Abstract
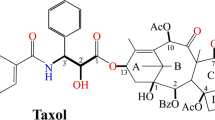
Taxol is an anticancer identified in both endophytic fungus and its host plant. Plant Taxol is a diterpenoid with geranylgeranyl diphosphate (GGPP) mediates the biosynthesis of its terpenoid moiety. Previous report has suggested that fungal Taxol may require terpenoid pathway for its biosynthesis. Here in this study, feeding a Taxol-producing endophytic fungus (Paraconiothyrium SSM001) with terpenoid precursors including isopentenyl pyrophosphate (IPP, isoprene) and GGPP enhanced Taxol production threefold and fivefold, respectively, compared to the control. Thus, we assumed that increasing the terpenoid pool size in particular GGPP by introducing a new copy number of GGPPS particularly from a Taxol-producing plant might increase the production level of fungal Taxol. Agrobacterium-mediated integration of Taxus canadensis geranylgeranyl diphosphate synthase (GGPPS) gene into the Paraconiothyrium SSM001 genome was successful and increased the terpenoid pool size indicated by an increase in carotenoid level and orange to red coloration of some GGPPS-transformed SSM001 colonies. Furthermore, the integration improved the level of Taxol production threefold. Feeding a GGPPS-transformed SSM001 fungus with a GGPP precursor increased the expression level of GGPPS transcript and Taxol production. The successful increase in both terpenoid and Taxol production levels due to GGPPS gene integration into the fungal genome might be a step forward in manipulating Taxol-producing endophytic fungi. Future control of the transformation time and the manipulation of the phenolic pathway could maximize the production level.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Taxol is a diterpene anticancer drug produced by both endophytic fungus and the host plant (Soliman et al. 2015). Taxol is reported as a product of the endophytic fungus Paraconiothyrium SSM001 (Soliman et al. 2011; Soliman et al. 2013). Chemical inhibitors confirmed that fungal Taxol could be produced by both terpenoids and shikimate pathways (Soliman et al. 2011). Terpenoids are present in all organisms (Singh and Sharma 2015). Terpenoids are necessary for the biosynthesis of essential compounds including gibberellins, carotenoids, and sterols and non-essential medicinal compounds such as Taxol (Roberts 2007). All diterpenoids including Taxol appear to be derived from geranylgeranyl diphosphate (GGPP), a 20-carbon terpenoid skeleton (four isoprene units) (Schütte 1997). The geranylgeranyl diphosphate synthase (GGPPS) enzyme catalyzes the biosynthesis of GGPP. The GGPP synthase has been found in a number of organisms, and its product, GGPP, also functions as a key intermediate in the biosynthesis of carotenoids, geranylgeranylated proteins, chlorophylls, gibberellins, diterpenes, and archael ether-linked lipids (Keeling and Bohlmann 2006).
The biosynthesis of GGPP can be summarized into two main steps: (1) biosynthesis of the isoprene unit (5C, IPP) which is the fundamental precursor; and (2) repetitive fusion of isoprene in a sequence of elongation reactions to produce the intermediate precursor geranylgeranyl in the form of diphosphate, geranylgeranyl diphosphate (20C, GGPP), either through geranyl diphosphate (10C, GPP) in case of plants (Zhu et al. 1997) or farnesyl diphosphate (15C, FPP) in case of fungi (Singkaravanit et al. 2010). GGPP is then cyclized by specific terpene synthases such as taxadiene synthase (TS) in case of plant Taxol biosynthesis (Koksal et al. 2011).
Some terpenoids are produced in large quantities such as essential oils and waxes; however, highly valuable terpenoids are found in low amounts from natural sources typically less than 2–3% of the total dry weight. This is especially true in the case of Taxol (Roberts 2007).
A common strategy for engineering terpenoid (including Taxol) biosynthesis has been focused on enhancing metabolic flux in order to increase the precursor pool size; these efforts have involved cloning and understanding the necessary genes and the early pathway enzymes that are rate-influencing in terpenoid accumulation (Kovacs et al. 2007; Roberts 2007). The first committed step in the biosynthesis of Taxol is the cyclization of GGPP to taxadiene. The low cyclization activity in Taxus plants and the low levels of taxadiene found in Taxus bark suggested that the cyclization of GGPP is slow relative to subsequent oxygenation steps leading researchers to conclude that this step is a good target for increasing Taxol production (Baloglu and Kingston 1999). With this hypothesis in mind and the benefits of producing Taxol in a non-tree system, considerable efforts have been made to reconstruct, and metabolically re-engineer the rate-limiting step in the Taxol pathway in model organisms such as Escherichia coli, yeast Saccharomyces cerevisiae, and Arabidopsis thaliana. But all of these experiments have thus far failed to produce even sufficient amounts of taxadiene, the first precursor in the Taxol pathway (Soliman and Tang 2015), mainly due to the insufficiency of precursor flux to GGPP biosynthesis (Besumbes et al. 2004; Huang et al. 2001). Also, Taxol is biosynthesized through putatively 19 enzymatic steps, but several genes involved in the oxygenation of its core nucleus have not been identified (Croteau et al. 2006). Therefore, Taxol metabolic engineering in a model organism via total reconstruction remains very difficult.
Some genes encoding enzymes, for example GGPP synthase, are specifically triggered by specific developmental stages (Hugueney et al. 1996). For example, the expression of plastid GGPP synthase is strongly induced during Capsicum annuum fruit ripening and during the chloroplast to chromoplast transition, and this induction is correlated with an increase in the activity of the enzyme (Kuntz et al. 1992). Furthermore, it is reported that feeding plant cell cultures with early precursors in the terpenoid pathway such as IPP or GGPP stimulated terpenoid production (Kang et al. 2006) and increased plant Taxol production (Croteau et al. 2006).
Taking all these in consideration, we assumed that either feeding Taxol-producing fungus with terpenoid precursor or increasing the GGPPS copy number by transformation of Taxol-producing fungus with the plant GGPP synthase gene might positively affect the production level of fungal Taxol. Recently, Alternaria alternata TPF6 endophytic fungus engineered with the modified mevalonate pathway increased the production of taxadiene (Bian et al. 2017). Agrobacterium tumefaciens-mediated transformation is known as successful transformation method of filamentous fungi by delivering the genetic components that are bordered by the left border (LB) and right border (RB) of its helper plasmid (Hood et al. 1993). In this report, increasing the GGPP pool size either by exogenous feeding of GGPP, by endogenous integration of the GGPP biosynthetic gene from plant by A. tumefaciens, or by both exogenous and endogenous feeding of GGPP increased fungal Taxol production.
Material and methods
Materials
The following reagents were purchased from Sigma (USA), including Taxol standard (# T7402), fungal nutrient media yeast-peptone-dextrose (YPD) (# Y1375) and potato-dextrose-agar (PDA) (# 70139), and terpenoid precursors isopentenyl pyrophosphate (isoprene, IPP; # I0503) and geranylgeranyl diphosphate (GGPP; # G6025).
Plasmids and fungal, yeast, and bacterial strains
Endophytic fungus Paraconiothyrium SSM001 (Accession # ATCC MYA-4697) is isolated from Taxus media plants and identified according to Soliman et al. 2011). Two-week-old pure fungal cultures grown on PDA plates were used for all subsequent experiments. Taxus canadensis full-length GGPPS complementary DNA (cDNA) in pET32 expression vector was kindly provided by Dr. Rodeny Croteau (Hefner et al. 1998). Yeast mutant strain lacking GGPPS activity (SFNY368) was kindly provided by Professor Susan Ferro-Novick (Department of Cell Biology and Howard Hughes Medical Institute, School of Medicine, Yale University, Connecticut, USA) (Jiang et al. 1995). The pBlueScript KS+ vector was used for cloning GGPPS. The vector pPK2 having hygromycin (hphR) and kanamycin resistance (KanR) genes with Aspergillus nidulans Pgpd promoter and TtrpC terminator was kindly provided by Sarah F. Covert (Warnell School of Forest Resources and Department of Botany, University of Georgia, Athens, GA 30602, USA) (Covert et al. 2001) was used for yeast and Paraconiothyrium transformation. E. coli DH5α (Thermo Fisher Scientific, # 18258012). Agrobacterium tumefaciens strain AGL-1 was kindly provided by Dr. Peter Romaine (Department of Plant Pathology, Spawn Science, 317 Buckhout Laboratory, Pennsylvania State University, University Park, PA) (Chen et al. 2000) and used for fungal transformation. All plasmid vectors and strains Are listed in Table 1.
Precursor feeding experiment
Ten-day-old fungal liquid YPD cultures were fed with either IPP or GGPP at concentrations 0.22 and 0.44 μM. The cultures were then incubated up to 21 days at 25 °C in the dark with shaking at 100 rpm. The mycelia were filtered from the liquid media using cheesecloth and squeezed and the liquid media extracted as described below. Methanol/DMSO (1:1) was employed as a negative control. Each experiment was repeated three times. Similarly, 10-day-old GGPPS-transformed fungus was fed with 0.44 μM GGPP followed by incubation in the dark at 25 °C with shaking for another 11 days prior to testing for carotenoids and Taxol production.
Construction of yeast and fungal expression plasmid
For the generation of recombinant plasmids, E. coli DH5α (competent cell, Thermo Fisher Scientific) was used as host. DNA fragments were generated by PCR, and the primers used are listed in Table 2. The 1182-bp fragment of T. canadensis GGPPS was amplified from the pET32 vector using primers GGPPSF4 and GGPPSR4 (Table 2) using high-fidelity Platinum Pfx DNA Polymerase enzyme (Thermo Fisher Scientific, # 11708). The following conditions were used for PCR amplification: 94 °C for 2 min, followed by 35 cycles of 94 °C for 45 s, 75 °C for 1 min, and 72 °C for 2 min, with a final extension cycle of 4 min at 72 °C. The fragment was purified using an Illustra GFX 96 PCR Purification Kit (GE Healthcare, USA). The amplified GGPPS fragment was digested with SmaI/HindIII and cloned into the pBlueScript KS+ vector to obtain plasmid pG2.The resulting plasmid was confirmed by restriction digestion using HindIII to afford approximately 4 kb and PCR using specific primers for GGPPS to afford approximately 1.2 kb.
For the insertion of terminator fragment into the expression cassette, the 700-bp TtrpC terminator was cut out from the pPK2 vector by BamHI/XbaI and ligated into the pBlueScript KS+ vector to yield plasmid pSS1. The resulting pSS1 plasmid was then cut by SmaI/XbaI and subcloned into the pG2 vector to produce vector pG2-T. The insertion of the 700-bp fragment of the terminator was confirmed by restriction digestion with BamHI/XbaI.
For adding the promoter sequence to the expression cassette, the 2-kb Pgpd promoter fragment was cut out from pPK2 by EcoRI and cloned into the pBlueScript KS+ vector to produce plasmid pSS2. The resulting pSS2 vector was digested by ScaI/KpnI, and the produced fragment containing the promoter was subcloned into the HincII/KpnI-digested pBlueScript KS+ vector producing the pSS3 vector. The promoter was cut out from pSS3 by HindIII/KpnI and subcloned into the pG2-T vector to produce pP-G2-T vector. Confirmation of the produced vector was done by restriction digestion using HindIII/KpnI to afford a fragment of 2 kb corresponding to the promoter and by XbaI/KpnI to afford a fragment of approximately 3.9 kb corresponding to the whole construct pP-G2-T.
The pPK2 vector was cut by HindIII/EcoRV to release Pgpd::hphR::TtrpC cassette, which is then cloned into pBlueScript KS+ to yield plasmid pSS4. The produced pSS4 plasmid was then cut by KpnI to release the Pgpd::hphR::TtrpC fragment. The Pgpd:GGPPS::TtrpC fragment was released by digestion of the pP-G2-T vector with XbaI/KpnI and subcloned into the pPK2 vector resulting in the pSS5 plasmid. The KpnI-digested Pgpd::hphR::TtrpC fragment was then subcloned into the KpnI-digested pP-G2_T plasmid to produce the pTFG plasmid that contains two cassettes, Pgpd::hphR::TtrpC and Pgpd::GGPPS::TtrpC. The last pTFG construct was confirmed by restriction digestion with KpnI to afford a fragment of approximately 4 kb.
Functional complementation of TFG vector in yeast
The yeast S. cerevisiae mutant strain (SFNY368) lacking GGPPS is cold-sensitive for growth because it lacks GGPPS activity and cannot survive at 14 °C (Liao et al. 2005). The confirmed pTFG plasmid was transformed into SFNY368 by the lithium acetate method (Gietz and Schiestl 2007) and incubated at 30 °C overnight. The transformants were then selected at 14 °C on the YPAD medium in a dark incubator. Yeast colonies started growing 3 days later, large colonies were picked up, and PCR was used to detect positive colonies using GGPPSFS4 and GGPPSRS4 primers. The growth of SFNY368 (mutant) and transgenic SFNY368 colonies containing pTFG at 14 and 30 °C was compared 7 days after plating yeast cells onto the YPAD medium. The same procedure was done for a control experiment using mutant yeast cells but with addition of the pPK2 plasmid to the transformation solution instead of the pTFG vector.
Hygromycin B selection experiment
Paraconiothyrium SSM001 hyphae tips were transferred onto PDA medium supplemented with different concentrations of hygromycin B: 0.25, 0.5, 1.0, 1.5, 2.0, 5.0, and 10 μg/mL. The fungal growth was then observed for up to 1 week.
T-DNA transfer
Agrobacterium tumefaciens-mediated transformation of Paraconiothyrium SSM001 spp. was carried out as described by Covert et al. (Covert et al. 2001) with the following modifications.
Preparation of electroportable Agrobacterium cells
Overnight (2%) LB-Agrobacterium cultures were grown at 30 °C to OD600 of 1.2 (30 h). The flask containing the Agrobacterium cultures were chilled on ice for 10 min and then centrifuged at 5000 rpm at 4 °C for 10 min. The pelts were then re-suspended in 50 mL of ice-cold sterile 10% glycerol two times and finally re-suspended in 0.75 mL 10% glycerol. Aliquots of 40 μL were dispensed into 1.5-mL eppendorf tubes and flash frozen in liquid nitrogen and stored at − 80 °C until electroporation. The plasmid, pTFG (Fig. 1, Table 1), was transformed into A. tumefaciens AGL-1 by electroporation at 2.2 kV, 400 Ω, and 25 μF for 5.9 ms. The electroporated cells were plated onto solid LB media supplemented with rifampicin (0.25 mg/mL) and kanamycin (1 mg/mL) and incubated at 30 °C in the dark for 2–3 days. The produced colonies were subcultured in liquid LB medium containing rifampicin (0.25 mg/mL), kanamycin (1 mg/mL), carbenicillin (1 mg/mL), and streptomycin (0.5 mg/mL) for 3 days. Glycerol stocks of the produced cultures were stored at − 80 °C. Another part of the culture was subjected for confirmation of transformants by extracting the plasmids followed by restriction digestion with KpnI to afford a fragment of approximately 3.9 kb.
Fungal transformation
Ten milliliters of liquid LB culture containing 0.5 mg/mL rifampicin, 1 mg/mL kanamycin, 1 mg/mL streptomycin, and 1 mg/mL carbenicillin was inoculated with a small tip of Agrobacterium strain AGL-1::pTFG glycerol stock and incubated in the dark at 29 °C at 150 rpm for 4 days until reaching OD660 of 0.6–0.8. The produced culture was then diluted with induction medium (IM) containing 200 μM acetosyringone (AS) (Aldrich Chemical, Milwaukee, WI) to achieve an OD660 of 0.2 to a final volume of 20 mL. The Agrobacterium cells were then grown for approximately 8 h at 29 °C, 150 rpm in the dark, until reaching OD660 of 0.6–0.8. Simultaneously, 50 mL YPD broth was inoculated with Paraconiothyrium SSM001 hyphal tip. After growing at room temperature in the dark for 5 days with shaking, the mycelia were collected, cut with a sharp sterile razor, and passed through a syringe needle (B-D, 22G1) several times to allow the separation of small pieces of hyphae. The hyphae pieces were then mixed with 15 mL A. tumefaciens AGL-1::TFG culture of LB media mixed with induction media containing 200 μM acetosyringone and incubated for 3 days in the dark at 29 °C and 100 rpm. Two-milliliter aliquots of this mixture were plated onto white filter paper (P8, # 09-795B, Fisher Scientific) which were laid upon IM with 200 μM AS and incubated in the dark at 28 °C for 3 days. After 3 days of growth, the filter papers were transferred to PDA plates containing 300 μg/mL cefotaxime (Merck) to kill the A. tumefaciens and 1.5 μg/mL hygromycin B (Calbiochem, La Jolla, CA) in the dark at 25 °C. Two days later, the filter papers were placed upside down on new PDA media containing 300 μg/mL cefotaxime and 3 μg/mL hygromycin B for another 1 day. The last two steps were repeated for 1 week increasing the concentration of hygromycin B for up to 5 μg/mL. Putative visible transformants (Fig. 4) were visible 3–5 days later as rapidly grown circular colonies. The produced colonies were transferred onto fresh PDA media containing 5 μg/mL hygromycin B. All transformation experiments were done on another Paraconiothyrium SSM001 culture using LB media containing A. tumefaciens AGL-1 alone or AGL-1 containing hygromycin-resistant gene and used as negative controls (Fig. 5). Another Paraconiothyrium culture was not treated with hygromycin and used as a negative control. Seven colonies were selected along with control fungus grown on filter paper laid upon PDA media mixed with LB media, and another fungus normally grown on PDA medium was used to inoculate 500 mL YPD media mixed with hygromycin B (3 μg/mL) in 1-L flasks at 25 °C with shaking at 100 rpm. All cultures were refreshed with another dose of hygromycin B every week. Two fungal controls were incubated in YPD media without hygromycin, one fungal culture taken from normally grown fungus on PDA and another one previously grown on filter paper laid upon PDA and mixed with plain LB.
PCR detection of the fungal transformants
The regenerated putative transformants were screened by PCR for the presence of the hph gene using primers hph-F and hph-R (Table 2). Meanwhile, GGPPS primers GGPPSF4S and GGPPSR4S were also used to amplify the fragment corresponding to the GGPPS gene. The fungal mycelia were disrupted by mortar and pestle under liquid nitrogen, and then fungal genomic DNA and RNA were isolated using the Plant DNeasy and RNeasy Kits, respectively (Qiagen, Germany), with additional adsorption of the DNA or RNA onto a silica membrane, followed by washing and centrifugation as previously described (Haugland et al. 2002). Fifty nanograms of the DNeasy-purified genomic DNA was added into a 20-μL PCR reaction, with 1.5 mM MgCl2, 0.2 mM dNTPs, 1× buffer (Green GoTaq Flexi Buffer, Promega, USA, # M8911), 0.5 U Taq DNA polymerase (New England Biolabs, USA), and 0.2 μM of each previously mentioned hph or GGPPS genes primers. The amplified fragment was purified, cloned, and sequenced. Each resulting sequence was then aligned with the sequence of strain SSM001 using the Align-BLAST search tool. DNA from untransformed wild-type fungus served as a negative control while the transformed pTFG was used as a positive control in PCR analysis. RNA was also extracted from fungal clones and converted to cDNA. Fungal cDNA was used similarly for PCR detection of GGPPS gene followed by cloning and sequencing.
Production, extraction, and identification of fungal Taxol
Fungal tips from 2-week-old pure PDA plate cultures were used for production and extraction of fungal Taxol as described previously (Soliman et al. 2011; Stierle et al. 1993). Fungal Taxol in the extracted liquid media was identified by LC-MS as previously described by Soliman et al. (2011) and quantified by HPLC according to Soliman et al. (2011) and Soliman et al. (2013).
Fungal carotenoid extraction and quantification
Carotenoid extraction procedure was performed as described by Rodriguez-Amaya (2001). Briefly, 3-g liquid N2-frozen samples were ground with mortar and pestle and extracted three times with 20 mL acetone. The mixture was then filtered in the dark. The acetone extracts were evaporated and partitioned between water and petroleum ether for three times. The petroleum ether layers were separated, filtered through anhydrous sodium sulfate, and evaporated till dryness. The residual extract was dissolved in chloroform and measured quantitatively by spectrophotometry at 450 nm. Phytoene (Sigma, Cat# 78903) standard was used to generate a calibration curve using 0.5, 1, 2, 5, 10, 20, 50, and 100 ng/μL. All analyses were performed in triplicate.
Results
Exogenous feeding of terpenoid precursors enhanced the production level of fungal Taxol
Supplementation of SSM001 fungal YPD liquid culture with both IPP (isoprene) and GGPP increased fungal Taxol production. GGPP caused a greater increase (fivefold) than IPP (threefold) relative to un-supplemented culture media (Fig. 2) indicating that GGPP may be a direct precursor of fungal Taxol. The results also indicated that the fungus can utilize more terpenoid precursors and hence can produce more Taxol. So we assumed that integration of the GGPPS gene from Taxol-producing plants to Taxol-producing fungi might complementarily multiply the production level.
The following are sequential steps of T. canadensis GGPPS gene integration into the SSM001 genome and its effect on fungal terpenoid production:
-
1.
Paraconiothyrium SSM001 fungus was sensitive to hygromycin B
The growth of wild-type Paraconiothyrium SSM001 strain on PDA media supplemented with hygromycin B at 0.25, 0.5, 1, and 1.5 μg/mL at 25 °C was assessed for 72 h. The fungal growth was significantly affected by hygromycin B in the medium, and the effect was continually observed for up to 1 week. Figure 3 shows that 1.0 μg/mL hygromycin B was enough to completely kill the fungal cells. Therefore, 1.5 μg/mL hygromycin B was used for the selection of SSM001 transformants during the transformation procedure.
-
2.
GGPPS was complementarily functional in mutant yeast strain
To identify the function of pTFG, yeast complementation assay was performed. The mutant (SFNY368) yeast lacking GGPPS activity is cold sensitive for growth and cannot grow at 14 °C. Thus, it could be used to identify if the GGPPS was active or not. The present study shows that pTFG-transgenic SFNY368 (SFNY368 + pTFG; YT) was able to grow at 14 °C, while SFNY368 (YS) could not survive, compared to control wild-type S. cerevisiae (YC) which could not survive too at 14 °C (Fig. 4a). However, both yeast strains (YT and YS) and yeast control (YC) were able to survive at 30 °C (Fig. 4b). These results proved that pTFG obtained in the present study could achieve complementation in SFNY368 yeast and was a functional GGPPS gene which did mediate the biosynthesis of GGPP. Verification of the transformation was confirmed using PCR (Fig. 4c). Compared to YS, YC, and control culture media (C), only YT showed positive band at ~ 1.2 kb corresponding to the GGPPS gene.
-
3.
GGPPS-mediated Agrobacterium transformation in SSM001 was successful
Yeast functional complementation assay: The assay showed that a pTFG-transgenic SFNY368 (SFNY368 + pTFG; YT) was able to grow at 14 °C, while SFNY368 (YS) and control wild-type S. cerevisiae (YC) could not survive. b Yeast strains (YT and YS) and yeast control (YC) were able to survive at 30 °C. c PCR confirmation of the transformation efficiency. Compared to YS, YC, and control culture media, only YT showed a positive band at ~ 1.2 kb corresponding to the GGPPS gene
SSM001 fungal hyphae were transformed by A. tumefaciens carrying pTFG as described in the “Material and methods” section. After hygromycin B selection, ~ 20 hygromycin-resistant colonies were regenerated (Fig. 5a) compared to three different negative controls, SSM001 fungus which was not treated with Agrobacterium (C1), SSM001 treated with wild-type A. tumefaciens (C2), and SSM001 treated with Agrobacterium harboring only the hygromycin-resistant gene (C3). All treated fungal strains were maintained on hygromycin B selection media. Few small and slow-growing colonies were grown from each plate 4–7 days post transformation. However, negative controls including C1 and C2 were dead, and C3 continued to grow. To investigate the stability of the transformants, they were successively cultured on PDA medium supplemented with aqueous solution containing 1.5 μg/mL hygromycin B for two rounds of selection, followed by immersion in solution containing 3 μg/mL hygromycin B and finally in a solution containing 5 μg/mL hygromycin B. All transformants continued to grow on the selection plates, compared to C1 and C2 negative controls which showed no growth (Fig. 5a). All transformants retained their resistance to hygromycin B, indicating that the transformants were stable. However, many transformants showed much slower growth rate than the wild-type strain (Fig. 5a). Transformants with various growth rates and colors were obtained in addition to SSM001 treated with Agrobacterium harboring only the hygromycin-resistant gene (C3) (Fig. 5b).
Agrobacterium-mediated transformation of Taxus GGPPS into SSM001. a Regeneration of hygromycin-resistant pTFG-transformed SSM001 colonies (Test) compared to three different negative controls, SSM001 fungus not treated with Agrobacterium (C1), SSM001 treated with wild-type Agrobacterium (C2), and SSM001 transformed with Agrobacterium harboring only hygromycin-resistant gene (C3). b Seven different TFG-SSM001 transformants with various growth rates and colors and C3 control were maintained on hygromycin-resistant media. c PCR confirmation of successful transformation using hygromycin (626 bp) gene primers/fungal genomic DNA (panel i), GGPPS (~ 1.2 kb) gene/fungal genomic DNA (panel ii), and GGPPS primers/fungal cDNA (panel iii). Panel iv represents stability testing of clone #7T culturing in liquid YPD media with and without supplementation of 0.44 μM GGPP using GGPPS gene primers/fungal cDNA
Seven different pTFG-transformed and hygromycin-resistant individuals were maintained and subjected to PCR screening for the detection of the presence of the GGPPS gene in the fungal genome and transcriptome. As shown in Fig. 5c, the anticipated 626-bp fragment corresponding to the hph gene appeared from all transformants and C3 control in panel i, while no product was amplified from negative controls including culture media (C), C1 and C2. The 1.2-kb band corresponding to the GGPPS gene appeared from 2T, 4T, 5T, and 7T transformants in panel ii, while no product was amplified from 1T, 3T, and 6T and negative controls including C, C1, C2, and C3. The sequencing result confirmed that the amplified product from the transformants had the same sequence with the anticipated GGPPS gene. Furthermore, running a PCR reaction using cDNA of the seven selected transformants showed that 2T, 4T, 5T, and 7T were positive (panel iii). Transformant 7T, orange to reddish in color, was re-subcultured to three plates followed by inoculation into YPD liquid media either supplemented with GGPP or not. All of them showed strong and stable positive bands before and after supplementation with GGPP; however, expression of GGPPS was higher from those supplemented with GGPP, compared to control (C) (panel iv). These results confirmed that the transgenic pTGF-SSM001fungi were generated by A. tumefaciens technique and the transformants were stable.
GGPPS transformation induced fungal terpenoid pool size and hence Taxol production
Fungal Taxol production was maintained with original fungus cultured during the whole experimental procedure compared to Taxol standard (lane 1) but at variable levels (Fig. 6a). Similarly, transformed SSM001 showed variation in Taxol production and only strains #5T and 7T showed Taxol production (indicated by black arrow) (Fig. 6b, c). On the other hand, pTFG-transformed fungus (#7T) showed threefold increase in Taxol production (black arrow) compared to control strains transformed with Agrobacterium-hygromycin (C3) (Fig. 6c, d) measured by Taxol immunoassay. Feeding 7T with GGPP precursor multiplies the production level of fungal Taxol two more folds measured by Taxol immunoassay (Fig. 6e). Furthermore, strain #7T showed a significant increase in carotenoid production compared to the original SSM001 strain (Fig. 6e). The results obtained indicated that transformation of Taxol-producing fungus with the plant GGPPS gene increased terpenoid pool size and hence enhanced the production of fungal Taxol.
The effect of GGPPS gene integration into fungal genome on fungal terpenoid production. a Detection of Taxol on TLC from different original SSM001 strains prior to transformation process. b, c Identification of fungal Taxol in seven selected SSM001 pTFG-transformants compared to three different negative control: SSM001 fungus not treated with Agrobacterium (C1), SSM001 treated with wild-type Agrobacterium (C2), and SSM001 transformed with Agrobacterium harboring only hygromycin-resistant gene (C3). d Effect of GGPPS transformation and GGPP supplementation of GGPPS-transformed fungus on total fungal Taxol production. e Carotenoid accumulation in pTFG-transformed SSM001 #7T compared to SSM001 transformed with Agrobacterium harboring only hygromycin-resistant genes
Discussion
Microbial production of metabolites in particular those with high value such as Taxol is an important target since it can easily manipulated. However, Taxol production by endophytic fungi is not the case because most elicitor studies failed to produce a sufficient amount of the compound. So we assumed that increasing the terpenoid pool size of the fungus either through feeding with terpenoid precursors or through increasing the GGPPS copy number in the fungal genome might enhance the fungal production level of Taxol. Fungal transformation with the plant GGPPS gene caused an increase in Taxol production and carotenoid level indicative of an increase in the terpenoid pool size.
Supplementation of fungal culture with terpenoid precursors including IPP and GGPP enhanced Taxol production threefold and fivefold, respectively, suggesting that the terpenoid pathway is of important contribution in the fungal Taxol biosynthesis, which is in consistence with the findings that terpenoid pathways are important in fungal Taxol biosynthesis (Soliman et al. 2011). Even though there is no study on the effect of feeding terpenoid precursors (IPP and GGPP) on Taxol production, there are several reports indicating that terpenoid precursors represent an important factor in compound production and precursor feeding enhanced their production (Moreno et al. 1993; Peebles et al. 2006).
On the other hand, integration of plant GGPP-producing genes into the fungal genome showed an increase in both carotenoid and Taxol production compared to fungus transformed with the hgh gene. The results obtained are in consistence with the notion that terpenoid pool size is high as in the case of TS-transformed tomato (Kovacs et al. 2007) and TS-transformed yeast (Ding et al. 2014; Engels et al. 2008) which enhanced taxadiene production compared to small terpenoid pool size organisms including TS-transformed Arabidopsis (Besumbes et al. 2004) and TS-transformed E. coli (Huang et al. 2001).
Similarly, supplementation of GGPPS-transformed fungus with GGPP increased GGPPS transcription and Taxol production. Generally, fungi including SSM001 have GGPPS for the biosynthesis of their own GGPP. GGPP is used by fungi for the production of their original metabolites such as carotenoids and gibberellins (Keeling and Bohlmann 2006). However, the amount of biosynthesized GGPP is only sufficient to produce a sufficient amount of essential fungal diterpenoids. On the other hand, introducing a new copy of GGPPS requires more precursors either GPP or FPP which are not available since it is used by the original fungal GGPPS (Singkaravanit et al. 2010; Zhu et al. 1997), and hence, exogenous addition of excess GGPP will be used by the original fungal terpenoid pathway which in its turn will allow enough terpenoid precursors including FPP and/or GPP to activate GGPPS and hence increase its transcription and Taxol production.
The Agrobacterium-mediated genome integration was successful, since it is a well-reported efficient technique in transformation of filamentous fungi as well as plants (de Groot et al. 1998). However, the production of Taxol was variable and colony-dependent similar to that of original un-transformed fungus. The differentiation in secondary metabolite production from colony to colony is a well-known feature in higher fungi, and there are several factors affecting fungal metabolite production including genetic, environmental, and developmental factors (Sim 2001). Furthermore, it is documented that fungal Taxol production is decreased because of successive culturing procedure and fungal aging (Soliman and Raizada 2013; Soliman et al. 2011; Stierle et al. 1993); thus, GGPPS-transformed SSM001 did not show huge production of Taxol since the transformation procedure takes 3 months. Future studies might be able to maximize the production level once the time used for transformation is reduced. On the other hand, in accordance to Soliman et al. (2011), phenolics contribute to the fungal Taxol biosynthesis (Soliman et al. 2011); thus, enhancing the phenolic part of the pathway could increase the production level of fungal Taxol.
As concluding remarks, a successful transformation method of endophytic fungus SSM001 based on Agrobacterium-mediated genome integration was developed. The plant GGPP-producing gene integration into fungal genome increased the terpenoid pool size of the fungus which was reflected by an increase in the level of fungal carotenoids. Fungal Taxol can be possibly maximized when overcoming the aging factor and/or enhancing the contribution by the phenolic pathway.
References
Baloglu E, Kingston DGI (1999) The taxane diterpenoids. J Nat Prod 62(10):1448–1472
Besumbes Ó, Sauret-Güeto S, Phillips MA, Imperial S, Rodríguez-Concepción M, Boronat A (2004) Metabolic engineering of isoprenoid biosynthesis in Arabidopsis for the production of taxadiene, the first committed precursor of Taxol. Biotechnol Bioeng 88(2):168–175
Bian G, Yuan Y, Tao H, Shi X, Zhong X, Han Y, Fu S, Fang C, Deng Z, Liu T (2017) Production of taxadiene by engineering of mevalonate pathway in Escherichia coli and endophytic fungus Alternaria alternata TPF6. Biotechnol J 12(1600697):1–10
Chen X, Stone M, Schlagnhaufer C, Romaine CP (2000) A fruiting body tissue method for efficient Agrobacterium-mediated transformation of Agaricus bisporus. Appl Environ Microbiol 66(10):4510–4513
Covert SF, Kapoor P, Lee M-h, Briley A, Nairn CJ (2001) Agrobacterium tumefaciens-mediated transformation of Fusarium circinatum. Mycol Res 105(3):259–264
Croteau R, Ketchum R, Long R, Kaspera R, Wildung M (2006) Taxol biosynthesis and molecular genetics. Phytochem Rev 5(1):75–97
de Groot MJA, Bundock P, Hooykaas PJJ, Beijersbergen AGM (1998) Agrobacterium tumefaciens-mediated transformation of filamentous fungi. Nat Biotech 16(9):839–842
Ding M-z, Yan H-f, Li L-f, Zhai F, Shang L-q, Yin Z, Yuan Y-j (2014) Biosynthesis of taxadiene in Saccharomyces cerevisiae: selection of geranylgeranyl diphosphate synthase directed by a computer-aided docking strategy. PLoS One 9(10):e109348
Engels B, Dahm P, Jennewein S (2008) Metabolic engineering of taxadiene biosynthesis in yeast as a first step towards Taxol (paclitaxel) production. Metab Eng 10(3):201–206
Gietz RD, Schiestl RH (2007) Large-scale high-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat Protocols 2(1):38–41
Haugland RA, Brinkman N, Vesper SJ (2002) Evaluation of rapid DNA extraction methods for the quantitative detection of fungi using real-time PCR analysis. J Microbiol Methods 50(3):319–323
Hefner J, Ketchum REB, Croteau R (1998) Cloning and functional expression of a cDNA encoding geranylgeranyl diphosphate synthase from Taxus canadensis and assessment of the role of this prenyltransferase in cells induced for Taxol production. Arch Biochem Biophys 360(1):62–74
Hood EE, Gelvin SB, Melchers LS, Hoekema A (1993) New Agrobacterium helper plasmids for gene transfer to plants. Transgenic Res 2(4):208–218. https://doi.org/10.1007/BF01977351
Huang Q, Roessner CA, Croteau R, Scott AI (2001) Engineering Escherichia coli for the synthesis of taxadiene, a key intermediate in the biosynthesis of Taxol. Bioorg Med Chem 9(9):2237–2242
Hugueney P, Bouvier F, Badillo A, Quennemet J, d’Harlingue A, Camara B (1996) Developmental and stress regulation of gene expression for plastid and cytosolic isoprenoid pathways in pepper fruits. Plant Physiol 111(61):9–626
Jiang Y, Proteau P, Poulter D, Ferro-Novick S (1995) BTS1 Encodes a geranylgeranyl diphosphate synthase in Saccharomyces cerevisiae. J Biol Chem 270(37):21793–21799
Kang S-M, Min J-Y, Kim Y-D, Park D-J, Jung H-N, Karigar CS, Ha Y-L, Kim S-W, Choi M-S (2006) Effect of supplementing terpenoid biosynthetic precursors on the accumulation of bilobalide and ginkgolides in Ginkgo biloba cell cultures. J Biotechnol 123(1):85–92
Keeling CI, Bohlmann J (2006) Genes, enzymes and chemicals of terpenoid diversity in the constitutive and induced defence of conifers against insects and pathogens. New Phytol 170(4):657–675
Koksal M, Jin Y, Coates RM, Croteau R, Christianson DW (2011) Taxadiene synthase structure and evolution of modular architecture in terpene biosynthesis. Nature 469(7328):116–120
Kovacs K, Zhang L, Linforth RST, Whittaker B, Hayes CJ, Fray RG (2007) Redirection of carotenoid metabolism for the efficient production of taxadiene [taxa-4(5),11(12)-diene] in transgenic tomato fruit. Transgenic Res 16(1):121–126
Kuntz M, Camara B, Weil J-H, Schantz R (1992) The psbL gene from bell pepper (Capsicum annuum): plastid RNA editing also occurs in non-photosynthetic chromoplasts. Plant Mol Biol 20(6):1185–1188
Liao Z, Gong Y, Kai G, Zuo K, Chen M, Tan Q, Wei Y, Guo L, Tan F, Sun X, Tang K (2005) An intron-free methyl jasmonate inducible geranylgeranyl diphosphate synthase gene from Taxus media and its functional identification in yeast. Mol Biol 39(1):11–17
Moreno P, Heijden R, Verpoorte R (1993) Effect of terpenoid precursor feeding and elicitation on formation of indole alkaloids in cell suspension cultures of Catharanthus roseus. Plant Cell Rep 12(12):702–705
Peebles CAM, Hong S-B, Gibson SI, Shanks JV, San K-Y (2006) Effects of terpenoid precursor feeding on Catharanthus roseus hairy roots over-expressing the alpha or the alpha and beta subunits of anthranilate synthase. Biotechnol Bioeng 93(3):534–540
Roberts S (2007) Production and engineering of terpenoids in plant cell culture. Nat Chem Biol 3(7):387–395
Rodriguez-Amaya DB (2001) A guide to carotenoid analysis in foods. ILSI press Washington, DC
Schütte HR (1997) Secondary plant substances. Diterpenes. In: Behnke HD, Lüttge U, Esser K, Kadereit JW, Runge M (eds) Progress in botany: structural botany physiology genetics taxonomy Geobotany/Fortschritte der Botanik Struktur Physiologie Genetik Systematik Geobotanik. Springer, Berlin, pp 255–277
Sim SC (2001) Characterization of genes in the sterigmatocystin gene cluster and their role in fitness of Aspergillus nidulans. Texas A & M University, College Station
Singh B, Sharma RA (2015) Plant terpenes: defense responses, phylogenetic analysis, regulation and clinical applications. 3. Biotech 5(2):129–151
Singkaravanit S, Kinoshita H, Ihara F, Nihira T (2010) Geranylgeranyl diphosphate synthase genes in entomopathogenic fungi. Adv Appl Microbiol 85(5):1463–1472
Soliman S, Greenwood John S, Bombarely A, Mueller Lukas A, Tsao R, Mosser Dick D, Raizada Manish N (2015) An endophyte constructs fungicide-containing extracellular barriers for its host plant. Curr Biol 25(19):2570–2576
Soliman S, Raizada MN (2013) Interactions between co-habitating fungi elicit synthesis of Taxol from an endophytic fungus in host Taxus plants. Front Microbiol 4:3
Soliman S, Tang Y (2015) Natural and engineered production of taxadiene with taxadiene synthase. Biotechnol Bioeng 112(2):229–235
Soliman S, Tsao R, Raizada MN (2011) Chemical inhibitors suggest endophytic fungal paclitaxel is derived from both mevalonate and non-mevalonate-like pathways. J Nat Prod 74(12):2497–2504
Soliman SSM, Trobacher CP, Tsao R, Greenwood JS, Raizada MN (2013) A fungal endophyte induces transcription of genes encoding a redundant fungicide pathway in its host plant. BMC Plant Biol 13:93–93. https://doi.org/10.1186/1471-2229-13-93
Stierle A, Strobel G, Stierle D (1993) Taxol and taxane production by Taxomyces andreanae, an endophytic fungus of Pacific yew. Science 260(5105):214–216
Zhu X, Suzuki K, Saito T, Okada K, Tanaka K, Nakagawa T, Matsuda H, Kawamukai M (1997) Geranylgeranyl pyrophosphate synthase encoded by the newly isolated gene GGPS6 from Arabidopsis thaliana is localized in mitochondria. Plant Mol Biol 35(3):331–341
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Informed consent
There are no human or animal subjects used in this study.
Rights and permissions
About this article
Cite this article
Soliman, S.S.M., Mosa, K.A., El-Keblawy, A.A. et al. Exogenous and endogenous increase in fungal GGPP increased fungal Taxol production. Appl Microbiol Biotechnol 101, 7523–7533 (2017). https://doi.org/10.1007/s00253-017-8509-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-017-8509-9