-accepting chemotaxis protein and phosphatase) linker (Anantharaman and Aravind 2000) and a cytosolic signaling domain at the C-terminus. Five chemoreceptors (Tar, Tsr, Tap, Trg, and Aer) have been identified in Escherichia coli, which respond to concentration gradients of amino acids, dipeptides, and sugars (Sourjik 2004). In the free-living environmental bacterium Pseudomonas putida, KT2440, McpG, McpU, McpH, McpA, and PcaY_PP are found to be chemoreceptors for γ-aminobutyric acid, polyamines, purine, amino acids, and C6-ring containing carboxylic acids, respectively (Reyes-Darias et al. 2015; Corral-Lugo et al. 2016; Fernández et al. 2016, 2017; Gavira et al. 2018). In both Bacillus amyloliquefaciens and Bacillus subtilis, McpB and McpC mediate chemotatic response to amino acids (Yssel et al. 2011). However, the ligands of the homologous MCP vary in different bacteria (Corral-Lugo et al. 2016). For example, in B. subtilis, McpA mediates the chemotaxis response towards glucose and α-methylglucoside and the repellent response to root exudates (Hanlon and Ordal 1994; Allard-Massicotte et al. 2016). However, McpA in Bacillus velezensis was a major chemoreceptor for a broad range of chemoattractants and is essential for chemotactic response to cucumber root exudates (Feng et al. 2018).
The interactions between the MCPs and the root exudates are important for the rhizosphere behavior of bacteria. But the MCPs that mediate the chemotaxis towards root exudates are different in bacteria. The chemotactic response to host of the alfalfa symbiont Sinorhizobium meliloti was mediated by 8 different MCPs, among which McpU is the chemoreceptor for sensing the amino acids, while McpX is the chemoreceptor for the quaternary ammonium compounds in root exudates of alfalfa (Medicago sativa) (Meier et al. 2007; Webb et al. 2016, 2017; Zatakia et al. 2017). Recently, multiple chemotaxis receptors (including McpB, McpC, and TlpC) were found to be involved in the chemotaxis of B. subtilis NCIB 3610 to Arabidopsis thaliana root exudates, and this process was required for the bacterial colonization (Allard-Massicotte et al. 2016). In a previous study, we comprehensively identified the chemoattractants in cucumber root exudates and their sensing chemoreceptors in a rhizospheric B. velezensis strain SQR9 (Feng et al. 2018). However, root exudates of plant always changed in response to the root colonization of the microbes, how the root-secreted chemoattractants changed and regulated the chemotaxis of the PGPR in response to inoculation has not been examined.
B. velezensis SQR9 is an efficient PGPR with close interactions with plant roots. Our previous study revealed several chemoattractants in root exudates and comprehensively illustrated their interaction with the MCPs in SQR9 (Feng et al. 2018). In this study, we identified a root-secreted chemoattractant, d-galactose, with strong activity and interesting properties. It was demonstrated that McpA is the only MCP responsible for chemotaxis towards d-galactose. Interestingly, d-galactose was shown to enhance the biofilm formation of SQR9 in an McpA-dependent manner. In addition, the d-galactose content is 3-fold higher in the root exudates of the SQR9-colonized cucumber than that of an uninoculated one.
Materials and methods
Bacterial strains, media, and growth conditions
Strains and plasmids are listed in Table 1. B. velezensis SQR9 (China General Microbiology Culture Collection Center, CGMCC accession no. 5808) was isolated from the cucumber rhizosphere. Wild-type or mutant strains were grown at 37 °C in low-salt Luria-Bertani (LLB) medium (peptone, 10 g L−1; yeast extract, 5 g L−1; and NaCl, 3 g L−1) solidified with 15 g L−1 agar. E. coli BL21 (DE3) was grown at 37 °C in Luria-Bertani (LB) medium (peptone, 10 g L−1; yeast extract, 5 g L−1; and NaCl, 5 g L−1) solidified with 15 g L−1 agar. When necessary, the final concentrations of antibiotics were added as follows: 5 mg L−1 chloramphenicol (Cm), 20 mg L−1 zeocin (Zeo), 100 mg L−1 spectinomycin (Spc), and 30 mg L−1 kanamycin (Kan).
Chemotaxis assay
The qualitative comparison chemotaxis assay was performed as described by Shen et al. (2014) using a simple and reusable microfluidic SlipChip device. Briefly, a 10 mg mL−1 bovine serum albumin (BSA) solution was injected into all the channels and used to wash the microwells for 5 min. After removing the BSA solution by using vacuum across the access holes, the solutions containing different chemoeffectors with the concentration of 1 mM (for qualitative detection only), bacterial cells suspended in phosphate-buffered saline (PBS; 8 g L−1 NaCl, 0.2 g L−1 KCl, 1.44 g L−1 Na2HPO4, and 0.24 g L−1 KH2PO4, pH 7.4), and PBS buffer (negative control) were loaded to the top, middle, and bottom microwells, respectively. Subsequently, the SlipChip device was slipped to ensure that the cells could migrate freely from the middle microwells to the ducts, and the microwells were loaded with the chemoeffector or PBS. The device was placed on an inverted fluorescence microscope (Ti-Eclipse, Nikon, Tokyo Metropolis, Japan) and stored for 30 min in a dark environment at room temperature. The cells in the top and bottom microwells were monitored. The chemotaxis index (It) was used to indicate the chemotactic ability of the bacterial cells using a chemoeffector at a certain concentration. It is defined as Ne/(Ne + Nc), where Ne is the number of cells that migrated into the chemoeffector-containing microwells and Nc is the number of cells that migrated into the control microwells over a certain time period.
In theory, an It value approximately equal to 0.5 means that the cells do not respond to the chemoeffector; an It value of more than 0.5 indicates that the cells are attracted by the chemoeffector; and an It value lower than 0.5 suggests that the cells are repelled by the chemoeffector. To ensure the stability and reliability of the assessment, we redefined the non-response interval of It to the range between 0.4 and 0.6, while > 0.6 and < 0.4 were indicated to be positive and negative responses to the chemoeffector, respectively.
In addition, a modified chemotaxis assay based on the Adler procedure (Adler 1973) was also performed for quantitative chemotaxis measurements. Bacterial strains were grown in LB media until they reached the log phase. The cells collected by centrifugation were washed twice with chemotaxis buffer and resuspended in the same buffer (OD600 nm of 0.6). A 200-μL pipette tip was filled with 100 μL of this cell suspension. A standard 1-mL syringe was loaded with 100 μL of various concentrations of d-galactose. The needle of the syringe was inserted to the narrow end of the pipette tip to connect the chemical solution and the cell suspension. After 1 h of static incubation at 37 °C, the contents in the syringe were transferred to an Eppendorf tube containing 1.0 mL of sterilized water. The suspension was then diluted and plated on LB plates. Colony-forming units (CFUs) were determined in the LB plates incubated for 24 h at 37 °C. Each treatment was repeated six times.
Protein expression and purification
The expression and purification of the ligand-binding domain (LBD) of McpA was performed as previously described (Feng et al. 2018). The DNA fragments encoding the LBD of McpA (Ala33-Pro278) were amplified with primer sets that contained restriction sites for NdeI and HindIII using SQR9 genomic DNA as a template. The PCR products were digested and cloned into the expression plasmid pET28a (+) linearized with the enzymes NdeI and HindIII. The insert and flanking regions of the reconstructed plasmid (pET28a-McpALBD) were verified by DNA sequencing. The expressed proteins contained an N-terminal His tag.
The constructed expression vector was individually transformed into E. coli BL21 (DE3). The strains obtained were grown in 1-L Erlenmeyer flasks containing 200 mL of Luria-Bertani (LB) medium supplemented with 30 mg L−1 kanamycin at 37 °C with 220-rpm (rounds per minute) shaking. When the OD600 of the culture reached 0.6, the growth temperature was lowered to 16 °C. After 30 min of incubation, protein expression was induced by adding 0.05 mM isopropyl-β-d-thiogalactopyranoside (IPTG). Growth was continued at 16 °C with 220-rpm shaking overnight prior to the harvest of the cells using centrifugation at 8000g for 10 min. The cells were washed at least twice with 0.01 M PBS. The cell pellets were resuspended in 50 mL 0.01 M PBS and disrupted by sonication (2-s pulse on, 3-s pulse off). The cellular lysates were centrifuged at 20,000 g for 50 min. Finally, for further clarification, the lysates were passed through a 0.22-μm filter (Merck KGaA, Darmstadt, Germany) to remove any other aggregates or insoluble particles followed by purification with His-affinity resin chromatography (Hua Chun Biotech, Tianjin, China) according to the manufacturer’s instructions. The purified protein was collected and stored in TKMDmod buffer (50 mM Tris, pH 8.0, 50 mM KCl, and 5 mM MgCl2) at − 80 °C.
Biolayer interferometry analysis
Experiments were performed on the Octet-RED 96 device (ForteBio, Inc., Menlo Park, USA). Data were generated automatically by the Octet User Software (version 9.0). All kinetic experiments were conducted at 25 °C. Prior to each assay, super streptavidin (SSA) biosensor tips (ForteBio, Inc., Menlo Park, USA) were pre-wet with PBS (pH 7.0) for 10 min. Buffer, protein, or samples were dispensed into 96-well black microtiter plates (Merck KGaA, Darmstadt, Germany) with 200 μL in each well. Firstly, baseline measurement was done in PBS for 60 s. Afterwards, the protein samples were immobilized on the biosensor tips by agitation for 120 s at 1000 rpm followed by equilibration with PBS for 60 s. Subsequently, association of McpA_LBD with the d-galactose in a concentration range of 0.10 μM, 0.39 μM 1.56 μM, 6.25 μM, 25.00 μM, and 100.00 μM was performed. Association at each tested concentration was carried out for 60 s. Finally, the dissociation was monitored with PBS for 60 s. All measurements were performed in triplicate. The binding profile of each sample was summarized as a “nm shift” (the wavelength/spectral shift in nanometers), which represented the association and disassociation of the protein and the potential ligands.
Biofilm formation assay
To determine the effects of d-galactose on the biofilm formation of B. velezensis SQR9 and the mutant strains, the biofilm formation assay was performed using 48-well microtiter plates as described by Hsueh et al. (2006). Bacteria were grown in LB medium at 37 °C until the log phase, washed with PBS, and finally resuspended in PBS to obtain a suspension of OD600 of 0.2. Each well was filled with 1 mL MSgg medium (Branda et al. 2001) inoculated with 10 μL suspension of bacteria. d-Galactose was added to obtain a final concentration of 1 to 100 μM, respectively. Four replicates were included for each treatment. The plates were incubated at 37 °C without shaking for 15 h. After then, liquid was discarded and the wells were washed with flowing water. Biofilms adhering to the well surface were proceeded for staining. Biofilm cells were stained using 1 mL of 0.1% crystal violet overnight. Subsequently, excess crystal violet was poured out, and the wells were washed twice with distilled water. The bound crystal violet was solubilized with 1 mL of 4:1 ethanol and acetone acid and measured at OD590.
Growth curve measurement
Growth curves of SQR9 and the mutant strains were measured with shaking at the same condition with biofilm formation assay. The measurement of absorbance at 600 nm was performed every 30 min. Six replicates were included for each treatment.
Root exudate collection in the split-root system
Cucumber seedlings were grown to a three-leaf stage in a chamber and transplanted into a split-root system as described by Liu et al. (2017). The systems were cultured at 23 °C with a 16-h light/8-h dark photoperiod, and the medium was replaced every day. Three days after transplantation, the split-root chamber was checked for contamination. Suspensions of SQR9 was inoculated to the left side of the split-root system and mixed with the sterilized water with the final concentration of 5 × 107 CFUs mL−1. The split-root system was cultured for two more days to allow for pre-exposure to the inoculants. Cultures in the chamber were collected after 3 days as shown in result. The treatments were repeated three times, including six plants for each replicate. Regular root exudates of the cucumber without inoculation were collected as the CK (control). A total of 0.72 L of root exudates containing water were collected for each replicate from six plants. Root exudate solutions were filtered through a 0.45-μm membrane (Merck KGaA, Darmstadt, Germany) and lyophilized. The sterility of the exudates after filtration was tested by plating 100 μL of each sample on an LB plate and incubating at 30 °C for contamination check. Subsequently, the exudates were concentrated to solid powder using a freezing dryer.
Quantification of galactose
Galactose was analyzed using ultrahigh-performance liquid chromatography-tandem mass-spectrometry (UHPLC-MS-MS). LC-MS/MS analyses were performed using an UHPLC system (1290, Agilent Technologies, Santa Clara, USA) with a Waters® ACQUITY UPLC HSS T3 column (100 × 2.1 mm, 1.8 μm, Waters, Miford, MA, USA) coupled to a 6460 Triple Quadrupole mass spectrometer. Briefly, 5 mg of each root exudate sample was dissolved in 1 mL of 50% (v/v) methanol and vortexed for 30 s, followed by sonication on ice for 15 min. The solution was incubated at − 20 °C for 1 h and centrifuged at 13,000g (4 °C) for 15 min. Samples were additionally treated for derivatization: a 100 μL aliquot of the clear supernatant (or standard solution) was mixed with 100 μL of reaction solution (2% acetic acid, 3% anthranilamide, and 2% sodium cyanoborohydride in methanol). The reaction mixtures were incubated at 80 °C for 60 min followed by the addition of 200 μL of water. Each tube was thoroughly mixed and centrifuged at 13,000×g (4 °C) for 15 min, and a 100 μL aliquot of the supernatant was prepared for further analysis. A 100 μL aliquot of the supernatant was transferred to an autosampler vial for the UHPLC-MS/MS analysis. The mobile phase A was 10 mM acetic acid and ammonium acetate in water, and the mobile phase B was acetonitrile. The elution gradients were the following: 0 min, 95%; 6 min, 75% A; 8 min, 5% A; and 12.5 min, 95% A, which were delivered at 0.35 mL min−1. The column temperature was set at 40 °C, and the injection volume was 1 μL. An Agilent 6460 triple quadrupole mass spectrometer (Agilent Technologies, Santa Clara, USA) equipped with an AJS electrospray ionization (AJS-ESI) interface was used. Typical ion source parameters were as follows: capillary voltage = + 4000/− 3500 V, nozzle voltage = + 500/− 500 V, gas (N2) temperature = 300 °C, gas (N2) flow = 5 L min−1, sheath gas (N2) temperature = 250 °C, sheath gas flow = 11 L min−1, and nebulizer = 45 psi. Absolute quantification of the compound in the root exudates was calculated based on their detected peak areas and the calibration curves of the relevant standard. The concentration was finally normalized by the original volumes in the root chambers.
Colonization assay
A suspension of SQR9, SQR9Δ8mcp and SQR9Δ8mcp/mcpA, was prepared in LLB media grown for 12 h at 37 °C with 200-rpm shaking. The bacterial cells were pelleted using centrifugation at 10,000g for 10 min and resuspended in 1/4 sucrose-free Murashige Skoog (MS, Murashige and Skoog 1962) medium to adjust the OD600 to approximately 1.0. At the three-leaf stage (approximately 20-day-old cucumber seedlings), the bacterial cells were inoculated in sterile conical flasks containing the 40 mL MS medium to obtain a final concentration of 5 × 107 CFU mL−1. After incubation for 3 days, the roots were cut and briefly washed with sterile distilled water. Subsequently, the mixed roots were weighed and ground with a mortar in sterile distilled water, and the suspensions acquired were diluted and plated on LB agar media. The bacterial cells colonized on the cucumber roots were quantified by counting the colonies after incubation for 12 h at 37 °C. Six replicates were included for each treatment. The results were normalized to the fresh weight of the root.
To view the colonization of B. velezensis SQR9-gfp on the root surfaces, the root tips were observed using a confocal laser scanning microscope system (Zeiss LSM 700, Carl Zeiss AG, Jena, Germany) with excitation wavelengths of 488 nm. The objective used was a Zeiss EC Plan-Neofluar × 10/0.30 Ph1. Emissions in the range of 490–555 nm were collected for the GFP visualization. Images of at least 20 root tips were obtained for each treatment.
Statistical analysis
Differences among the treatments were calculated and statistically analyzed using an analysis of variance (ANOVA) and Duncan’s multiple range tests (P < 0.05) for colonization. SPSS version 22.0 was used for statistical analysis (SPSS Inc., Chicago, IL).
Results
d-Galactose attracts B. velezensis SQR9
By using a microfluidic SlipChip device, we screened the root-secreted low molecular weight compounds for an attractant of the rhizospheric B. velezensis SQR9. The SlipChip includes three chambers in line. One side chamber was filled with the 1 mM of pending chemical, while the other side chamber was filled with PBS to indicate the control. The bacterial suspension was spread in the middle chamber, and the cells were allowed to swim to either of the side chambers that were filled with PBS or the pending chemical through the diffused gradient. In addition, due to the dilution effect of the SlipChip, the concentration used in this method does not have quantitative significance. To normally compare the attractant, the chemotaxis index (It), showing the ratio of cell number in the chemoattractant chamber to the sum of the cell numbers in the PBS control chamber and the chemoattractant chamber, was introduced. The subscript “t” indicates the incubation time; I30 indicates the chemotaxis index with incubation for 30 min. A higher I30 indicates a stronger attraction to the bacteria cell (Supplementary Table S1). When compared with the other attractants previously identified, such as d-maltose (I30 = 0.94), malic acid (I30 = 0.98), succinic acid (I30 = 0.98), gluconic acid (I30 = 0.97), citric acid (I30 = 0.95), threonine (I30 = 0.98), cystine (I30 = 0.98), and asparagine (I30 = 0.97) (Feng et al. 2018), d-galactose is much stronger (I30 = 0.99) to attract SQR9 than the previously described attractants (Fig. 1, Feng et al. 2018). Moreover, the result showed that the number of cells in the galactose chamber was approximately threefold higher than that of the secondly efficient attractant, and the I30 of d-galactose was the highest (0.99) (Supplementary Table S1). These findings suggested that d-galactose could be an important signal for the chemotaxis of SQR9 in the rhizosphere.
Chemotaxis of B. velezensis SQR9 to root-secreted compounds. Cell counts were performed in a SlipChip device. Cells suspensions were dropped into the middle of the highly concentrated chemoattractant solution (treatment, 1 mM) and the chemotaxis buffer (control) and allowed to swim to either side. Cells in the chemotaxis chamber and the chemotaxis buffer chamber of the SlipChip device were counted and plotted. For data of chemotaxis to the compounds other than d-galactose, please refer to Feng et al. (2018). Black and gray columns indicate the cell numbers in the chemoattractant chamber and the chemotaxis buffer chamber, respectively
Chemoreceptor for d-galactose in SQR9
Based on the strongly attractive activity of d-galactose, it is of interest to explore how bacteria sense the signal transduced from the plant. SQR9 has eight MCPs for chemotactic signal perception, including McpA, McpB, McpC, McpR, TlpA, TlpB, HemAT, and YfmS (Feng et al. 2018). A mutant that is deficient in all eight genes encoding the MCPs (SQR9Δ8mcp) has been created, and the SQR9Δ8mcp has been complemented with the individual receptors in a previous study (Feng et al. 2018). The chemotaxis of the wild-type SQR9 and all the mutants were evaluated using a microfluidic SlipChip device. The results showed that the chemotaxis indexes (I30) of SQR9Δ8mcp, SQR9Δ8mcp/mcpB, SQR9Δ8mcp/mcpC, SQR9Δ8mcp/mcpR, SQR9Δ8mcp/tlpA, SQR9Δ8mcp/tlpB, SQR9Δ8mcp/hemAT, or SQR9Δ8mcp/yfmS were significantly lower than that of the wild-type strain and SQR9Δ8mcp/mcpA. In addition, no significant difference was observed between the wild-type strain and SQR9Δ8mcp/mcpA (Fig. 2a). A classic capillary assay was performed for a range of the chemotactic concentrations of d-galactose. The attraction of d-galactose to SQR9 was the highest at the final concentration of 1 μM, while it decreased as the concentration increased to 10 μM, as did that of SQR9Δ8mcp/mcpA (Fig. 2b). Similar to the SlipChip chemotaxis assay, SQR9Δ8mcp did not show chemotaxis to either concentration of d-galactose. These findings suggest that McpA is the chemoreceptor responsible for sensing d-galactose. In comparison with malic acid, another strong chemoattractant that sensed by McpA, the concentration needed for best chemoattraction for d-galactose is much lower; in addition, chemotaxis response of SQR9 to d-galactose is stronger than malic acid at their best concentration for chemotaxis.
Effect of methyl-accepting chemotaxis protein (MCP) mutations on the chemotaxis to d-galactose. a The chemotaxis index (I30) of the wild-type SQR9, SQR9Δ8mcp, SQR9Δ8mcp/mcpA, SQR9Δ8mcp/mcpB, SQR9Δ8mcp/mcpC, SQR9Δ8mcp/mcpR, SQR9Δ8mcp/tlpA, SQR9Δ8mcp/tlpB, SQR9Δ8mcp/hemAT, and SQR9Δ8mcp/yfmS to d-galactose was calculated from the SlipChip chemotaxis experiment. An It value approximately equal to 0.5 means that the cells do not respond to the chemoeffector; an It value of more than 0.5 indicates that the cells are attracted by the chemoeffector; and an It value lower than 0.5 suggests that the cells are repelled by the chemoeffector. Error bars represent the standard errors of 13 biological replicates. One asterisk (*) indicates P < 0.05 and two asterisks (**) indicates P < 0.01 (Student’s t test). b Capillary assay with the gradient concentration of d-galactose and malic acid. d-Galactose and malic acid solutions with the concentration of 10−7 M, 10−6 M, 10−5 M, 10−4 M, and 10−3 M were tested for their attractions to SQR9, SQR9Δ8mcp and SQR9Δ8mcp/mcpA. Error bars represent the standard errors of six biological replicates. Asterisks indicate that the mean value is significantly different between the strains
Binding activity of d-galactose and McpA
To further verify the interaction between d-galactose and McpA, the amino terminal-sensing domain of McpA (Ala36-Pro277) was expressed in E. coli BL21 (DE3) and purified (Supplementary Fig. S1). Its binding of d-galactose was investigated using Biolayer interferometry (BLI) analysis. But the BLI result showed that McpA could not directly bind d-galactose in vitro (data not shown). This result suggests that McpA would sense d-galactose by an indirect manner. As similar cases have been found in E. coli and Azospirillum brasilense (Scholle et al. 1987; Van Bastelaere et al. 1999), it is proposed that a galactose-binding protein might capture d-galactose in environment and subsequently bind with McpA.
Biofilm formation induced by d-galactose is McpA-dependent
Biofilm formation on the root surface is another important step for the colonization of a PGPR. In this study, the effect of d-galactose on biofilm formation was tested. Both the qualitative and quantitative experiments showed that d-galactose could significantly induce biofilm formation in SQR9 at a final concentration of 100 μΜ (Fig. 3a, b). Interestingly, deletion of all eight MCPs caused a deficiency of biofilm formation response to d-galactose induction. However, the reintroduction of McpA, the MCP that is responsible for chemotaxis to d-galactose, restored the biofilm formation mediated by d-galactose (Fig. 3a, b). This observation suggested that an McpA-dependent biofilm formation regulation pathway could be activated by d-galactose.
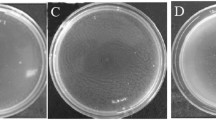
Effects of d-galactose on biofilm formation. a Images of biofilm induced by d-galactose with concentrations of 1 μM, 10 μM, and 100 μM. b Quantification of biofilm formation in response to d-galactose. d-Galactose was applied to the medium with concentration of 1 μM, 10 μM, and 100 μM. Biofilm was stained with crystal violet overnight. Excess crystal violet was poured out, and the biofilm was washed with water. The bound crystal violet was solubilized with 1 mL of 4:1 ethanol:acetone and measured at OD590. Error bars indicate the standard deviations based on four different replicated experimental values. c Images of biofilm for wild-type SQR9, Δ8mcp and Δ8mcp/tlpB, when maltose was added to the medium. TlpB in B. velezensis SQR9 is the chemoreceptor for maltose. d Quantification of biofilm formation in response to maltose. e Growth curve of the wild-type strain and the mutant in MSgg medium with or without additives. Uppercase letters indicate the group for statistics. Different lowercase letters indicate significant differences between treatments (Duncan’s least significant difference test, P < 0.05, mean ± standard deviation). Two asterisks (**) indicates P < 0.01 (Student’s t test), while “ns” indicates P > 0.05
This experiment was also repeated with another chemoattractant of SQR9, maltose, and the relevant chemoreceptor TlpB (Feng et al. 2018) to confirm whether the enhanced biofilm formation by d-galactose is generally resulted from the chemotactic response. However, no difference was observed with SQR9Δ8mcp/tlpB and SQR9Δ8mcp (Fig. 3c, d). This result indicated that the participation in biofilm formation of McpA is not related to its function on chemotaxis.
A growth curve was recorded to ensure that the effect on biofilm formation is not simply due to its effect on bacterial growth (Fig. 3e). Because the biofilm was formed at 13 h post-inoculation, the growth curve was measured before 13 h. In MSgg medium, SQR9Δ8mcp, SQR9Δ8mcp/mcpA, and the wild-type strain all grow well and no significant differences were observed (Fig. 3e). At 1 μΜ, 10 μΜ, and 100 μΜ, d-galactose showed no effect on growth of all strains (Fig. 3e).
Induction of d-galactose production from cucumber roots
The concentration of d-galactose in the rhizosphere is a dominant factor to provide its chemoattractant and biofilm inducer functions. We measured the concentration of d-galactose in the root chamber after incubation for 1 day. In this experiment, cucumbers were grown in a split-root system; one side was inoculated with B. velezensis SQR9, while another side of the root chamber was used for collecting the root exudates (Fig. 4a). The result showed that 1 g of root from the uninoculated cucumber secreted 0.161 μg of d-galactose in 24 h (Fig. 4b); however, 1 g of root from the cucumber inoculated with SQR9 secreted 0.478 μg of d-galactose in 24 h (Fig. 4b), which is nearly 3 times higher than that of control. It indicated that secretion of d-galactose was induced by B. velezensis SQR9. This finding suggests d-galactose might be an inducible signal secreted by plant root.
Quantification of d-galactose. a Sample collection. b Quantification of d-galactose in root exudates. Quantification of d-galactose was performed by LC-MS/MS. The concentration was calculated based on the standard curved made from pure d-galactose. The root secretion of d-galactose was normalized by the root fresh weight. Error bars represent the standard errors of three biological replicates with six plants for each replicate
d-Galactose promotes the colonization of SQR9 on cucumber roots
Since we found d-galactose participated in biofilm formation, pure d-galactose was further tested for its effect on the root colonization of SQR9. As a homogeneous concentration of d-galactose in the liquid MS medium was formed, the influence of chemotaxis towards d-galactose on colonization was avoided in this experiment. The green fluorescent protein labeled SQR9 (SQR9-GFP) was used for detecting the root colonization of B. velezensis SQR9 under a confocal laser scanning microscope. The images showed that more SQR9-GFP cells were bound to the roots with exogenous d-galactose in a dose-dependent manner (Fig. 5a). The quantitative measurement of its root colonization was performed. The root colonization of SQR9 increased significantly in parallel with the increasing of concentration of d-galactose in rhizosphere (Fig. 5b).
Root colonization of B. velezensis SQR9. a Confocal scanning laser microscopy images of the root tips colonized by B. velezensis. d-Galactose was applied to the growth medium with concentrations of 10−6 M, 10−5 M, 10−4 M, and 10−3 M. b Colony-forming unit (CFU) counting for root colonization in response to exogenous d-galactose with concentrations of 10−6 M, 10−5 M, 10−4 M, and 10−3 M. Error bars represent the standard errors of six biological replicates. c Bacterial cell growth in root chamber supplied with d-galactose without plant. d-Galactose was applied to the growth medium with concentration of 10−6 M, 10−5 M, 10−4 M, and 10−3 M. Uppercase letters above the bar indicate the group for statistics. Different lowercase letters indicate significant differences between treatments (Duncan’s least significant difference test, P < 0.05, mean ± standard deviation)
In addition, to exclude the possibility that the effect of d-galactose on colonization is simply due to its stimulation of bacterial growth, the growth of SQR9 in the same condition without plant at 24, 48, and 72 h after inoculation was measured (Fig. 5c). The results showed that d-galactose did not stimulate growth at all the tested concentrations; 100 μM of d-galactose even showed inhibition of SQR9 growth at 24 h. Because the colonization assay was sampled at 72 h, it is believed the differences observed in colonization assay have not been interfered by the growth effect.
Discussion
B. velezensis SQR9 is a beneficial plant rhizobacterium with a strong ability to colonize roots and to chemotactically swim towards cucumber root exudates (Weng et al. 2013). McpA, the primary chemoreceptor of SQR9, has been predicted to have a conserved ligand-binding domain (LBD), a dCACHE domain, to sense environmental signals. Our previous study showed that the McpA of SQR9 was responsible for sensing 20 root-secreted compounds, including five amino acids (aspartic acid, glutamic acid, isoleucine, lysine, and tyrosine), ten organic acids (citric acid, malic acid, oxalic acid, fumaric acid, succinic acid, phthalic acid, adipic acid, dehydroascorbic acid, glyceric acid, and 3-hydroxypropionic acid), and five other compounds (hydroxycarbamate, mannose, ribose, fucose, and ribitol) (Feng et al. 2018). In this study, it was shown that cucumber root-secreted d-galactose was a more efficient chemotactic signal with significant attraction to SQR9 at the concentration of 1 μM. Our data revealed that McpA is the only MCP responsible for sensing d-galactose. This study also found that d-galactose could induce biofilm formation of SQR9 in an McpA-dependent manner. Another finding is that when the cucumber was inoculated with SQR9, the root secreted more d-galactose to attract SQR9. These observations demonstrated the importance of the root-secreted d-galactose in root colonization of the beneficial microbes.
However, d-galactose could not bind to McpA directly. It has reported that bacteria could sense galactose by direct receptor-ligand binding or by indirect way. Phytopathogen Campylobacter jejuni uses the chemoreceptor Tlp11(CcrG), to sense d-galactose directly (Rahman et al. 2014; Day et al. 2016). But in E. coli, an extracellular galactose-binding protein MglB, is necessary for the MCP-mediated chemotaxis to galactose (Scholle et al. 1987). When galactose binds to MglB, the protein undergoes a conformational change and interacts with the chemotaxis receptor Trg to initiate a chemotactic signal (Scholle et al. 1987). It was suggested that a galactose-binding protein might occur in B. velezensis to directly capture d-galactose and interact with McpA in order to initial the chemotaxis to galactose. Further identification of the galactose-binding protein in B. velezensis would help to clarify the galactose chemotaxis signaling pathway. Similar cases have also been found in A. brasilense, the sugar binding protein SbpA is involved in the uptake of d-galactose and functions in the chemotaxis towards d-galactose (Van Bastelaere et al. 1999).
In addition, the function of McpA in different bacteria varies, the homologous McpA in other bacteria senses different chemicals, for example, McpA in P. putida KT2440R senses l-alanine, l-methionine, l-proline, and l-serine (Corral-Lugo et al. 2016). In B. subtilis, McpA mediates the attraction towards glucose and α-methylglucoside (Hanlon and Ordal 1994) and may sense repellent molecule(s) secreted by A. thaliana (Allard-Massicotte et al. 2016).
d-Galactose is frequently found in mammals and some microbes (Seifert et al. 2002; Conklin et al. 2006). In the plant, galactose is generally present in other forms, such as polymers or UDP-galactose, which is a component of the plant cell wall (Seifert et al. 2002; Conklin et al. 2006). For regular growth, development, and metabolism, the plant evolves a series of cell wall–degrading enzymes, including polygalacturonase, pectinesterase, and β-galactosidase, to degrade the polymers of cell wall to galactose and other oligosaccharides (Brett and Waldron 1990; Marin-Rodriguez 2002; Wolf et al. 2012). In addition to the plant itself, some soil-borne pathogens have been reported to bind the hydrolase of the plant cell wall to combat the plant’s physical defenses (Aro et al. 2005). In this study, SQR9 inoculation on the split-root cucumber caused increased root secretion of d-galactose in the medium. This phenomenon demonstrated that a long-distance signal transduction from the local tissue to the distal tissue should occur during this induction. Considering the systemic resistance induced by SQR9 in a previous study (Wu et al. 2018), it would be interesting to test whether this systemic induction of exudation of galactose employs the same signaling pathway or not.
We found McpA was involved in the exogenous d-galactose-induced enhancement of biofilm formation. However, another ligand-receptor pair, maltose-TlpB, did not affect biofilm formation. These results indicated that McpA is involved in biofilm formation stimulation by exogenous signals and its function is to not generally rely on chemotaxis. Both chemotaxis and biofilm formation have been shown to be important for microorganisms to colonize surfaces (Yaryura et al. 2008). DegU is a known regulator for both biofilm formation and chemotaxis in B. subtilis and Listeria monocytogenes (Kobayashi 2007; Gueriri et al. 2008). In some other cases, cyclic di-GMP could be a convergent pass of the regulation of biofilm formation and chemotaxis (O’Connor et al. 2012; Russell et al. 2013). There is similar evidence in Campylobacter jejuni that Tlp3, a chemoreceptor that is responsible for sensing lysine and glucosamine, suppressed biofilm formation (Rahman et al. 2014). A deficiency of tlp3 led to a 1.5-fold increase in biofilm formation (Rahman et al. 2014). Two chemoreceptors, McpU and Wsp, in P. putida KT2440 have been reported to modulate biofilm formation (Corral-Lugo et al. 2016). As recently reported in Comamonas testosterone, the kinase CheA, autophosphorylation of which is controlled by ligand sensing by MCPs (Szurmant and Ordal 2004), was found to phosphorylate not only its cognate response regulator CheY but also one of the response regulators from the pathway mediating biofilm formation, FlmD (Huang et al. 2019). We proposed whether CheA in Bacillus could phosphorylate any response regulators for the biofilm formation pathway; the future results would explain how McpA exert its dual role in biofilm formation and chemotaxis.
In conclusion, root-secreted d-galactose was found to be an inducible signal that regulates chemotaxis and biofilm formation of B. velezensis SQR9 in an McpA-dependent manner. These finding would strengthen the knowledge of how the two processes, biofilm formation and chemotaxis towards plant roots, are integrated in the root colonization.
References
Adler J (1973) A method for measuring chemotaxis and use of the method to determine optimum conditions for chemotaxis by Escherichia coli. J Gen Microbiol 74:77–91. https://doi.org/10.1099/00221287-74-1-77
Allard-Massicotte R, Tessier L, Lécuyer F, Lakshmanan V, Lucier J-F, Garneau D, Caudwell L, Vlamakis H, Bais HP, Beauregard PB (2016) Bacillus subtilis early colonization of Arabidopsis thaliana roots involves multiple chemotaxis receptors. mBio 7:e01664–e01616. https://doi.org/10.1128/mBio.01664-16
Anantharaman V, Aravind L (2000) Cache–a signaling domain common to animal Ca2± channel subunits and a class of prokaryotic chemotaxis receptors. Trends Biochem Sci 25:535–537. https://doi.org/10.1016/S0968-0004(00)01672-8
Aro N, Pakula T, Penttilä M (2005) Transcriptional regulation of plant cell wall degradation by filamentous fungi. FEMS Microbiol Rev 29:719–739. https://doi.org/10.1016/j.femsre.2004.11.006
Badri DV, Chaparro JM, Zhang R, Shen Q, Vivanco JM (2013) Application of natural blends of phytochemicals derived from the root exudates of Arabidopsis to the soil reveal that phenolic-related compounds predominantly modulate the soil microbiome. J Biol Chem 288:4502–4512. https://doi.org/10.1074/jbc.M112.433300
Belas R (2014) Biofilms, flagella, and mechanosensing of surfaces by bacteria. Trends Microbiol 22:517–527. https://doi.org/10.1016/j.tim.2014.05.002
Branda SS, Gonzalez-Pastor JE, Ben-Yehuda S, Losick R, Kolter R (2001) Fruiting body formation by Bacillus subtilis. Proc Natl Acad Sci 98:11621–11626. https://doi.org/10.1073/pnas.191384198
Brett C, Waldron K (1990) Cell-wall degradation. In: Brett C, Waldron K (eds) Physiology and biochemistry of plant cell walls. Chapman & Hall, London, UK, pp 168–179
Conklin PL, Gatzek S, Wheeler GL, Dowdle J, Raymond MJ, Rolinski S, Isupov M, Littlechild JA, Smirnoff N (2006) Arabidopsis thaliana VTC4 encodes L-galactose-1-P phosphatase, a plant ascorbic acid biosynthetic enzyme. J Biol Chem 281:15662–15670. https://doi.org/10.1074/jbc.M601409200
Corral-Lugo A, De la Torre J, Matilla MA, Fernández M, Morel B, Espinosa-Urgel M, Krell T (2016) Assessment of the contribution of chemoreceptor-based signalling to biofilm formation. Environ Microbiol 18:3355–3372. https://doi.org/10.1111/1462-2920.13170
Day CJ, King RM, Shewell LK, Tram G, Najnin T, Hartley-Tassell LE, Wilson JC, Fleetwood AD, Zhulin IB, Korolik V (2016) A direct-sensing galactose chemoreceptor recently evolved in invasive strains of Campylobacter jejuni. Nat Commun 7:1–8. https://doi.org/10.1038/ncomms13206
de Weert S, Vermeiren H, Mulders IHM, Kuiper I, Hendrickx N, Bloemberg GV, Vanderleyden J, De Mot R, Lugtenberg BJJ (2002) Flagella-driven chemotaxis towards exudate components is an important trait for tomato root colonization by Pseudomonas fluorescens. Mol Plant-Microbe Interact 15:1173–1180. https://doi.org/10.1094/MPMI.2002.15.11.1173
Feng H, Zhang N, Du W, Zhang H, Liu Y, Fu R, Shao J, Zhang G, Shen Q, Zhang R (2018) Identification of chemotaxis compounds in root exudates and their sensing chemoreceptors in plant-growth-promoting rhizobacteria Bacillus amyloliquefaciens SQR9. Mol Plant-Microbe Interact 31:995–1005. https://doi.org/10.1094/MPMI-01-18-0003-R
Fernández M, Morel B, Corral-Lugo A, Krell T (2016) Identification of a chemoreceptor that specifically mediates chemotaxis toward metabolizable purine derivatives. Mol Microbiol 99:34–42. https://doi.org/10.1111/mmi.13215
Fernández M, Matilla MA, Ortega Á, Krell T (2017) Metabolic value chemoattractants are preferentially recognized at broad ligand range chemoreceptor of Pseudomonas putida KT2440. Front Microbiol 8:1–14. https://doi.org/10.3389/fmicb.2017.00990
Gavira JA, Ortega Á, Martín-Mora D, Conejero-Muriel MT, Corral-Lugo A, Morel B, Matilla MA, Krell T (2018) Structural basis for polyamine binding at the dCACHE domain of the McpU chemoreceptor from Pseudomonas putida. J Mol Biol 430:1950–1963. https://doi.org/10.1016/j.jmb.2018.05.008
Glekas GD, Mulhern BJ, Kroc A, Duelfer KA, Lei V, Rao CV, Ordal GW (2012) The Bacillus subtilis chemoreceptor McpC senses multiple ligands using two discrete mechanisms. J Biol Chem 287:39412–39418. https://doi.org/10.1074/jbc.M112.413518
Gueriri I, Bay S, Dubrac S, Cyncynatus C, Msadek T (2008) The Pta-AckA pathway controlling acetyl phosphate levels and the phosphorylation state of the DegU orphan response regulator both play a role in regulating Listeria monocytogenes motility and chemotaxis. Mol Microbiol 70:1342–1357. https://doi.org/10.1111/j.1365-2958.2008.06496.x
Haichar F e Z, Santaella C, Heulin T, Achouak W (2014) Root exudates mediated interactions belowground. Soil Biol Biochem 77:69–80. https://doi.org/10.1016/j.soilbio.2014.06.017
Hanlon DW, Ordal GW (1994) Cloning and characterization of genes encoding methyl-accepting chemotaxis proteins in Bacillus subtilis. J Biol Chem 269:14038–14046
Hess JF, Oosawa K, Kaplan N, Simon MI (1988) Phosphorylation of three proteins in the signaling pathway of bacterial chemotaxis. Cell 53:79–87. https://doi.org/10.1016/0092-8674(88)90489-8
Hsueh YH, Somers EB, Lereclus D, Wong ACL (2006) Biofilm formation by Bacillus cereus is influenced by PlcR, a pleiotropic regulator. Appl Environ Microbiol 72:5089–5092
Huang Z, Wang Y-H, Zhu H-Z, Andrianova EP, Jiang C-Y, Li D, Ma L, Feng J, Liu Z-P, Xiang H, Zhulin IB, Liu S-J (2019) Cross talk between chemosensory pathways that modulate chemotaxis and biofilm formation. mBio 10:1–15. https://doi.org/10.1128/mBio.02876-18
Kobayashi K (2007) Gradual activation of the response regulator DegU controls serial expression of genes for flagellum formation and biofilm formation in Bacillus subtilis. Mol Microbiol 66:395–409. https://doi.org/10.1111/j.1365-2958.2007.05923.x
Liu Y, Chen L, Wu G, Feng H, Zhang G, Shen Q, Zhang R (2017) Identification of root-secreted compounds involved in the communication between cucumber, the beneficial Bacillus amyloliquefaciens, and the soil-borne pathogen Fusarium oxysporum. Mol Plant-Microbe Interact 30:53–62. https://doi.org/10.1094/MPMI-07-16-0131-R
Lugtenberg BJJ, Kravchenko LV, Simons M (1999) Tomato seed and root exudate sugars: composition, utilization by Pseudomonas biocontrol strains and role in rhizosphere colonization. Environ Microbiol 1:439–446. https://doi.org/10.1046/j.1462-2920.1999.00054.x
Marin-Rodriguez MC (2002) Pectate lyases, cell wall degradation and fruit softening. J Exp Bot 53:2115–2119. https://doi.org/10.1093/jxb/erf089
Meier VM, Muschler P, Scharf BE (2007) Functional analysis of nine putative chemoreceptor proteins in Sinorhizobium meliloti. J Bacteriol 189:1816–1826. https://doi.org/10.1128/JB.00883-06
Milburn MV, Prive GG, Milligan DL, Scott WG, Yeh J, Jancarik J, Koshland DE Jr, Kim SH (1991) Three-dimensional structures of the ligand-binding domain of the bacterial aspartate receptor with and without a ligand. Science 254:1342–1347. https://doi.org/10.1126/science.1660187
Murashige T, Skoog F (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiologia 15:473–497. https://doi.org/10.1111/j.1399-3054.1962.tb08052.x
O’Connor JR, Kuwada NJ, Huangyutitham V, Wiggins PA, Harwood CS (2012) Surface sensing and lateral subcellular localization of WspA, the receptor in a chemosensory-like system leading to c-di-GMP production. Mol Microbiol 86:720–729. https://doi.org/10.1111/mmi.12013
Philippot L, Raaijmakers JM, Lemanceau P, Van Der Putten WH (2013) Going back to the roots: the microbial ecology of the rhizosphere. Nat Rev Microbiol 11:789–799. https://doi.org/10.1038/nrmicro3109
Rahman H, King RM, Shewell LK, Semchenko EA, Hartley-Tassell LE, Wilson JC, Day CJ, Korolik V (2014) Characterisation of a multi-ligand binding chemoreceptor CcmL (Tlp3) of Campylobacter jejuni. PLoS Pathog 10:1–14. https://doi.org/10.1371/journal.ppat.1003822
Rao CV, Glekas GD, Ordal GW (2008) The three adaptation systems of Bacillus subtilis chemotaxis. Trends Microbiol 16:480–487. https://doi.org/10.1016/j.tim.2008.07.003
Reyes-Darias JA, García V, Rico-Jiménez M, Corral-Lugo A, Lesouhaitier O, Juárez-Hernández D, Yang Y, Bi S, Feuilloley M, Muñoz-Rojas J, Sourjik V, Krell T (2015) Specific gamma-aminobutyrate chemotaxis in Pseudomonads with different lifestyle. Mol Microbiol 97:488–501. https://doi.org/10.1111/mmi.13045
Rudrappa T, Czymmek KJ, Pare PW, Bais HP (2008) Root-secreted malic acid recruits beneficial soil bacteria. Plant Physiol 148:1547–1556. https://doi.org/10.1104/pp.108.127613
Russell MH, Bible AN, Fang X, Gooding JR, Campagna SR, Gomelsky M, Alexandrea G (2013) Integration of the second messenger c-di-GMP into the chemotactic signaling pathway. mBio 4:1–11. https://doi.org/10.1128/mBio.00001-13
Scholle A, Vreemann J, Blank V, Nold A, Boos W, Manson MD (1987) Sequence of the mglB gene from Escherichia coli K12: comparison of wild-type and mutant galactose chemoreceptors. MGG Mol Gen Genet 208:247–253. https://doi.org/10.1007/BF00330450
Seifert GJ, Barber C, Wells B, Dolan L, Roberts K (2002) Galactose biosynthesis in Arabidopsis: genetic evidence for substrate channeling from UDP-D-galactose into cell wall polymers. Curr Biol 12:1840–1845. https://doi.org/10.1016/S0960-9822(02)01260-5
Shen C, Xu P, Huang Z, Cai D, Liu SJ, Du W (2014) Bacterial chemotaxis on SlipChip. Lab a Chip - Miniaturisation Chem Biol 14:3074–3080. https://doi.org/10.1039/c4lc00213j
Sourjik V (2004) Receptor clustering and signal processing in E. coli chemotaxis. Trends Microbiol 12:569–576. https://doi.org/10.1016/j.tim.2004.10.003
Szurmant H, Ordal GW (2004) Diversity in chemotaxis mechanisms among the bacteria and archaea. Microbiol Mol Biol Rev 68:301–319. https://doi.org/10.1128/MMBR.68.2.301-319.2004
Van Bastelaere E, Lambrecht M, Vermeiren H, Van Dommelen A, Keijers V, Proost P, Vanderleyden J (1999) Characterization of a sugar-binding protein from Azospirillum brasilense mediating chemotaxis to and uptake of sugars. Mol Microbiol 32:703–714. https://doi.org/10.1046/j.1365-2958.1999.01384.x
Webb BA, Helm RF, Scharf BE (2016) Contribution of individual chemoreceptors to Sinorhizobium meliloti chemotaxis towards amino acids of host and nonhost seed exudates. Mol Plant-Microbe Interact 29:231–239. https://doi.org/10.1094/MPMI-12-15-0264-R
Webb BA, Karl Compton K, Castañeda Saldaña R, Arapov TD, Keith Ray W, Helm RF, Scharf BE (2017) Sinorhizobium meliloti chemotaxis to quaternary ammonium compounds is mediated by the chemoreceptor McpX. Mol Microbiol 103:333–346. https://doi.org/10.1111/mmi.13561
Weng J, Wang Y, Li J, Shen Q, Zhang R (2013) Enhanced root colonization and biocontrol activity of Bacillus amyloliquefaciens SQR9 by abrB gene disruption. Appl Microbiol Biotechnol 97:8823–8830. https://doi.org/10.1007/s00253-012-4572-4
Wolf S, Hématy K, Höfte H (2012) Growth control and cell wall signaling in plants. Annu Rev Plant Biol 63:381–407. https://doi.org/10.1146/annurev-arplant-042811-105449
Wu G, Liu Y, Xu Y, Zhang G, Shen Q, Zhang R (2018) Exploring elicitors of the beneficial rhizobacterium Bacillus amyloliquefaciens SQR9 to induce plant systemic resistance and their interactions with plant signaling pathways. Mol Plant-Microbe Interact 31:560–567. https://doi.org/10.1094/MPMI-11-17-0273-R
Yan X, Yu H-J, Hong Q, Li S-P (2008) Cre/lox system and PCR-based genome engineering in Bacillus subtilis. Appl Environ Microbiol 74:5556–5562. https://doi.org/10.1128/AEM.01156-08
Yaryura PM, León M, Correa OS, Kerber NL, Pucheu NL, García AF (2008) Assessment of the role of chemotaxis and biofilm formation as requirements for colonization of roots and seeds of soybean plants by Bacillus amyloliquefaciens BNM339. Curr Microbiol 56:625–632. https://doi.org/10.1007/s00284-008-9137-5
Yssel A, Reva O, Tastan Bishop O (2011) Comparative structural bioinformatics analysis of Bacillus amyloliquefaciens chemotaxis proteins within Bacillus subtilis group. Appl Microbiol Biotechnol 92:997–1008. https://doi.org/10.1007/s00253-011-3582-y
Zatakia HM, Arapov TD, Meier VM, Scharf BE (2017) Cellular stoichiometry of methyl-accepting chemotaxis proteins in Sinorhizobium meliloti. J Bacteriol 200:1–14. https://doi.org/10.1128/JB.00614-17
Zhou C, Shi L, Ye B, Feng H, Zhang J, Zhang R, Yan X (2017) pheS*, an effective host-genotype-independent counter-selectable marker for marker-free chromosome deletion in Bacillus amyloliquefaciens. Appl Microbiol Biotechnol 101:217–227. https://doi.org/10.1007/s00253-016-7906-9
Funding
This work was funded by the National Natural Science Foundation of China grants 31672232, 31600088, and 31572214, and the National Key R & D Program of China grant 2018YFD0500201.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(PDF 53 kb)
Rights and permissions
About this article
Cite this article
Liu, Y., Feng, H., Fu, R. et al. Induced root-secreted d-galactose functions as a chemoattractant and enhances the biofilm formation of Bacillus velezensis SQR9 in an McpA-dependent manner. Appl Microbiol Biotechnol 104, 785–797 (2020). https://doi.org/10.1007/s00253-019-10265-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-019-10265-8