Abstract
Lasso peptides are ribosomally synthesized and post-translationally modified natural products with a characteristic slipknot-like structure, which confers these peptides remarkable stability and diverse pharmacologically relevant bioactivities. Among all the reported lasso peptides, lassomycin and lariatins are unique lasso peptides that exhibit noticeable anti-tuberculosis (TB) activity. Due to the unique threaded structure and the unusual bactericidal mechanism toward Mycobacterium tuberculosis, these peptides have drawn considerable interest, not only in the field of total synthesis but also in several other fields including biosynthesis, bioengineering, and structure-activity studies. During the past few years, significant progress has been made in understanding the biosynthetic mechanism of these intriguing compounds, which has provided a solid foundation for future work. This review highlights recent achievements in the discovery, structure elucidation, biological activity, and the unique anti-TB mechanism of lasso peptides. Moreover, the discovery of their biosynthetic pathway has laid the foundation for combinatorial biosynthesis of their analogs, which provides new perspectives for the production of novel anti-TB lasso peptides.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Long gone are the days when tuberculosis (TB) would kill about one-fourth of the infected population during the nineteenth century. However, TB is still a leading cause of death and is ranked as the ninth leading cause of the world’s mortality from a single infectious disease for humans (Cano-Muniz et al. 2018). According to the World Health Organization, in 2017, approximately 10.0 million individuals were diagnosed with TB infection, with 1.3 million of them dying of this life-threatening disease (WHO 2018). Moreover, it is estimated that 23% of the world’s population is infected with latent TB infection (WHO 2018).
With the development of diverse antibiotics against TB, the currently practiced TB treatment includes consuming four medicines (ethambutol (EMB), pyrazinamide (PZA), isoniazid (INH), and rifampin (RIF)) for a 2-month period, followed by a consumption of two antibiotics (INH and RIF) for a 4-month period (Cano-Muniz et al. 2018). The long treatment duration is challenging in terms of patient adherence. Worse still, the multi-drug-resistant (MDR) and extensively drug-resistant (XDR) M. tuberculosis are a continuing threat (Hanumunthadu et al. 2016). In most cases, treatment of MDR/XDR TB infection takes 9–20 months or even longer periods of time (Xu et al. 2017). This is a global public health concern since most first-line antibiotics have been used in clinical practice for over 40 years and antibiotic resistance is rapidly increasing (Falzon et al. 2017). For example, there were 558,000 new TB cases reported with resistance to rifampicin in 2017 (WHO 2018). Therefore, there is a pressing need to develop more effective and shorter TB therapy regimens to tackle this problem.
The presence of dormant mycobacterial cells in patients is the main reason for long treatment durations. These cells can resuscitate after standard antibiotic therapy, leading to relapse (Cano-Muniz et al. 2018). Therefore, the discovery of new drugs targeting particularly dormant mycobacterial cells with novel mechanism is critical for the development of new TB therapies. Traditionally, most of the antibiotics were discovered from natural sources, especially during the 1950s–1960s, which is recognized as the golden age of natural products research (Harvey et al. 2015; Shen 2015). However, natural product programs have been gradually reduced by pharmaceutical companies, which have led to the overall reduction in the pipeline for the development of new antibiotics (Newman and Cragg 2016). Despite this, natural products, especially microbial natural products, remain an ideal source of antibiotic and other medicinal candidates (Newman and Cragg 2016). During the past few years, progress in microbial genomics, fundamental understanding of natural product biosynthesis, and development of novel high-throughput screening (HTS) methods have led to the discovery of several highly promising anti-TB drugs from soil bacteria (Gavrish et al. 2014; Ling et al. 2015). Some of these molecules, including teixobactin and lassomycin, can target the dormant M. tuberculosis cells, thereby providing several highly promising groups of anti-TB drugs (Gavrish et al. 2014; Ling et al. 2015).
Lassomycin belongs to a subclass of ribosomally synthesized and post-translationally modified peptides (RiPPs) known as lasso peptides (Arnison et al. 2013; Challis 2008; Letzel et al. 2014; Velásquez and van der Donk 2011). These are characterized by an extraordinarily stable knot structure that consists of an N-terminal 7–9-membered macrolactam ring through which C-terminal peptide tail threads, giving them the appearance of a lasso fold (Hegemann et al. 2015; Maksimov and Link 2014; Maksimov et al. 2012a). Apart from lassomycin, other lasso peptides, including lariatin A and lariatin B, also showed notable anti-TB activity (Iwatsuki et al. 2006). Because of their intriguing three-dimensional structures and notable bioactivity, these molecules have drawn considerable attention from academia (Dit Fouque et al. 2018; Fouque et al. 2017; Metelev et al. 2017; Tietz et al. 2017; Zong et al. 2017, 2018). However, the total synthesis of these molecules remains an open challenge (Lear et al. 2016; Martin-Gomez and Tulla-Puche 2018). On the other hand, the biosynthetic pathways for lariatins and lassomycin-like peptides have been elucidated recently which has opened up new avenues for the successful heterologous biosynthesis of these intriguing compounds and has also provided a technical foundation for further bioengineering studies. Herein, we summarize the isolation, structural elucidation, bioactivity, biosynthesis, and bioengineering of known anti-TB lasso peptides. Perspectives on the discovery and combinatorial biosynthesis of anti-TB lasso peptides and their analogs are also discussed.
Discovery, structure, and biological activity of anti-TB lasso peptides
Lasso peptides exhibit a diverse range of biologically relevant properties, ranging from antimicrobial function to receptor antagonistic activities (Bayro et al. 2003; Hegemann et al. 2013a, 2014; Knappe et al. 2008; Ogawa et al. 1995). Most of the lasso peptides with a known pharmaceutical activity were discovered in the course of activity-driven compound screening (Hegemann et al. 2015; Maksimov and Link 2014; Maksimov et al. 2012a, b). Actinobacteria, especially those from the genus Streptomyces, are the main producers of novel bioactive lasso peptides. Among them, only lariatins and lassomycin have been shown to exhibit anti-TB activity (Gavrish et al. 2014; Iwatsuki et al. 2006, 2007).
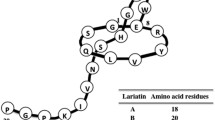
Lariatins were discovered in 2005 during the screening of antibiotics specifically targeting Mycobacterium smegmatis. Two compounds, named lariatin A and lariatin B, were successfully identified from the same Rhodococcus sp. K01-B0171 strain (renamed as Rhodococcus jostii K01-B0171) (Iwatsuki et al. 2006, 2007). Upon isolation, the solution structure of lariatin A was then successfully elucidated by 2D NMR according to the method previously established by Wüthrich et al. Final structural models were built based on the distance and dihedral angle constraints acquired by different NMR spectra (Iwatsuki et al. 2006). It was discovered that lariatin A exhibits a “lasso” fold where the C-terminal tail (Trp9-Pro18) loops back and threads through the N-terminal 8-residue macrolactam ring linked by an isopeptide bond formed through the amino group of Gly1 and the carboxylic acid side chain of Glu8 (Fig. 1a) (Iwatsuki et al. 2006). Lariatin A has three main hydrophobic regions. One particular region, consisting of residues Tyr6 and Ile16, is important for its antibacterial activity. Lariatins belong to the class II lasso peptide group according to the number of disulfide bonds present. In the absence of a disulfide bond to hold the lasso structure, lariatin relies on residues His12 and Asn14 to form a “steric lock” (so-called plug) to prevent the unthreading of the tail (Fig. 1a). The C terminus of the tail is very flexible and disordered in solution. Based on the sequence of the biosynthetic pathway, lariatin A and lariatin B were found to originate from the same precursor peptide. Two additional residues (Gly19, Pro20) were observed in lariatin B and could be easily hydrolyzed by unspecific peptidases in the strains producing these peptides.
Representative structure of anti-TB lasso peptides. A NMR structure of lariatin A drawn from coordinates in reference (Iwatsuki et al. 2006) (left); the tail amino acids are colored in blue, the ring forming Glu8 in red, two plug side chains are colored in green, and the remaining ring amino acids in yellow; interactions between the macrocyclic ring and the C-terminal tail of lariatin A (right). Plug side chains His12 and Asn14 and the macrolactam ring are shown as solvent-accessible surfaces. The macrolactam ring is colored in orange and the surface of the plugs is colored by elements. B Proposed NMR structure of lassomycin (PDB code: 2MAI, left); the tail amino acids are colored in blue, the ring forming Asp8 in red, two plug side chains are colored in green, the methylated position are colored in cyan, and the remaining ring amino acids in yellow; proposed interactions between the macrocyclic ring and the C-terminal tail of lassomycin (right). Arg13 and Asn15 are proposed to be the potential Plug residues. These two residues and the macrolactam ring are shown as solvent-accessible surfaces. The macrolactam ring is colored in orange and the surface of the plugs is colored by elements
Lariatins show unique antimicrobial activity only against mycobacterial species. Lariatin A showed notable inhibition against M. smegmatis with an MIC (minimum inhibitory concentration) value of about 3.13 μg/ml (Table 1) (Iwatsuki et al. 2007). Moreover, lariatin A displayed an MIC value of 0.39 μg/ml against M. tuberculosis growth measured by liquid microdilution methods. Lariatin B exhibited inhibition against M. smegmatis with an MIC value of 6.25 μg/ml (Iwatsuki et al. 2007). The C-terminal tail of lariatins could affect their bioactivity and the Lys17 in lariatin A has been proved to be essential for its anti-mycobacterial activity (Iwatsuki et al. 2009). However, the bactericidal mechanism of lariatins remains elusive. It was proposed that the target of lariatins might lie within the mycobacterial cell wall and they might inhibit cell wall biosynthesis in M. tuberculosis (Iwatsuki et al. 2006). However, further investigation is necessary to uncover this mystery.
Lassomycin is another anti-TB lasso peptide discovered by the Lewis group in 2014 (Gavrish et al. 2014; Parish 2014). By developing a novel in situ cultivation methods, lassomycin was discovered from an uncultured strain Lentzea kentuckyensis sp. The structure of lassomycin was also elucidated after its isolation (Gavrish et al. 2014). Based on the NOE distance restraints obtained from NOESY NMR spectra, a structural model for lassomycin was proposed (Gavrish et al. 2014). Unlike other lasso peptides with a characteristic threaded lasso structure, this model shows that lassomycin has an unthreaded form in solution. The peptide consists of 16 residues in which an eight-residue macrolactam ring was linked through an isopeptide bond between the N terminus Gly1 and Asp8. Instead of passing through the N terminus ring, the C-terminal tail packs tightly with the loop (Fig. 1b) (Gavrish et al. 2014). The Arg13 and Asn15 directly interact with Arg3, which forms a sandwich structure. However, a recent study on the total chemical synthesis of unthreaded lassomycin by the same group revealed that the synthesized lassomycin lacks bioactivity and demonstrates an NMR spectrum different from the naturally occurring lassomycin (Lear et al. 2016). These results suggest that the native lassomycin should have a threaded conformation and that the reported structural model was inappropriate. Nevertheless, based on the preliminary NMR analysis, Arg13 could be a suitable plug in lassomycin and revision of the structure is in progress.
Lassomycin exhibits strong bactericidal activity towards a variety of mycobacterial species, including MDR and XDR M. tuberculosis, with an MIC value of 0.8–3 μg/ml (Gavrish et al. 2014). Unlike other broad-spectrum antibiotics, lassomycin showed a very narrow activity spectrum as it specifically targets mycobacterial species and had no obvious activity against other species and symbionts of the human microbiota (Gavrish et al. 2014). In addition, lassomycin had low cytotoxicity against human cells and high stability against diverse serum proteases (Gavrish et al. 2014; Lee and Suh 2016). Compared with the most potent existing antibiotic for TB, rifampicin, lassomycin not only showed excellent killing activity against M. tuberculosis in the exponential growth phase but also had greater killing activity against M. tuberculosis in the stationary phase (Gavrish et al. 2014). Hence, lassomycin has great potential as a first-line anti-TB drug.
The bactericidal mechanism of lassomycin has also been revealed. M. tuberculosis that spontaneously developed resistance against lassomycin was screened, and ClpC1 was identified as the sole target through genome sequencing of the resistant mutants (Gavrish et al. 2014). ClpC1 is a hexameric ATPase that belongs to the caseinolytic protease (Clp) complex (Lee and Suh 2016; Weinhaupl et al. 2018). It directly interacts with the two heptameric ring proteases ClpP1/P2 and catalyzes the unfolding of the protein substrate in an ATP-dependent fashion and the subsequent translocation to the ClpP1/P2 complexes for further breakdown. Interestingly, lassomycin could stimulate the ATPase activity of ClpC1 upon binding, while uncoupling ATP hydrolysis from the ClpP1/P2-mediated proteolysis (Fig. 2) (Gavrish et al. 2014). This process is highly detrimental to M. tuberculosis because a functional ClpC1/ClpP1/P2 complex is essential for the survival of the organism. Based on the mutations observed in the resistant strains, a highly acidic region including the residues Gln17, Arg21, and Pro79 located at the N terminus of ClpC1 was proposed to be the interaction site for lassomycin, which has been further confirmed by docking studies (Gavrish et al. 2014). However, since the structural elucidation of lassomycin was incorrect, further studies including the analysis of the solution structure of ClpC1 with a threaded lassomycin is necessary to understand the molecular mechanism in detail.
Schematic diagram of bactericidal mechanism of lassomycin. Hexameric ClpC1 recognizes, unfolds, and translocates the dysfunctional protein into the chamber of the heptameric ClpP1/P2 proteolytic complex depending on the ATPase activity of ClpC1 (top). Lassomycin could stimulate the ATPase activity of ClpC1 upon binding, while uncoupling ATP hydrolysis from the ClpP1/P2-mediated proteolysis (down)
Biosynthesis of lariatins, lassomycin, and structurally related lasso peptides
Taking advantage of the expedient molecular biology technologies developed in the past three decades and the emerging genome mining technologies, significant progress has been made in understanding the biosynthesis of lasso peptides. Currently, several different families of lasso peptides have been studied based on the organization of their biosynthetic pathways.
Generally, lasso peptides require three essential components for their biosynthesis: one gene encoding a precursor peptide A, one gene encoding a cysteine protease B for cleavage of the leader peptide, and another gene for an ATP-dependent lactam synthetase C for macrolactam ring formation (Cheung et al. 2016; Hegemann et al. 2013a, b; Li et al. 2015; Metelev et al. 2015; Piscotta et al. 2015; Yan et al. 2012). However, several subsets of lasso peptide biosynthetic pathways feature a “split” B protein (B1 and B2) including lassomycin, lariatin, paeniondin, chaxapeptin, sviceucin and fusilassin, or fuscanodin (DiCaprio et al. 2018; Elsayed et al. 2015; Gavrish et al. 2014; Inokoshi et al. 2012; Koos and Link 2018; Li et al. 2015; Zhu et al. 2016b, c). The lariatin biosynthetic pathway was the first lasso peptide system to be discovered to have a “split” B protein. Pioneering work by Inokoshi et al. identified all the genes necessary for the assembly of lariatins and proved that the B1 protein is essential for lasso peptide biosynthesis (Cheung et al. 2016; Inokoshi et al. 2012). The biosynthetic pathway for lassomycin was first identified by the Lewis Group but not publicly released (Gavrish et al. 2014). Nevertheless, two lassomycin-like gene clusters have been successfully discovered via a genome mining strategy (Su et al. 2019).
As of now, despite the absence of any report on the in vitro total biosynthesis of lariatins and lassomycin, research on other systems provide several clues for their biosynthesis. For example, past in vitro studies on the Mccj25 system have been performed, which revealed that the McjB protein is a type of cysteine protease and McjC is responsible for the isopeptide bond formation (Duquesne et al. 2007; Yan et al. 2012). Further investigation showed that McjB and McjC are interdependent and formed a synthetase complex that assembles McjA into the final mature MccJ25 (Duquesne et al. 2007; Yan et al. 2012). Though the lariatin and lassomycin systems are different from the Mccj25 system, the C protein might have a similar function. On the other hand, recent in vitro studies on the “split” B protein containing systems have unveiled its roles. In the biosynthetic pathways of streptomonomicin, paeniondin, and lariatin, the B1 protein was shown to be a PqqD-like chaperone specifically binding to the leader peptide known as the RiPP precursor peptide recognition element (RRE) (Burkhart et al. 2015; Cheung et al. 2016; Zhu et al. 2016a). Furthermore, in the paeniondin system, the B1 protein was shown to be able to deliver its peptide substrate to the B2 protein for processing (Zhu et al. 2016a). More recently, in vitro total biochemical synthesis of a lasso peptide from Thermobifida fusca was achieved (DiCaprio et al. 2018; Koos and Link 2018). Based on these studies, we propose that all the “split” B containing systems might follow the same rules for biosynthesis. For lassomycin biosynthesis, there is an additional O-methyltransferase located downstream of the ABC transporter that is responsible for C-terminal methylation (Gavrish et al. 2014). We have recently identified two rare lassomycin-like biosynthetic gene clusters in Actinobacteria via genome mining (Su et al. 2019). Through in vitro studies, the methyltransferase from Streptomyces sp. Amel2xC10 was shown to be a novel C-terminal peptide carboxyl methyltransferase that specifically modified the precursor peptide (Su et al. 2019). Therefore, a biosynthetic scheme for lariatins and lassomycin could be proposed using the above information. The precursor peptide was first synthesized and directly modified by the tailoring methyltransferase, in the case of lassomycin. The B1 protein then binds to the leader peptide and transfers the precursor peptides to B2 and C proteins, which assemble the precursor into the lasso fold and remove the leader peptide (Su et al. 2019). Finally, the D protein is responsible for the export of the folded products, which function as antibiotics to kill the competitors in the environment (Fig. 3).
Gene clusters of anti-TB lasso peptides (top). Lasso peptide biosynthetic gene cluster for lassomycin and a potential one from Streptomyces sp. Amel2xC10 contains a methyltransferase while the gene cluster for lariatin lacks the tailoring enzyme; proposed biosynthetic pathway of lariatin and lassomycin (down). The precursor peptide was methylated in the case of methyltransferase containing gene clusters. The B1 protein then binds to the leader peptide and transfers the precursor peptides to B2 and C proteins, which assembles the precursor into the lasso fold and removes the leader peptide. The D protein is responsible for the export of the mature products
Precursor-directed biosynthesis of anti-TB lasso peptide analogs
Total chemical synthesis of lassomycin was attempted owing to its bioactivity and unusual 3D structure. Though unthreaded lassomycin and lassomycin-amide have been generated through total synthesis, no native lasso peptides have been synthesized via chemical methods (Lear et al. 2016). The structural complexity is a major challenge and a practical method for the chemical synthesis of lasso peptide analogs has not been achieved so far (Chekan et al. 2016; Piscotta et al. 2015; Tietz et al. 2017; Zong et al. 2016). However, progress in the biosynthesis of lasso peptides has significantly expedited the process of obtaining new analogs (Piscotta et al. 2015). The repertoire of lasso peptide analogs could be significantly expanded by simple site-directed mutagenesis on the precursor since lasso peptides are of ribosomal origin (Hegemann et al. 2013a, 2014; Inokoshi et al. 2016; Pan and Link 2011). Currently, a convergent expression system for the precursor larA was established in the larA-deficient R. jostii strain (Inokoshi et al. 2016). The larA-carrying plasmid was readily used to generate up to 36 variants and enabled parallel biosynthesis of lariatin mutants for structure-activity relationship (SAR) studies. Firstly, the mutational study identified four residues (Gly1, Arg7, Glu8, and Trp9) in the core peptide that are critical for the biosynthesis of lariatin A (Inokoshi et al. 2016). Furthermore, all the generated lariatin A analogs were used for systematic SAR studies, which demonstrated that three residues (Tyr6, Gly11, and Asn14) are important for anti-TB activity (Fig. 4a) (Inokoshi et al. 2016). Remarkably, tyrosine at the macrolactam ring and the last four residues at the C terminus proved to be essential for enhancing the activity. The region is likely to be involved in the binding of the specific target. A previous study showed that lariatin A without the C terminus completely loses its anti-mycobacterial peptide activity, which further confirmed this conclusion (Iwatsuki et al. 2009). Compared with the native lariatin A (Fig. 4b), lariatin A-Y6F (Fig. 4c), lariatin A-V15A/I16A (Fig. 4d), lariatin A-K17R (Fig. 4e), and lariatin A-P18A (Fig. 4f) exhibited much higher anti-mycobacterial peptide activity (Inokoshi et al. 2016). Notably, only an aromatic amino acid present at position 6 of lariatin exhibits anti-mycobacterial activity. These new lariatin analogs and their effects on bioactivity have provided valuable information regarding the SAR of lariatin and demonstrate that it is possible to generate therapeutically effective lariatin analogs with higher activity by engineering the potential binding site.
Structure-activity relationship (SAR) studies of lariatin. a Three residues (Tyr6, Gly11, and Asn14) are essential for anti-TB activity of lariatin. b Wild type of lariatin; five positions essential for enhancing the activity are shown as solvent-accessible surfaces. c Structure of the lariatin A-Y6F, d lariatin A-V15A/I16A, e lariatin A-K17R, f lariatin A-P18A. Selected mutated residues are shown in different colors as solvent-accessible surfaces which could enhance the anti-TB activity of lariatin
For lassomycin, there are no analogs that have been reported thus far. This is due to the lack of biosynthetic information and the absence of any heterologous expression systems (Gavrish et al. 2014). Coincidently, we have recently identified two lassomycin-like gene clusters (Su et al. 2019). Notably, the gene cluster from Streptomyces sp. Amel2xC10 is closely related to lassomycin (Su et al. 2019). The core peptides are very similar and amino acid substitutions were observed only at 5 out of 16 positions. Thus, the identified gene clusters provide valuable information on lassomycin biosynthesis and the construction of genetically engineered strains for lassomycin derivatives might be feasible (Su et al. 2019). Though an E. coli-based heterologous system has not been established, we propose that a Streptomyces-based production system might be an ideal biosynthetic platform to generate diverse lassomycin analogs, among which the ones with enhanced activity might be further developed as promising anti-TB medicines (Mevaere et al. 2018; Su et al. 2019).
Conclusion and perspectives
Herein, we highlight recent achievements and useful approaches regarding anti-TB lasso peptides. The progressive advances in the fundamental understanding of RiPP biosynthesis and genomics studies as well as emerging biotechnological techniques have reinvigorated the development of novel anti-TB lasso peptide analogs. With these developments, we suppose that there might be three main directions for the discovery of novel anti-TB lasso peptides. First, the exploration of novel lasso peptide biosynthetic pathways is still an efficient way for the discovery of these compounds. It is worth noting that the lassomycin lasso peptide is very close to the lariatins, based on the phylogenetic analysis of the C protein, from the biosynthetic gene clusters (Fig. 5a) (Su et al. 2019). Previous studies have shown that antimicrobial lasso peptides normally exhibit activity against closely related strains (Knappe et al. 2008). It is reasonable to check other closely related lasso peptide biosynthetic gene clusters that might also target M. tuberculosis. Through homology-based searches, Rhodococcus rhodochrous, Streptomyces sp. Amel2xC10, Streptomyces radiopugnans, and Sanguibacter keddieii were identified to be potential resources for finding novel anti-TB lasso peptides as they all show high similarity with the lariatin or lassomycin biosynthetic systems (Su et al. 2019). However, since the sequence of the final products showed little similarity, the future investigation might lead to promising drug candidates.
Lasso peptide biosynthetic gene clusters that are closely related to lassomycin or lariatin. a Phylogenetic tree of C proteins from different lasso peptide biosynthetic gene clusters closely related to lariatin and lassomycin. The sequence of the predicted lasso peptides is shown on the right. b Combinatorial biosynthesis of a set of modified lasso peptide analogs employing diverse tailoring enzymes
The second strategy for the generation of anti-TB lasso peptide analogs is precursor-directed mutagenesis. The lasso peptide libraries can be generated rapidly and predictably as demonstrated by the lariatin system. The associated “split” B and C proteins showed a wide substrate specificity and the precursor peptides can be extensively mutated (Inokoshi et al. 2016). The generated variants can be used for screening activity and the variants with enhanced activity could be further evaluated for clinical application. Generally, this strategy requires a heterologous expression system for biosynthesis. In the case of lariatin, a convergent expression system also worked well. However, no such platform for lassomycin has been reported so far due to the lack of biosynthetic information at the gene level (Su et al. 2019). We think that the discovery of the lassomycin-like gene clusters from S. keddieii and Streptomyces sp. Amel2xC10 is perhaps an important step towards achieving such goals (Su et al. 2019). In the future, the elucidation of the lassomycin biosynthetic pathway could also significantly contribute to research in this area (Gavrish et al. 2014). Further structure-activity relationships revealed by the analogs could lay a solid foundation towards rational design of more promising anti-TB drugs.
The third strategy is the application of combinatorial biosynthesis to generate lasso peptide analogs, and this approach has been supported by several recent successful attempts. For instance, Burkhart et al. successfully generated several non-natural hybrid RiPP products through artificial leader peptides that enable recognition and processing by diverse tailoring enzymes from unrelated RiPP pathways (Burkhart et al. 2017). In the past, further tailoring of lasso peptides has rarely been observed. However, recently discovered paeniondin, lassomycin, MS-271, and albusnodin have greatly changed our understanding of lasso peptide biosynthesis (Feng et al. 2018; Gavrish et al. 2014; Su et al. 2019; Zhu et al. 2016b, 2016c; Zong et al. 2018). These tailoring enzymes showed interesting promiscuous activities towards diverse substrates (Su et al. 2019; Zhu et al. 2016b, c). For example, the kinase from the paeniondin pathway and the methyltransferase from the lassomycin-like pathway showed wide substrate specificity regardless of the length and sequence of the precursor peptide, which makes them very promising candidates for combinatorial biosynthesis (Su et al. 2019; Zhu et al. 2016b, c). Additionally, an acetyltransferase from the albusnodin pathway, a potential epimerase in the MS-271 lasso peptide pathway and potential nucleotidyltransferases and sulfotransferases in lasso peptide pathways from Firmicutes have also been identified (Feng et al. 2018; Zhu et al. 2016c; Zong et al. 2018). All these tailoring enzymes provide diverse tools for the design of artificial lasso peptide biosynthetic pathways, which could lead to the production of various modified lasso peptides. Moreover, other tailoring enzymes from different RiPP pathways also have the potential to be incorporated into the new lasso peptide biosynthetic pathway via rational design. With these strategies, it could be possible to generate diverse modified lasso peptide analogs for further SAR studies.
In summary, lariatins, lassomycin, and their analogs represent an important family of anti-TB drugs with novel bactericidal mechanisms. Considering the devastating impact of TB on human health, research on these compounds should be prioritized. We recognize that lassomycin has already attracted the attention of the pharmaceutical industry as a grant from the Bill & Melinda Gates Foundation has been awarded to NovoBiotic Pharmaceuticals, to evaluate its potential as a novel anti-TB medicine. Furthermore, with more microbial genomes being sequenced, novel biosynthetic pathways combined with unprecedented tailoring enzymes will provide an essential genetic basis for the combinatorial biosynthesis of these interesting compounds. This biosynthetic approach will open a new avenue towards the synthesis of novel anti-TB lasso peptides through rational design.
References
Arnison PG, Bibb MJ, Bierbaum G, Bowers AA, Bugni TS, Bulaj G, Camarero JA, Campopiano DJ, Challis GL, Clardy J, Cotter PD, Craik DJ, Dawson M, Dittmann E, Donadio S, Dorrestein PC, Entian K-D, Fischbach MA, Garavelli JS, Goransson U, Gruber CW, Haft DH, Hemscheidt TK, Hertweck C, Hill C, Horswill AR, Jaspars M, Kelly WL, Klinman JP, Kuipers OP, Link AJ, Liu W, Marahiel MA, Mitchell DA, Moll GN, Moore BS, Muller R, Nair SK, Nes IF, Norris GE, Olivera BM, Onaka H, Patchett ML, Piel J, Reaney MJT, Rebuffat S, Ross RP, Sahl H-G, Schmidt EW, Selsted ME, Severinov K, Shen B, Sivonen K, Smith L, Stein T, Sussmuth RD, Tagg JR, Tang G-L, Truman AW, Vederas JC, Walsh CT, Walton JD, Wenzel SC, Willey JM, van der Donk WA (2013) Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature. Nat Prod Rep 30(1):108–160. https://doi.org/10.1039/C2NP20085F
Bayro MJ, Mukhopadhyay J, Swapna GVT, Huang JY, Ma L-C, Sineva E, Dawson PE, Montelione GT, Ebright RH (2003) Structure of antibacterial peptide Microcin J25: a 21-residue lariat protoknot. J Am Chem Soc 125(41):12382–12383. https://doi.org/10.1021/ja036677e
Burkhart BJ, Hudson GA, Dunbar KL, Mitchell DA (2015) A prevalent peptide-binding domain guides ribosomal natural product biosynthesis. Nat Chem Biol 11(8):564–570. https://doi.org/10.1038/nchembio.1856
Burkhart BJ, Kakkar N, Hudson GA, van der Donk WA, Mitchell DA (2017) Chimeric leader peptides for the generation of non-natural hybrid RiPP products. ACS Cent Sci 3(6):629–638. https://doi.org/10.1021/acscentsci.7b00141
Cano-Muniz S, Anthony R, Niemann S, Alffenaar JC (2018) New approaches and therapeutic options for Mycobacterium tuberculosis in a dormant state. Clin Microbiol Rev 31(1). https://doi.org/10.1128/CMR.00060-17
Challis GL (2008) Genome mining for novel natural product discovery. J Med Chem 51(9):2618–2628. https://doi.org/10.1021/jm700948z
Chekan JR, Koos JD, Zong C, Maksimov MO, Link AJ, Nair SK (2016) Structure of the lasso peptide isopeptidase identifies a topology for processing threaded substrates. J Am Chem Soc 138(50):16452–16458. https://doi.org/10.1021/jacs.6b10389
Cheung WL, Chen MY, Maksimov MO, Link AJ (2016) Lasso peptide biosynthetic protein LarB1 binds both leader and core peptide regions of the precursor protein LarA. ACS Cent Sci 2(10):702–709. https://doi.org/10.1021/acscentsci.6b00184
DiCaprio AJ, Firouzbakht A, Hudson GA, Mitchell DA (2018) Enzymatic reconstitution and biosynthetic investigation of the lasso peptide fusilassin. J Am Chem Soc 141:290–297. https://doi.org/10.1021/jacs.8b09928
Dit Fouque KJ, Moreno J, Hegemann JD, Zirah S, Rebuffat S, Fernandez-Lima F (2018) Identification of lasso peptide topologies using native nanoelectrospray ionization-trapped ion mobility spectrometry-mass spectrometry. Anal Chem 90:5139–5146. https://doi.org/10.1021/acs.analchem.7b05230
Duquesne S, Destoumieux-Garzón D, Zirah S, Goulard C, Peduzzi J, Rebuffat S (2007) Two enzymes catalyze the maturation of a lasso peptide in Escherichia coli. Chem Biol 14(7):793–803. https://doi.org/10.1016/j.chembiol.2007.06.004
Elsayed SS, Trusch F, Deng H, Raab A, Prokes I, Busarakam K, Asenjo JA, Andrews BA, van West P, Bull AT, Goodfellow M, Yi Y, Ebel R, Jaspars M, Rateb ME (2015) Chaxapeptin, a lasso peptide from extremotolerant Streptomyces leeuwenhoekii strain C58 from the hyperarid Atacama Desert. J Org Chem 80(20):10252–10260. https://doi.org/10.1021/acs.joc.5b01878
Falzon D, Schunemann HJ, Harausz E, Gonzalez-Angulo L, Lienhardt C, Jaramillo E, Weyer K (2017) World Health Organization treatment guidelines for drug-resistant tuberculosis, 2016 update. Eur Respir J 49(3):1602308. https://doi.org/10.1183/13993003.02308-2016
Feng Z, Ogasawara Y, Nomura S, Dairi T (2018) Biosynthetic gene cluster of a d-tryptophan-containing lasso peptide, MS-271. ChemBioChem 19:2045–2048. https://doi.org/10.1002/cbic.201800315
Fouque KJD, Lavanant H, Zirah S, Hegemann JD, Zimmermann M, Marahiel MA, Rebuffat S, Afonso C (2017) Signatures of mechanically interlocked topology of lasso peptides by ion mobility–mass spectrometry: lessons from a collection of representatives. J Am Soc Mass Spectrom 28(2):315–322. https://doi.org/10.1007/s13361-016-1524-8
Gavrish E, Sit CS, Cao S, Kandror O, Spoering A, Peoples A, Ling L, Fetterman A, Hughes D, Bissell A, Torrey H, Akopian T, Mueller A, Epstein S, Goldberg A, Clardy J, Lewis K (2014) Lassomycin, a ribosomally synthesized cyclic peptide, kills Mycobacterium tuberculosis by targeting the ATP-dependent protease ClpC1P1P2. Chem Biol 21(4):509–518. https://doi.org/10.1016/j.chembiol.2014.01.014
Hanumunthadu B, Harrison T, Mathew D, Cotter M (2016) Multidrug-resistant (MDR) and extensively drug-resistant (XDR) tuberculosis: successes and complications on outpatient parenteral antimicrobial therapy at a London teaching hospital between 2009 and 2016. Open Forum Infect Dis 3(suppl_1):558–558. https://doi.org/10.1093/ofid/ofw172.421
Harvey AL, Edrada-Ebel R, Quinn RJ (2015) The re-emergence of natural products for drug discovery in the genomics era. Nat Rev Drug Discov 14(2):111–129. https://doi.org/10.1038/nrd4510
Hegemann JD, Zimmermann M, Xie X, Marahiel MA (2013a) Caulosegnins I–III: a highly diverse group of lasso peptides derived from a single biosynthetic gene cluster. J Am Chem Soc 135(1):210–222. https://doi.org/10.1021/ja308173b
Hegemann JD, Zimmermann M, Zhu S, Klug D, Marahiel MA (2013b) Lasso peptides from proteobacteria: genome mining employing heterologous expression and mass spectrometry. Pept Sci 100(5):527–542. https://doi.org/10.1002/bip.22326
Hegemann JD, Zimmermann M, Zhu S, Steuber H, Harms K, Xie X, Marahiel MA (2014) Xanthomonins I-III: a new class of lasso peptides with a seven-residue macrolactam ring. Angew Chem Int Ed Engl 53(8):2230–2234. https://doi.org/10.1002/anie.201309267
Hegemann JD, Zimmermann M, Xie X, Marahiel MA (2015) Lasso peptides: an intriguing class of bacterial natural products. Acc Chem Res 48(7):1909–1919. https://doi.org/10.1021/acs.accounts.5b00156
Inokoshi J, Matsuhama M, Miyake M, Ikeda H, Tomoda H (2012) Molecular cloning of the gene cluster for lariatin biosynthesis of Rhodococcus jostii K01-B0171. Appl Microbiol Biotechnol 95(2):451–460. https://doi.org/10.1007/s00253-012-3973-8
Inokoshi J, Koyama N, Miyake M, Shimizu Y, Tomoda H (2016) Structure-activity analysis of gram-positive bacterium-producing lasso peptides with anti-mycobacterial activity. Sci Rep 6:30375. https://doi.org/10.1038/srep30375
Iwatsuki M, Tomoda H, Uchida R, Gouda H, Hirono S, Omura S (2006) Lariatins, antimycobacterial peptides produced by Rhodococcus sp. K01-B0171, have a lasso structure. J Am Chem Soc 128(23):7486–7491. https://doi.org/10.1021/ja056780z
Iwatsuki M, Uchida R, Takakusagi Y, Matsumoto A, Jiang CL, Takahashi Y, Arai M, Kobayashi S, Matsumoto M, Inokoshi J, Tomoda H, Omura S (2007) Lariatins, novel anti-mycobacterial peptides with a lasso structure, produced by Rhodococcus jostii K01-B0171. J Antibiot 60(6):357–363. https://doi.org/10.1038/ja.2007.48
Iwatsuki M, Koizumi Y, Gouda H, Hirono S, Tomoda H, Omura S (2009) Lys17 in the ‘lasso’ peptide lariatin A is responsible for anti-mycobacterial activity. Bioorg Med Chem Lett 19(10):2888–2890. https://doi.org/10.1016/j.bmcl.2009.03.033
Knappe TA, Linne U, Zirah S, Rebuffat S, Xie X, Marahiel MA (2008) Isolation and structural characterization of capistruin, a lasso peptide predicted from the genome sequence of Burkholderia thailandensis E264. J Am Chem Soc 130(34):11446–11454. https://doi.org/10.1021/ja802966g
Koos JD, Link AJ (2018) Heterologous and in vitro reconstitution of fuscanodin, a lasso peptide from Thermobifida fusca. J Am Chem Soc 141:928–935. https://doi.org/10.1021/jacs.8b10724
Lear S, Munshi T, Hudson AS, Hatton C, Clardy J, Mosely JA, Bull TJ, Sit CS, Cobb SL (2016) Total chemical synthesis of lassomycin and lassomycin-amide. Org Biomol Chem 14(19):4534–4541. https://doi.org/10.1039/c6ob00631k
Lee H, Suh JW (2016) Anti-tuberculosis lead molecules from natural products targeting Mycobacterium tuberculosis ClpC1. J Ind Microbiol Biotechnol 43(2–3):205–212. https://doi.org/10.1007/s10295-015-1709-3
Letzel A-C, Pidot SJ, Hertweck C (2014) Genome mining for ribosomally synthesized and post-translationally modified peptides (RiPPs) in anaerobic bacteria. BMC Genomics 15(1):983. https://doi.org/10.1186/1471-2164-15-983
Li Y, Ducasse R, Zirah S, Blond A, Goulard C, Lescop E, Giraud C, Hartke A, Guittet E, Pernodet J-L, Rebuffat S (2015) Characterization of Sviceucin from Streptomyces provides insight into enzyme exchangeability and disulfide bond formation in lasso peptides. ACS Chem Biol 10(11):2641–2649. https://doi.org/10.1021/acschembio.5b00584
Ling LL, Schneider T, Peoples AJ, Spoering AL, Engels I, Conlon BP, Mueller A, Schaberle TF, Hughes DE, Epstein S, Jones M, Lazarides L, Steadman VA, Cohen DR, Felix CR, Fetterman KA, Millett WP, Nitti AG, Zullo AM, Chen C, Lewis K (2015) A new antibiotic kills pathogens without detectable resistance. Nature 517(7535):455–459. https://doi.org/10.1038/nature14098
Maksimov MO, Link AJ (2014) Prospecting genomes for lasso peptides. J Ind Microbiol Biotechnol 41(2):333–344. https://doi.org/10.1007/s10295-013-1357-4
Maksimov MO, Pan SJ, James Link A (2012a) Lasso peptides: structure, function, biosynthesis, and engineering. Nat Prod Rep 29(9):996–1006. https://doi.org/10.1039/C2NP20070H
Maksimov MO, Pelczer I, Link AJ (2012b) Precursor-centric genome-mining approach for lasso peptide discovery. Proc Natl Acad Sci U S A 109(38):15223–15228. https://doi.org/10.1073/pnas.1208978109
Martin-Gomez H, Tulla-Puche J (2018) Lasso peptides: chemical approaches and structural elucidation. Org Biomo Chem 16(28):5065–5080. https://doi.org/10.1039/c8ob01304g
Metelev M, Tietz Jonathan I, Melby Joel O, Blair Patricia M, Zhu L, Livnat I, Severinov K, Mitchell Douglas A (2015) Structure, bioactivity, and resistance mechanism of Streptomonomicin, an unusual lasso peptide from an understudied halophilic Actinomycete. Chem Biol 22(2):241–250. https://doi.org/10.1016/j.chembiol.2014.11.017
Metelev M, Arseniev A, Bushin LB, Kuznedelov K, Artamonova TO, Kondratenko R, Khodorkovskii M, Seyedsayamdost MR, Severinov K (2017) Acinetodin and klebsidin, RNA polymerase targeting lasso peptides produced by human isolates of Acinetobacter gyllenbergii and Klebsiella pneumoniae. ACS Chem Biol 12(3):814–824. https://doi.org/10.1021/acschembio.6b01154
Mevaere J, Goulard C, Schneider O, Sekurova ON, Ma H, Zirah S, Afonso C, Rebuffat S, Zotchev SB, Li Y (2018) An orthogonal system for heterologous expression of actinobacterial lasso peptides in Streptomyces hosts. Sci Rep 8(1):8232. https://doi.org/10.1038/s41598-018-26620-0
Newman DJ, Cragg GM (2016) Natural products as sources of new drugs from 1981 to 2014. J Nat Prod 79(3):629–661. https://doi.org/10.1021/acs.jnatprod.5b01055
Ogawa T, Ochiai K, Tanaka T, Tsukuda E, Chiba S, Yano K, Yamasaki M, Yoshida M, Matsuda Y (1995) RES-701-2, -3 and -4, novel and selective endothelin type B receptor antagonists produced by Streptomyces sp. I. Taxonomy of producing strains, fermentation, isolation, and biochemical properties. J Antibiot 48(11):1213–1220
Pan SJ, Link AJ (2011) Sequence diversity in the lasso peptide framework: discovery of functional microcin J25 variants with multiple amino acid substitutions. J Am Chem Soc 133(13):5016–5023. https://doi.org/10.1021/ja1109634
Parish T (2014) Targeting mycobacterial proteolytic complexes with natural products. Chem Biol 21(4):437–438. https://doi.org/10.1016/j.chembiol.2014.04.002
Piscotta FJ, Tharp JM, Liu WR, Link AJ (2015) Expanding the chemical diversity of lasso peptide MccJ25 with genetically encoded noncanonical amino acids. Chem Commun 51(2):409–412. https://doi.org/10.1039/C4CC07778D
Shen B (2015) A new golden age of natural products drug discovery. Cell 163(6):1297–1300. https://doi.org/10.1016/j.cell.2015.11.031
Su Y, Han M, Meng X, Feng Y, Luo S, Yu C, Zheng G, Zhu S (2019) Discovery and characterization of a novel C-terminal peptide carboxyl methyltransferase in a lassomycin-like lasso peptide biosynthetic pathway. Appl Microbiol Biotechnol. https://doi.org/10.1007/s00253-019-09645-x
Tietz JI, Schwalen CJ, Patel PS, Maxson T, Blair PM, Tai HC, Zakai UI, Mitchell DA (2017) A new genome-mining tool redefines the lasso peptide biosynthetic landscape. Nat Chem Biol 13(5):470–478. https://doi.org/10.1038/nchembio.2319
Velásquez JE, van der Donk WA (2011) Genome mining for ribosomally synthesized natural products. Curr Opin Chem Biol 15(1):11–21. https://doi.org/10.1016/j.cbpa.2010.10.027
Weinhaupl K, Brennich M, Kazmaier U, Lelievre J, Ballell L, Goldberg A, Schanda P, Fraga H (2018) The antibiotic cyclomarin blocks arginine-phosphate-induced millisecond dynamics in the N-terminal domain of ClpC1 from Mycobacterium tuberculosis. J Biol Chem 293(22):8379–8393. https://doi.org/10.1074/jbc.RA118.002251
WHO (2018) WHO World Health Organization. 2018. Global tuberculosis report 2018. World Health Organization, Geneva
Xu Y, Wu J, Liao S, Sun Z (2017) Treating tuberculosis with high doses of anti-TB drugs: mechanisms and outcomes. Ann Clin Microbiol Antimicrob 16(1):67. https://doi.org/10.1186/s12941-017-0239-4
Yan K-P, Li Y, Zirah S, Goulard C, Knappe TA, Marahiel MA, Rebuffat S (2012) Dissecting the maturation steps of the lasso peptide microcin J25 in vitro. ChemBioChem 13(7):1046–1052. https://doi.org/10.1002/cbic.201200016
Zhu S, Fage CD, Hegemann JD, Mielcarek A, Yan D, Linne U, Marahiel MA (2016a) The B1 protein guides the biosynthesis of a lasso peptide. Sci Rep 6:35604. https://doi.org/10.1038/srep35604
Zhu S, Fage CD, Hegemann JD, Yan D, Marahiel MA (2016b) Dual substrate-controlled kinase activity leads to polyphosphorylated lasso peptides. FEBS Lett 590(19):3323–3334. https://doi.org/10.1002/1873-3468.12386
Zhu S, Hegemann JD, Fage CD, Zimmermann M, Xie X, Linne U, Marahiel MA (2016c) Insights into the unique phosphorylation of the lasso peptide Paeninodin. J Biol Chem 291(26):13662–13678. https://doi.org/10.1074/jbc.M116.722108
Zong C, Maksimov MO, Link AJ (2016) Construction of lasso peptide fusion proteins. ACS Chem Biol 11(1):61–68. https://doi.org/10.1021/acschembio.5b00745
Zong C, Wu MJ, Qin JZ, Link AJ (2017) Lasso peptide benenodin-1 is a thermally actuated [1] rotaxane switch. J Am Chem Soc 139(30):10403–10409. https://doi.org/10.1021/jacs.7b04830
Zong C, Cheung-Lee WL, Elashal HE, Raj M, Link AJ (2018) Albusnodin: an acetylated lasso peptide from Streptomyces albus. Chem Commun 54(11):1339–1342. https://doi.org/10.1039/c7cc08620b
Funding
This work was supported by the Fundamental Research Funds for the Central Universities (XK1802-8 and XK1803-06), National Natural Science Foundation of China (NSFC, Grant No. 21706005), and National Great Science and Technology Projects (2018ZX09721001). Dedicated to Renzhi Zhu.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical statement
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Zhu, S., Su, Y., Shams, S. et al. Lassomycin and lariatin lasso peptides as suitable antibiotics for combating mycobacterial infections: current state of biosynthesis and perspectives for production. Appl Microbiol Biotechnol 103, 3931–3940 (2019). https://doi.org/10.1007/s00253-019-09771-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-019-09771-6