Abstract
D-p-hydroxyphenylglycine (D-HPG) functions as an intermediate and has important value in antibiotic industries. The high pollution and costs from chemical processes make biotechnological route for D-HPG highly desirable. Here, a whole-cell transformation process by D-hydantoinase(Hase) and D-carbamoylase(Case) was developed to produce D-HPG from DL-hydroxyphenylhydantoin(DL-HPH) in Escherichia coli. The artificially designed ribosome binding site with strong intensity significantly facilitated the protein expression of limiting step enzyme Case. Next, the cell wall permeability was improved by disturbing the peptidoglycan structure by overproduction of D,D-carboxypeptidases without obviously affecting cell growth, to increase the bioavailability of low soluble hydantoin substrate. By fine-tuning regulation of expression level of D,D-carboxypeptidase DacB, the final production yield of D-HPG increased to 100% with 140 mM DL-HPH substrate under the optimized transformation conditions. This is the first example to enhance bio-productivity of chemicals by cell wall engineering and creates a new vision on biotransformation of sparingly soluble substrates. Additionally, the newly demonstrated ‘hydroxyl occupancy’ phenomenon when Case reacts with hydroxyl substrates provides a referential information for the enzyme engineering in future.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The optical D-amino acids as intermediate for antibiotics, peptide hormones, pyrethroids, and other insecticides (Andreas et al. 1998; Park et al. 2000) are widely used in pharmacy, cosmetics, and agriculture (Liu et al. 2017). Among them, D-p-hydroxyphenylglycine (D-HPG) is mainly used for semi-synthesis of β-lactam antibiotics, such as cephalexin, cefadroxil, amoxicillin, etc (Nanba et al. 1998). The chemical methods to produce D-HPG have many problems in the process of decarbamoylation, such as large disposal of waste and low yield. Therefore, biocatalytic processes have gained enormous attentions.
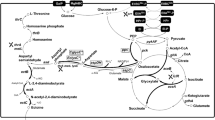
The hydantoin-transforming reaction is a classical double enzyme tandem reaction (Olivieri et al. 1981), starting from substrate DL-hydroxyphenylhydantoin(DL-HPH) (Fig. 1). In the first step, the DL-HPH is converted to the corresponding N-carbamoyl-D-amino acid by D-hydantoinase (Hase). And the intermediate N-carbamoyl-D-p-hydroxyphenylglycine (CpHPG) is further hydrolysed to D-HPG by N-carbamoyl-D-amino acid amidohydrolase (Case). To establish an efficient reaction, screening of high enantioselectivity, specific enzyme activity, protein solubility, and reaction compatibility of Hase and Case is critical. Hase has been identified and characterized in a variety of microorganisms (Lapointe et al. 1995), including Agrobacterium (Runser and Meyer 1993), Bacillus stearothermophilus (Cheon et al. 2002), Burkholderia pickettii (Xu et al. 2003), Bacillus thermocatenulatus(Park et al. 1998), Jannaschia sp. (Cai et al. 2009), and Brevibacillus parabrevis (Nandanwar et al. 2013). Case was first isolated from Agrobacterium radiobacter (Olivieri et al. 1979) and later found in Arthrobacter (Möller et al. 1988), Pseudomonas (Ikenaka et al. 1998), Sinorhizobium morelens (Wu et al. 2005), and Comamonas(Ogawa et al. 1993). Based on the identified enzymes, most of studies selected Escherichia coli as host for co-expression of homologous Hase and Case to produce D-HPG by transformation processes. Yu et al. (2009) also over-produced endogenous enzymes in Ralstonia pickettii to further improve the production of D-HPG.
Case normally has lower activity and functional expression level compared to Hase. To improve the reaction efficiency, efforts have been made to engineer enzyme stability (Jiang et al. 2007) and increased the reaction efficiency. However, there is still a big problem needed to be solved, the low solubility of the hydantoin substrates, which is also a key factor to limit the transformation rate. Reduction of the mass transfer resistance across the cell envelope should improve the uptake efficiency of the hydantoin substrate. In the previous publications, adding co-solvent to the reaction system (Liu et al. 2017), implementing aqueous two-phase system (Qian et al. 2012), and disruption of bacterial cells (Liu et al. 2008) were performed to increase the bioavailability of the hydantoin substrate for enzymatic reactions. However, adding organic solvent or cell broken process will obviously increase the costs of operation and downstream treatments, which is not satisfactory to the industrial application.
In this work, the enzymes with highest activities were chosen from the database, and their production in E. coli was optimized by artificial high intensity ribosome binding site (RBS) and changing the gene order as well. To solve the problem from sparing solubility of hydantoin substrate, the cell wall permeability of E. coli was engineered by fine-tuning the regulation of native D,D-carboxypeptidase gene expression level. Combination with process optimization, a biotransformation process with high yield and productivity of D-HPG, was developed.
Materials and methods
Bacterial strains, plasmids, and cultivation conditions
E. coli TOP10 was used for plasmid construction and E. coli MG1655 was the host for biocatalytic production of D-HPG. Plasmid pTrc99a was used for Hase and Case gene expression, while the plasmid pYB1s was used for expressing dacA and dacB. All the plasmids and strains used in this study are listed in Table 1.
The strains E. coli TOP10 and E. coli MG1655 were cultivated in Luria-Bertani (LB; 10 g/L tryptone, 5 g/L yeast extract, 10 g/L NaCl) medium for 12 h at 37 °C in an orbital shaker. All strains harboring recombinant plasmids were maintained with antibiotics as selective pressure such as ampicillin (100 μg/mL) and streptomycin (50 μg/mL). The corresponding strains for protein expression were first cultured at 37 °C with shaking. When the culture reached an optical density at 600 nm of 0.8, the inducer was added, and then, the strains were further cultivated for 14 h at 25 °C and 200 rpm. Gene expression in strain with plasmid pTrc99a was induced with 0.5 mM isopropyl-β-d-thiogalactoside(IPTG) while the recombinant strains harboring plasmid pYB1s were induced with 0.2% (v/v) arabinose.
Construction of expression vector for production of HPG
The Hase and Case genes were synthesized with E. coli codon optimization and the sequences were deposited in NCBI database under the accession numbers of MN073199 and MN073200, respectively. For constructing the expression plasmid pT01, the gene of Hase was amplified by PCR with primers Hase-F1 and Hase-R1 using the codon-optimized Hase gene as the template, and gene of Case was amplified with primers Case-F1 and Case-R1 based on the codon-optimized Case gene as well. Hase and Case fusion proteins were connected by four different linkers. For plasmid pT011/pT012/pT013/pT014 construction, primers for gene of Hase were used with Hase-F1 and Hase-RG1/Hase-RG2/Hase-RG2/Hase-RG4; for Case, primers were used with Case-FG1/Case-FG2/Case-FG3/Case-FG4 and Case-R1, separately. Separate expression of the two proteins, the gene of Hase, was cloned with Hase F/R02, while the gene of Case was cloned with Case F/R02. To construct plasmid pT04, plasmid pT03 with Case F02/R03 and Hase F/R03 was firstly constructed; then, MAX-F and MAX-R were used to change the RBS sequence. PCR amplification was performed using PrimeSTAR HS (Premix) polymerase (TaKaRa, Shanghai, China). The PCR product was purified from agarose gel (Omega Bio-Tek, Inc. USA), and then ligated into the pTrc99a or pYB1s plasmid digested with the appropriate restriction enzymes (New England BioLabs, Beverly, MA). The recombinant plasmids were transformed into E. coli TOP10 and cultivated at 37 °C on LB agar plate with 100 μg/mL ampicillin or 50 μg/mL streptomycin overnight. The positive recombinant strains were verified by DNA sequencing and were further transformed into E. coli MG1655 for protein expression and testing the capacity for production of D-HPG from DL-HPH. All primers used in this study are listed in Table 2.
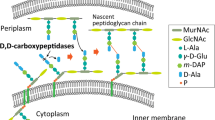
Engineering cell wall permeability by overproduction of D,D-carboxypeptidases
To overexpress the native genes dacA and dacB which encode the D,D-carboxypeptidases, the genomic DNA of E. coli MG1655, which was extracted by TIANamp Bacteria DNA Kit (TIANGEN Biotech, Co. Ltd., Beijing, China), was used as the PCR template. The pelB signal peptide sequence was annealed by primer pelB-F and pelB-R at 95 °C for 10 min and put on ice for 30 min. The primers A/B-F/R1 used to overexpress the dacA or dacB and the primers A-F1/R2, B-F2/R1 used for combined expression of dacA and dacB are listed in Table 2. The primers were designed based on the sequences from NCBI with accession numbers of NP_415165.1 (dacA) and NP_417649.1 (dacB), respectively. Recombinant plasmids were introduced into competent E. coli MG1655 cells containing recombinant pT04 plasmids. The positive strains were selected on LB agar plates with 100 μg/mL ampicillin and 50 μg/mL streptomycin.
Whole-cell bioconversion for D-HPG production
All the strains were grown at 37 °C in LB medium to the post-exponential growth phase (OD600 = 0.8); then, 0.5 mM IPTG was added and the cultivation was performed at 25 °C. The strains containing pYB1s required additional 0.2% (v/v) arabinose as an inducer. After cultivation for 14 h, the induced cells were harvested by centrifugation at 10,000×g for 5 min and the pellets were resuspended in 50 mM Tris-HCl buffer (pH 8.0). The reaction mixture contained 20 mM DL-HPH (Aladdin, Shanghai, China), 1.0 mM DTT, and 1.0 mM MnCl2 in 1-mL 50 mM Tris-HCl buffer (pH 8.0).
Molecular dynamics simulation of Case with N-carbamoyl-D-amino acids
The molecular dynamics (MD) simulations were performed via Gromacs-5.1.4 with force field AMBER-03(Abraham et al. 2015; Sorin and Pande 2005). The starting conformation for the protein N-carbamyl-D-amino acid amidohydrolase with RCSB code 1erz (Nakai et al. 2000) was immersed in a periodic box of TIP3P water, having a size of 13 nm × 13 nm × 13 nm (Jorgensen et al. 1983). A total of 40 Na+ ions and 20 Cl− were added to the box to neutralize the negative charges of the protein. A cut-off of 1 nm was applied for the Lennard-Jones interaction and the real space portion of electrostatic interactions. The PME method was used to calculate the reciprocal space portion of electrostatic interactions (Darden et al. 1993). The water molecules and counter ions were relaxed for 5 ns with the position of the protein backbone atoms position restrained. Subsequently, the system was equilibrated for 5 ns with constant pressure and temperature conditions (NPT) of 300 K and 1 bar. The NPT condition was obtained by coupling the system to a Parrinello-Rahman barostat and the Nóse-Hoover thermostat, with relaxation time of 0.5 ps (Parrinello and Rahman 1981; Nose 1984). The last 1 ns of NPT simulation results was used to calculate an average structure for substrate docking analysis. The substrate molecules N-carbamoyl-D-p-hydroxyphenylglycine and N-carbamoyl-D-phenylglycine were built by PRODRG (Schüttelkopf and van Aalten 2004). AutoDock performs the molecular dockings by pre-calculating energy grids around a site of interest on the target (Morris et al. 2009). A stochastic search algorithm utilizing the Lamarkian Genetic Algorithm (LGA) for exploring the grid space is used to perform energy evaluations of the position of the substrate with respect to the target energy grids. This algorithm explores the various orientations and conformations of the whole substrate relative to the energy grids for the defined number of energy evaluations and returns the lowest energy conformation in the target site. According to the previous report (Nakai et al. 2000), the target docking pocket of the tetramer was explored by two substrates, respectively.
Analytical methods
To detect the cell permeability, the engineered strains were harvested by centrifugation at 10,000×g for 5 min after cultivation for 14 h, and the pellets were resuspended in 50 mM Tris-HCl buffer (pH 8.0) to OD600 = 0.5. For inner membrane permeability assessment, 140-μL cell suspension was mixed with 60-μL 30 mM o-nitrophenylβ-D-galactopyranoside (Aladdin Co., Shanghai, China). The reactions were performed at 37 °C for 30 min and the absorbance was detected at 420 nm (Lehrer et al. 1988). For outer membrane permeability assessment, 180-μL cell suspension was mixed with 20-μL 100 μM N-phenyl-α-naphthylamine (Bidepharm Co., Shanghai, China). The UV emission was measured at 420 nm and the excitation wavelength was set at 350 nm. The fluorescence was recorded (Loh et al. 1984).
The amount of product was detected by high-performance liquid chromatography (HPLC) (Kim and Kim 1995; Lee et al. 1995). The HPLC mobile phase was 10% acetonitrile: 90% ddH2O (v/v) and the solvent flow rate was 0.1 mL/min. The column Poroshell 120 EC18-2.7 m (4.6 mm × 50 mm) (Agilent, California, USA) was used. The column eluent was detected at 230 nm. The retention times of DL-HPH, intermediate CpHPG, and D-HPG were 13.48, 14.12, and 24.16 min, respectively. The yield was calculated according to the product/substrate (mol/mol). All experiments were independently performed in triplicate times, and data are shown as mean ± standard deviation (SD).
Results
Database screening of enzymes for D-HPG production in E. coli
To establish an efficient hydantoin-transforming reaction, we first screened suitable enzymes in BRENDA database (https://www.brenda-enzymes.org/) and literatures. Hase from Bacillus stearothermophilusSD-1 with mutant of M63I/F159S (accession number is 1K1D_A) and Case from Agrobacterium sp. strain KNK712 (accession number is 1ERZ_A) were finally selected due to their highest activities in the database (Louwrier and Knowles 1996). Unexpectedly, the initial strain with plasmid of pT01 showed low activity towards D-HPG production, in which the yield was 19.4% in 0.5 h with a titer of less than 4.0 mM. Even we extended the reaction time to 20 h, the results were still unsatisfied and there was an accumulation of intermediate CpHPG. SDS-PAGE analysis revealed that the Hase gene expression was quite well while the expression of the Case remained at low level (data not shown). Previous reports also showed that the enzyme Case was the limiting step setting the pace of the hydantoin-transforming reaction (Chao et al. 1999; Park et al. 2000). Given the above fact, we first focused on the optimization of the gene expression.
Optimization of the D-carbamoylase gene expression in E. coli
As the gene expression of Hase was good, we first tried the fusion of Case after Hase to drive Case gene expression accordingly. The linker (GGGGS)n with different repeat number (n) is most widely used in fusion engineering because it provides flexibility and allows for mobility of the fused functional domains (Chen et al. 2013). Then, four flexible linkers (GGGGS)1 to (GGGGS)4 were used to construct Hase-Case fusion proteins. After the whole-cell bioconversion, the strains which added protein linker from linkers (GGGGS)1 to (GGGGS)4, the product yield of D-HPG in 0.5 h were 15.1%, 23.2%, 21%, and 21.2%, respectively. The production of D-HPG was slightly increased compared to the strain without fusion expression (19.4%), except that the production of strain with (GGGGS)1 was declined (Fig. 2). None of the fusion expression strains gave satisfactory results by measuring the production of D-HPG. The gene expression was also investigated on two separate plasmids, while it only gave a yield of 24.3% in 0.5 h.
Optimizing gene expression of the rate-limiting enzyme Case in E. coli. Trc, Trc promoter. (GGGGS)n, linker sequence and n represents the repeat number of GGGGS. MAX, strongest RBS intensity for Case designed by the RBS Calculator and the sequence is shown in Table 3. Gray bars represent the yield (mM produced D-HPG/mM consumed DL-HPH). Black bars represent D-HPG titer (mM). All the experiments were performed in triplicate. The SDS-PAGE analyses on crude enzymes showed the gene expression of Hase and Case. M, protein maker. BC, control strain MG with empty plasimd pTrc99a. Lane 01, strain MG01, and lane 02, strain MG04
Many researches revealed that the protein expression levels are related to RBS strength (Nowroozi et al. 2014; Zhang et al. 2015). In order to increase the expression of the Case, we selected the strongest RBS intensity to express Case gene (Table 3), which designed based on the gene sequence by the RBS Calculator (https://salislab.net/software/). The gene order was also changed, in which the Case gene was put in front adjacent to Ptrc promoter to maximize the Case gene expression. Consequently, the resultant strain carrying plasmid pT04 can produce D-HPG with the yield reached to 46.9% in 0.5 h which increased the yield of D-HPG by 1.42-fold as compared to the strain with plasmid of pT01, and the intermediate CpHPG remained at low concentration of 2.59 ± 0.31 mM. SDS-PAGE analysis also confirmed that Case gene expression was dramatically improved in E. coli (Fig. 2).
Engineering cell wall permeability to increase the productivity
The substrate is poorly soluble and appears precipitation of heat-dissolved DL-HPH in a period time, which severely limits the efficiency of reaction (Liu et al. 2017). We first added different kinds of solubilizers (PEG 400, Tween 80, methanol, and DMSO) in the reaction system to increase the bioavailability of DL-HPH, but the effect was not obvious (data not shown). To verify if cell wall is indeed the barrier for D-HPG production, the cells carrying plasmid pT04 were disrupted by ultrasonication and the supernatant was used as cell-free system to transform DL-HPH. The yield of D-HPG significantly increased to 75.6% in 0.5 h, which did indicate that the barrier of cell wall is a key factor. Since cell disruption is costly for industrial use, and additionally, substrate particles and titrant NaOH solution were reported to deactivate free enzymes (Park et al. 2000). Therefore, whole cell instead of free enzymes could be more applicable in industrial scenes.
Recently, the cell wall engineering by disturbing peptidoglycan structure was a new direction for enhancing cell wall permeability (Yang et al. 2018a, b). PBP5 (DacA) is one of the main D,D-carboxypeptidases, and PBP4 (DacB) has both D,D-endopeptidase and D,D-carboxypeptidase activities. Both of them are significant in peptidoglycan synthesis network (Kishida et al. 2006; Nelson and Young 2001), and importantly, none is essential for bacterial viability (Denome et al. 1999). To investigate the effects of overexpressing dacA and dacB on D-HPG production, the two genes were overexpressed separately and combinedly. Since the site of action of D,D-carboxypeptidase is in the periplasm (Kishida et al. 2006), each gene was added a signal peptide of pelB. As shown in Fig. 3a, even at reduced OD600 by 1.0 after overexpression of dacA and dacB, the production of D-HPG was increased by 20%. The D-HPG production of recombinant strains MG04A (overexpression of dacA with RBS intensity of 27,000 in the plasmid, designed by the RBS Calculator) and MG04B (overexpression of dacB with RBS intensity of 50) were 14.24 mM and 15.56 mM in 4 h from 20 mM substrates, respectively. The strains that overexpressed both genes, MG04AB1, showed the best D-HPG production titer and yield (Fig. 3b). These results clearly demonstrated that the cell wall engineering strategy is useful for improving D-HPG production performance in E. coli.
The effect of D,D-carboxypeptidase gene overexpression on cell growth (a) and D-HPG production (b). Black bars represent D-HPG production (mM) in 0.5 h. Gray bars represent D-HPG production (mM) in 4 h. CK, control strain MG04 without overexpression of carboxypeptidase. a Strain MG04A with overexpression of DacA. b MG04B with overexpression of DacB. AB1, strain MG04AB1 with overexpression of DacA and DacB. All the experiments were performed in triplicate
Fine-tuning regulation of dacB expression on D-HPG production
The above studies showed that overexpression of dacB gene seemed more important to increase the production of D-HPG. At initial construction of plasmid for overexpressing both genes, the RBS intensity of the dacB gene was adjusted to a lower level, considering to avoid adverse effects on cell growth. The recombinant strain MG04AB1 (the RBS intensity of the dacB gene was 50) resulted in 16.3 mM in 4 h from 20 mM substrate. To further increase the yield of D-HPG, the RBS intensities for expression of dacB gene were increased to 250 and 500, respectively, while the production performance was the same with MG04AB1. Then, the effects of DacB by fine-tuning regulation of its expression level with strong RBS intensity were investigated. The plasmids pYAB2 and pYAB3 (the RBS intensities of the dacB gene were 10,000 and 60,000, respectively; 60,000 is the maximum strength that could be designed) were constructed to enhance the production of protein DacB. As shown in Fig. 4a, the constructed strains with higher RBS intensity of dacB gene had higher production of D-HPG than the low RBS intensity one. And the one with modest RBS intensity of 10,000 for dacB gene, MG04AB2, had better performance for D-HPG production, in which the titers were increased to 17.65 mM at 4 h from 20 mM substrate. Meanwhile, the yield of D-HPG reached 88.25% in 4 h while the yield of strain without cell wall engineering was only 63.00%. Notably, the yield of cell wall engineered strain in 0.5 h was 69.2%, which reached to 91% of the outcome from cell disruption reaction.
Effects on D-HPG production and cell permeability by fine-turning regulation of DacB gene expression. a The effects on D-HPG production. Black bars represented D-HPG production (mM) in 0.5 h and gray bars represented D-HPG production (mM) in 4 h. b The effects on cell permeability. c The transcriptional analysis of dacA and dacB in the best strain MG04AB2. CK, control strain without cell wall engineering; AB1, strain with co-expression of dacA and dacB with calculated RBS intensity of 50; AB2, strain with co-expression of dacA and dacB with calculated RBS intensity of 10,000; and AB3, strain with co-expression of dacA and dacB with calculated RBS intensity of 60,000. All the experiments were performed in triplicate
The cell permeability of the engineered cells was also determined. As shown in Fig. 4b, the cell permeability increased accordingly with the increase of RBS intensity of dacB and the best strain MG04AB2 gave the highest cell membrane permeability. Additionally, the transcriptional analysis of dacA and dacB with strain MG04AB2 also well supported the conclusion, in which the transcriptional level of dacA and dacB in MG04AB2 was much higher than those of the control strain (Fig. 4b). These results indicated the contributory role of overexpression of dacA and dacB for increase of D-HPG production.
Optimization of whole cell transformation conditions for D-HPG production
The whole cell transformation by strain MG04AB2 was optimized to develop an efficient D-HPG production process. The effects of reaction temperature on hydantoin-transforming reaction were first studied. It was found that reaction temperature had significantly effective D-HPG production. A decrease in reaction temperature leads to a significant decrease in D-HPG production. The titers of D-HPG in 45 °C and 50 °C were identically highest in 0.5 h while the reaction at 45 °C gave the better performance since the yield reached almost 100% in 4 h (Fig. 5a). To demonstrate the optimal pH of this double enzyme tandem reaction, different buffers with pH from 6.5 to 9.0 were evaluated at 45 °C. The hydantoin-transforming reaction displayed highest production of D-HPG at pH 8.5 with a yield of 100% in 4 h, respectively (Fig. 5b). The optimization of cell reaction concentration was also evaluated under the optimized pH and temperature condition. As shown in Fig. 5c, the yield of D-HPG in OD600 from 10 to 30 reached 100% in 4 h with the highest specific productivity of 1.23 (g/g DCW/h), obtained with cell mass of 10 OD/mL.
Optimization of transformation conditions for D-HPG production by strain MG04AB2. a The effect of reaction temperature optimization on D-HPG production. Black bars represented D-HPG production (mM) in 0.5 h. Gray bars represented D-HPG production (mM) in 4 h. b The effect of reaction pH optimization on D-HPG production. Square pattern represented phosphate buffer (pH 6.5–8.0, 50 mM). Circular pattern represented Tris-HCl buffer (pH 7.0–9.0, 50 mM). c The effect of cell reaction concentration optimization on D-HPG production. Gray bars represented specific productivity (g/g DCW/h) in 4 h. Black plots represented the molar yield of D-HPG (%) in 4 h. d Biotransformation production of D-HPG with 100 and 140 mM substrate DL-HPH under optimized conditions, respectively
After optimizing the reaction conditions, a high substrate concentrations of respective 100 and 140 mM were tested, in which the concentration of 140 mM is expected to reach the upper solubility concentration limit of D-HPG in aqueous phase (Fan et al. 2000), if all substrate was transformed. As shown in Fig. 5d, all 140 mM DL-HPH could be transformed to D-HPG in 32 h with a yield of 100%, although the specific productivity decreased reasonably. Normally, the lower concentration should lead a shorter reaction time. While even a low concentration was tested, the complete conversion time for 100 mM substrate was consistent with that of 140 mM substrate.
MD simulations revealed catalytic disadvantage of Case for hydroxyl substrate
With intent to explore the possible mechanisms for the above phenomenon, molecular dynamics (MD) simulation was carried out to reveal the catalytic mechanism between enzyme and substrate. Hase used in this study is already mutated (M63I/F159S) to increase the activity towards DL-HPH(Louwrier and Knowles 1996). As Case was the limiting-step enzyme, we focused on Case to find the cause. When docking the intermediate N-carbamoyl-D-p-hydroxyphenylglycine (CpHPG) to the active pocket of Case, the lowest energy structure of docking is − 4.91 kcal/mol. And we unexpectedly discovered that the hydroxyl group of CpHPG is more easily to channel into the active center, instead of N-carbamoyl group (Fig. 6a). The hydroxyl group of CpHPG forms a hydrogen bond with the amino acids near the docking interface, hinders the substrate from entering the catalytic reaction center in a normal posture, and thereby lowering the activity of the enzyme. When the non-hydroxyl substrate, N-carbamoyl-D-phenylglycine, was used, the group of N-carbamoyl is correctly set at the active pocket of Case and the lowest energy structure of docking is − 5.69 kcal/mol (Fig. 6b), which is consistent with the previous analysis with another non-hydroxyl substrate N-carbamoyl-phenylalanine (Nakai et al. 2000). It sounds like that a ‘hydroxyl occupancy’ occurred when CpHPG was reacted with Case. The ‘wet’ experiments were also investigated by the strain MG04AB2 with hydroxyl and non-hydroxyl substrates, respectively. The experimental data clearly supported the prediction from MD simulations, which the substrate phenylhydantoin reacted much faster than p-hydroxyphenylhydantoin at 20 mM concentration and the difference was more obvious when 60 mM substrates were investigated (Fig. 6c, d). This indicated a congenital deficiency of Case to react with hydroxyl substrates and hindered to easily engineer the enzyme with high activity.
Molecular dynamics simulation of Case with N-carbamoyl-D-amino acids and experimental verification. a The molecular dynamics simulation of Case with N-carbamoyl-D-amino acid substrates of N-carbamyl-D-p-hydroxyphenylglycine and bN-carbamyl-D- phenylglycine, respectively. The hydroxyl group of N-carbamyl-D-p- hydroxyphenylglycine was indicated in black circle and N-carbamoyl group was in red. The catalytic efficiencies were tested by whole cells of strain MG04AB2 with substrates of p-hydroxyphenylhydantoin and phenylhydantoin at concentrations of 20 mM (c) and 60 mM (d), respectively
Discussion
D-HPG is an important building block in the fine chemicals industry while its current production is mainly based on chemical process. Since enzymatic decarbamoylation offers several advantages over the chemical method, how to establish a cost-effective biocatalyst is the first to provide an economically feasible process for production of D-HPG. E. coli is the most commonly used as the host for the generation of D-HPG in the reports of the past decade. However, the second step enzyme Case expressed in E. coli often resulted in low expression or formation of inactive inclusion bodies. Many studies showed that the production rates of D-HPG are affected to a large extent by the enzyme Case (Yu et al. 2009; Zhang et al. 2011). In this study, selecting the strongest artificial RBS intensity and putting the gene adjacent to the promoter successfully improved the gene expression and increased the yield of D-HPG.
Besides the point of rate-limiting enzyme, there is still a problem to be solved: the sparing solubility of the hydantoin substrate. In order to enhance the absorption of the substrate DL-HPH to the cells, the strategy of disturbing cell wall peptidoglycan structure was used in this study to increase the permeability of the E. coli MG1655. In most bacterial cell walls, the peptidoglycan sacculus are made from a short peptide cross-linked glycan chain to form a unique structure surrounding the membrane of the cell (Mengin-Lecreulx and Lemaitre 2005; Nanninga 1998; Vollmer et al. 2008). And Gram-negative bacteria harbor a more tightly arranged structural form than Gram-positive bacteria (Beveridge 1999). In E. coli, the penicillin-binding proteins (PBPs) are crucial in peptidoglycan synthesis network (Typas et al. 2011; Yang et al. 2018b). The activity of D,D-carboxypeptidase is hydrolysis of terminal amino acid residues, and the activity of endopeptidase is the cross-linking of the cleaved peptide (Typas et al. 2011). In present study, overexpression of dacB gene alone was better than that of dacA, probably because the DacA is one of the major D,D-carboxypeptidases while DacB has both D,D-carboxypeptidase and endopeptidase activities (Kishida et al. 2006; Nelson and Young 2001). Overexpression of dacA and dacB can disrupt the peptidoglycan synthesis network and the cell wall structure subsequently, which enhances the permeability of the inner and outer membranes of E. coli to let more substrate enter into cells for D-HPG production. Liu et al. (2008) used sonication-treated cells to conquer the barrier of cell wall, in which the yield of D-HPG was 96.3% in 4 h with 15 mM DL-HPH as substrate. By fine-tuning regulation of protein DacB expression level, the production yield of D-HPG increased obviously in our study. The yield of 100% with 20 mM DL-HPH as substrate was obtained in 4 h by using cell wall-engineered bacteria without cell disruption. Changing cell morphology by cell wall engineering strategy to improve production performance has been reported in other literatures. Overexpression or deletion of D,D-carboxypeptidase genes (dacA and dacB) to interrupt peptidoglycan structure has been proven to be an efficient way to boost secretion of extracellular proteins in E. coli (Yang et al. 2018a, b). Inhibition of peptidoglycan synthesis in Streptococci strain also improved the lipid secretion (Horne et al. 1977). Wu et al. (2016) improved PHB production by engineering E. coli cell wall with an elongated shape. Beyond secretion of enzymes or lipids and internal production of biopolymer, this study is the first example to enhance productivity of chemicals by disrupting cell wall structure and provided a new vision on improvement of biotransformation efficiency from sparingly soluble substrates.
After engineering a biocatalyst with good performance, the process optimization is vital to obtain an economical biotechnology. The conversion rate for DL-HPH to D-HPG at pH 8.5 was much higher than that at neutral pH conditions, although the optimal pH for Case was found to be pH 7.0 while Hase showed the maximum activity at pH 8.5 (Park et al. 2000). It was reported that the rate of chemical racemization of substrate DL-HPH is related to alkaline condition (Yu et al. 2009). The best performance obtained at pH 8.5 indicated that the spontaneous chemical racemization of substrate may also play an important role in the bioconversion process, although the enzyme of Case is not happy under this condition. From an economical standpoint of view, the substrate concentration is expected to feed as high as possible in order to increase the productivity and facilitate the downstream product recovery processes. However, the starting substrate DL-HPH is just about 60 mM at 45 °C (Lee and Kim 1998). Considering that the product D-HPG also has a solubility concentration of less than 140 mM in aqueous phase (Fan et al. 2000) and more D-HPG will be lost during downstream separation process, the substrate concentration at 140 mM was tested. Using the cell wall-engineered cells, the high yield of 100% was achieved in this study. The above data clearly demonstrated the great potential of cell wall engineering strategy for low soluble substrate reaction.
At our initial purpose, MD simulation was expected to find key residues for further improving the activity of Case, which is a limiting step enzyme. However, we unexpectedly revealed that the hydroxyl, but not N-carbamoyl group, is more easily channeled into the active center of enzyme Case. The reaction speed was much faster when the non-hydroxyl substrate phenylhydantoin was used, which well supported the computational analysis. This new finding indicated a congenital deficiency of Case to react with hydroxyl substrates and might explain why lower substrate concentration did not result in a shorter reaction time. The high initial concentration of substrate provides increasing probability of correctly positioning into active center and hence increased productivity. When the concentration decreased, the correct probability also decreased and resulted in the lower productivity, as shown in Fig. 5d. Additionally, the ‘hydroxyl occupancy’ deficiency of Case could also explain why there were no positive achievements to date on engineering of Case for increased activities, as compared to those of Hase. Thus, this study also provided the referential information for the enzyme engineering in future, from simple design of increasing activities to emphasis on elimination of ‘hydroxyl occupancy’ deficiency.
In conclusion, we developed an efficient D-HPG-producing process based on optimization of catalytic enzyme expression and destabilizing cell wall structure by fine-tuning regulation of D,D-carboxypeptidase DacB gene expression in E. coli. The new finding in limiting-step enzyme corrects the protein engineering direction for further improving catalytic activity of D-carbamoylases and hence developing an economical bioprocess for D-HPG production in future. The rational design of Case to eliminating ‘hydroxyl occupancy’ is under investigation.
References
Abraham MJ, Murtola T, Schulz R, Páll S, Smith JC, Hess B, Lindahl E (2015) GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1-2:19–25
Andreas SB, Michael S, Karlheinz D (1998) Biocatalysis to amino acid-based chiral pharmaceuticals-examples and perspectives. J Mol Catal B-Enzym 5(1-4):1–11
Beveridge TJ (1999) Structures of gram-negative cell walls and their derived membrane vesicles. J Bacteriol 181(16):4725–4733
Cai YH, Trodler P, Jiang S, Zhang W, Wu Y, Lu YH, Yang S, Jiang WH (2009) Isolation and molecular characterization of a novel D-hydantoinase from Jannaschia sp. CCS1. FEBS J 276(13):3575–3588
Cescau S, Debarbieux L, Wandersman C (2007) Probing the in vivo dynamics of Type I protein secretion complex association through sensitivity to detergents. J Bacteriol 189(5):1496–1504
Chao YP, Fu H, Lo TE, Chen PT, Wang JJ (1999)One-step production of D-p-hydroxyphenylglycine by recombinant Escherichia coli strains. Biotechnol Prog 15(6):1039–1045
Chen XY, Zaro JL, Shen WC (2013) Fusion protein linkers: property, design and functionality. Adv Drug Deliv Rev 65(10):1357–1369
Cheon YH, Kim HS, Han KH, Abendroth J, Niefind K, Schomburg D, Wang J, Kim Y (2002) Crystal structure of D-hydantoinase from Bacillus stearothermophilus: insight into the stereochemistry of enantioselectivity. Biochemistry 41(30):9410–9417
Darden T, York D, Pedersen L (1993) Particle mesh Ewald: An N•log(N) method for Ewald sums in large systems. J Chem Phys 98:10089–10092
Denome SA, Elf PK, Henderson TA, Nelson DE, Young KD (1999)Escherichia coli mutants lacking all possible combinations of eight penicillin binding proteins: viability, characteristics, and implications for peptidoglycan synthesis. J Bacteriol 181(13):3981–3993
Fan CH, Lee CK, Chao YP (2000) Recombinant Escherichia coli cell for D-p-hydroxyphenylglycine production from D-N-carbamoyl-p- hydroxyphenylglycine. Enzym Microb Technol 26(2-4):222–228
Horne D, Hakenbeck R, Tomasz A (1977) Secretion of lipids induced by inhibition of peptidoglycan synthesis in streptococci. J Bacteriol 132(2):704–717
Ikenaka Y, Nanba H, Yamada Y, Yajima K, Takano M, Takahashi S (1998) Screening, characterization, and cloning of the gene for N-carbamyl-D-amino acid amidohydrolase from thermotolerant soil bacteria. Biosci Biotechnol Biochem 62(5):882–886
Jiang S, Li CH, Zhang WW, Cai YH, Yang YL, Yang S, Jiang WH (2007) Directed evolution and structural analysis of N-carbamoyl-D-amino acid amidohydrolase provide insights into recombinant protein solubility in Escherichia coli. Biochem J 402(3):429–437
Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML (1983) Comparison of simple potential functions for simulating liquid water. J Chem Phys 79:926–935
Kim GJ, Kim HS (1995) Optimization of the enzymatic synthesis of D-p-hydroxyphenylglycine from DL-5-substituted hydantoin using D-hydantoinase and N-carbamoylase. Enzym Microb Technol 17(1):63–67
Kishida H, Unzai S, Roper DI, Lloyd A, Park SY, Tame JR (2006) Crystal structure of penicillin binding protein 4 (dacB) from Escherichia coli, both in the native form and covalently linked to various antibiotics. Biochemistry 45(3):783–792
Lapointe G, Leblanc D, Morin A (1995) Use of a polymerase-chain-reaction-amplified DNA probe from Pseudomonas putida to detect D-hydantoinase-producing microorganisms by direct colony hybridization. Appl Microbiol Biotechnol 42(6):895–900
Lee DC, Kim HS (1998) Optimization of a heterogeneous reaction system for the production of optically active D-amino acids using thermostable D-hydantoinase. Biotechnol Bioeng 60(6):729–738
Lee SG, Lee DC, Hong SP, Sung MH, Kim HS (1995) Thermostable D-hydantoinase from thermophilic Bacillus stearothermophilus SD-1: characteristics of purified enzyme. Appl Microbiol Biotechnol 43(2):270–276
Lehrer RI, Barton A, Ganz T (1988) Concurrent assessment of inner and outer membrane permeabilization and bacteriolysis in E. coli by multiple-wavelength spectrophotometry. J Immunol Methods 108(1-2):153–158
Liu YQ, Li Q, Hu XJ, Yang JC (2008) Construction and co-expression of polycistronic plasmid encoding D-hydantoinase and D-carbamoylase for the production of D-amino acids. Enzym Microb Technol 42(7):589–593
Liu YF, Xu GC, Han RZ, Dong JJ, Ni Y (2017) Identification of D-carbamoylase for biocatalytic cascade synthesis of D-tryptophan featuring high enantioselectivity. Bioresour Technol 249:720–728
Liu B, Xiang SM, Zhao G, Wang BJ, Ma YH, Liu WF, Tao Y (2019) Efficient production of 3-hydroxypropionate from fatty acids feedstock in Escherichia coli. Metab Eng 51:121–130
Loh B, Grant C, Hancock RE (1984) Use of the fluorescent probe 1-N-phenylnaphthylamine to study the interactions of aminoglycoside antibiotics with the outer membrane of Pseudomonas aeruginosa. Antimicrob Agents Chemother 26(4):546–551
Louwrier A, Knowles CJ (1996) The purification and characterization of a novel D-specific carbamoylase enzyme from an Agrobacterium sp. Enzym Microb Technol 19(8):562–571
Mengin-Lecreulx D, Lemaitre B (2005) Structure and metabolism of peptidoglycan and molecular requirements allowing its detection by the Drosophila innate immune system. J Endotoxin Res 11(2):105–111
Möller A, Syldatk C, Schulze M, Wagner F (1988)Stereo- and substrate-specificity of a D-hydantoinase and a D-N-carbamyl-amino acid amidohydrolase of Arthrobacter crystallopoietes AM 2. Enzym Microb Technol 10(10):618–625
Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ (2009) Autodock4 and AutoDockTools4: automated docking with selective receptor flexiblity. J Comput Chem 30(16):2785–2791
Nakai T, Hasegawa T, Yamashita E, Yamamoto M, Kumasaka T, Ueki T, Nanba H, Ikenaka Y, Takahashi S, Sato M, Tsukihara T (2000) Crystal structure of N-carbamyl-D-amino acid amidohydrolase with a novel catalytic framework common to amidohydrolases. Structure 8(7):729–739
Nanba H, Ikenaka Y, Yamada Y, Yajima K, Takano M, Takahashi S (1998) Isolation of Agrobacterium sp. strain KNK712 that produces N-carbamyl-D-amino acid amidohydrolase, cloning of the gene for this enzyme, and properties of the enzyme. Biosci Biotechnol Biochem 62(5):875–881
Nandanwar H, Prajapati R, Hoondal GS (2013) (D)-p-Hydroxyphenylglycine production by thermostable D-hydantoinase from Brevibacillus parabrevis-PHG1. Biocatal Biotransform 31(1):22–32
Nanninga N (1998) Morphogenesis of Escherichia coli. Microbiol Mol Biol Rev 62(1):110–129
Nelson DE, Young KD (2001) Contributions of PBP 5 and D,D-carboxypeptidase penicillin binding proteins to maintenance of cell shape in Escherichia coli. J Bacteriol 183(10):3055–3064
Nose S (1984) A unified formulation of the constant temperature molecular-dynamics methods. J Chem Phys 81:511–519
Nowroozi FF, Baidoo EE, Ermakov S, Redding-Johanson AM, Batth TS, Petzold CJ, Keasling JD (2014) Metabolic pathway optimization using ribosome binding site variants and combinatorial gene assembly. Appl Microbiol Biotechnol 98(4):1567–1581
Ogawa J, Shimizu S, Yamada H (1993)N-carbamoyl-D-amino acid amidohydrolase from Comamonas sp. E222c purification and characterization. Eur J Biochem 212(3):685–691
Olivieri R, Fascetti E, Angelini L, Degen L (1979) Enzymatic conversion of N- carbamoyl-D-amino acids to D-amino acids. Enzym Microb Technol 1(3):201–204
Olivieri R, Fascetti E, Angelini L, Degen L (1981) Microbial transformation of racemic hydantoins to D-amino acids. Biotechnol Bioeng 23(10):2173–2183
Park JH, Kim GJ, Lee SG, Kim HS (1998) Biochemical properties of thermostable D-hydantoinase from Bacillus thermocatenulatus GH-2. Ann N Y Acad Sci 864(1):337–340
Park JH, Kim GJ, Kim HS (2000) Production of D-amino acid using whole cells of recombinant Escherichia coli with separately and coexpressed D-hydantoinase and N-carbamoylase. Biotechnol Prog 16(4):564–570
Parrinello M, Rahman A (1981) Polymorphic transitions in single-crystals - a new molecular-dynamics method. J Appl Physiol 52:7182–7190
Qian JQ, Chen CC, Liu M, Lv BF (2012) Bioconversion production D-p-hydrophenyglycine from DL-p-hydroxyphenylhydantoin by Pseudomonas putida in aqueous two-phase system. Indian J Biotechnol 11:445–452
Runser SM, Meyer PC (1993) Purification and biochemical characterization of the hydantoin hydrolyzing enzyme from Agrobacterium species. A hydantoinase with no 5,6-dihydropyrimidine amidohydrolase activity. Eur J Biochem 213(3):1315–1324
Schüttelkopf AW, van Aalten DM (2004) PRODRG - a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr D60:1355–1363
Sorin EJ, Pande VS (2005) Exploring the Helix-Coil transition via all-atom equilibrium ensemble simulations. Biophys J 88(4):2472–2493
Typas A, Banzhaf M, Gross CA, Vollmer W (2011) From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat Rev Microbiol 10(2):123–136
Vollmer W, Blanot D, de Pedro MA (2008) Peptidoglycan structure and architecture. FEMS Microbiol Rev 32(2):149–167
Wu S, Liu Y, Liu YB, Zhao GG, Wang JJ, Sun WR (2005) Enzymatic production of D-p-hydroxyphenylglycine from DL-5-p-hydroxyphenylhydantoin by Sinorhizobium morelens S-5. Enzym Microb Technol 36(4):520–526
Wu H, Chen J, Chen GQ (2016) Engineering the growth pattern and cell morphology for enhanced PHB production by Escherichia coli. Appl Microbiol Biotechnol 100(23):9907–9916
Xu Z, Liu YQ, Yang YL, Jiang WH, Arnold E, Ding JP (2003) Crystal structure of D-hydantoinase from Burkholderia pickettii at a resolution of 2.7 Angstroms: insights into the molecular basis of enzyme thermostability. J Bacteriol 185(14):4038–4049
Yang HQ, Lu X, Hu JY, Chen Y, Shen W, Liu L (2018a) Boosting secretion of extracellular protein by Escherichia coli via cell wall perturbation. Appl Environ Microbiol 84(20):e01382–e01318
Yang HQ, Hu JY, Lu X, Wang FX, Shen W, Hu W, Wang LL, Chen XZ, Liu L (2018b) Improving extracellular protein production in Escherichia coli by overexpressing D,D-carboxypeptidase to perturb peptidoglycan network synthesis and structure. Appl Microbiol Biotechnol 103(2):793–806
Yu H, Yang S, Jiang W, Yang Y (2009) Efficient biocatalytic production of D-4-hydroxyphenylglycine by whole cells of recombinant Ralstonia pickettii. Folia Microbiol 54(6):509–515
Zhang DL, Zhu FY, Fan WC, Tao RS, Yu H, Yang YL, Jiang WH, Yang S (2011) Gradually accumulating beneficial mutations to improve the thermostability of N-carbamoyl-d-amino acid amidohydrolase by step-wise evolution. Appl Microbiol Biotechnol 90(4):1361–1371
Zhang SS, Zhao XJ, Tao Y, Lou CB (2015) A novel approach for metabolic pathway optimization: Oligo-linker mediated assembly (OLMA) method. J Biol Eng 9:23
Funding
The work was supported by Beijing Natural Science Foundation, China (5182021), National Science Foundation of China (11604359), and Ministry of Science & Technology, China (KY201701011).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interests.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Liu, Y., Zhu, L., Qi, W. et al. Biocatalytic production of D-p-hydroxyphenylglycine by optimizing protein expression and cell wall engineering in Escherichia coli. Appl Microbiol Biotechnol 103, 8839–8851 (2019). https://doi.org/10.1007/s00253-019-10155-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-019-10155-z