Abstract
The biosynthetic pathway for hyaluronic acid (HA) has been proposed; however, a thorough genetic and functional analysis is required to further elucidate the roles of genes involved in HA production. Previously, we developed a markerless gene-deletion system for Streptococcus zooepidemicus and confirmed that hasA is essential for HA synthesis. Here, we constructed a comprehensive set of deletion mutants and investigated the roles of ten additional predicted genes in the HA synthetic pathway. Phenotypic assays revealed that all ten genes play a role in cell growth and/or HA synthesis. As expected, the deletion of hasA or hasB abolished HA production with little effect on growth, while the deletion of genes that are also required for peptidoglycan biosynthesis (hasE, glmM, and glmS) significantly reduced cell growth and HA production. Either of the glmU homologues (hasD and gcaD) was sufficient for optimal growth and the mucoid phenotype, while no double mutant could be isolated. Of the two UDP-glucose pyrophosphorylase (UGPase) paralogues, the operon-encoded hasC1 was responsible for 65 % of the activity, while hasC2 was responsible for the remaining 35 %. The deletion of hasC1 had no effect on cell growth and caused only a moderate decrease in the UDP-glucose level and HA production. The deletion of both hasC1 and hasC2 resulted in a severe growth defect and negligible UDP-glucose accumulation, HA production, and pyrophosphorylase activity. Of the two phosphoglucomutase paralogues, pgm1 and pgm2, the former is responsible for around 10 % of activity, while the latter is responsible for 90 %. The deletion of pgm1 showed no apparent effect on HA synthesis and growth, while the deletion of pgm2 resulted in the abolishment of HA synthesis and a significantly slower growth. These results should guide the metabolic engineering of S. zooepidemicus to improve HA productivity and quality.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Hyaluronic acid (HA) is a linear polysaccharide consisting of repeating disaccharide units of d-glucuronic acid (GlcUA) and N-acetylglucosamine (GlcNAc) linking alternatively by β-1,3 and β-1,4 glycosidic bonds (Chong and Nielsen 2003). Its distinct physical-chemical properties and biocompatibility have seen HA and its derivatives being widely applied in the biomedical, cosmetic, and food industries (Liu et al. 2011). HA is found in the connective tissues of animals as well as in the capsules of various bacteria such as streptococci and Pasteurella (Wessels et al. 1991). Currently, the fermentation of group C Streptococcus zooepidemicus is the most widely used strategy to produce HA commercially (Chen et al. 2014).
The biosynthetic pathway for HA production has been proposed, and enzymes involved in the catalytic processes were suggested (Chong and Nielsen 2003) (Fig. 1a). Using glucose-6-P as a substrate, one metabolic branch catalyzed sequentially by a phosphoglucomutase (PGM), a uridyltransferase, and a dehydrogenase leads to the synthesis of the first precursor, GlcUA, while the parallel branch catalyzed in order by phosphoglucose isomerase, glutamine amidotransferase, phosphoglucosaminemutase, acetyl-CoA acetyltransferase, and UTP uridyltransferase yields the second precursor, GlcNAc. Finally, the HA polymer is synthesized by the membrane-associated hyaluronan synthase through the alternative addition of GlcUA and GlcNAc.
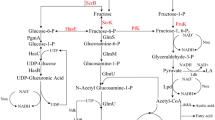
Genes involved in hyaluronic acid (HA) biosynthetic pathway in Streptococcus zooepidemicus. a The predicted HA biosynthetic pathway. b The names and chromosomal loci of 11 genes in S. zooepidemicus are depicted. Arrows represent the direction of transcription of these genes. c Total RNA was prepared from wild-type S. zooepidemicus culture and used for RT-PCR assay. The numbers 1–11 represent the genes hasA, hasB, hasC1, hasC2, hasD, gcaD, hasE, pgm1, pgm2, glmS, and glmM in order. M represents the DNA ladder
A has operon containing a cluster of three genes (hasA, hasB, and hasC), which encode hyaluronan synthase, UDP-glucose dehydrogenase (UDPGDH), and UDP-glucose pyrophosphorylase, respectively, was identified in S. pyogenes and extensively characterized (Crater et al. 1995; DeAngelis et al. 1993; Dougherty and van de Rijn 1993). The has operon is conserved in all strains of group A streptococci and encapsulated group C streptococci (Crater et al. 1995). In contrast to the essential role of hasC for HA production in S. pyogenes, the hasC located on has operon is not necessary for HA synthesis in S. pneumoniae (Crater et al. 1995), while the non-operon galU was identified as an essential UDP-glucose pyrophosphorylase-encoding gene for HA production (Mollerach et al. 1998). In S. zooepidemicus, the has operon contains additional two genes involved in HA biosynthesis, glmU (hasD) and pgi (hasE), downstream of hasC (Blank et al. 2008). hasD encodes a putative dual function enzyme acetyl-CoA acetyltransferase and pyrophosphorylase, while hasE encodes a phosphoglucoisomerase. Overexpression of the has operon genes resulted in the production of HA with varying molecular weight, suggesting that all the five genes in the has operon are involved in HA synthesis in S. zooepidemicus (Chen et al. 2009). Overexpression of the non-operon genes (pgm, glmS, and glmM) had less of an effect on molecular weight, but the latter two affected HA yield (Chen et al. 2014). However, a comprehensive analysis of individual gene deficient strains is still necessary to fully elucidate the role of has operon and non-operon HA pathway genes in cell growth, metabolism, and HA synthesis. In particular, homologues exist for several of the genes and their relative importance has not been established. Recently, the release of complete genome sequences of several S. zooepidemicus strains and the development of a markerless gene-deletion system for this bacterium have enabled the prediction and confirmation of the role, if any, of the genes involved in of HA production (Beres et al. 2008; Izawa et al. 2011; Sun et al. 2013).
In this paper, we describe the identification of genes in the genome of S. zooepidemicus and the investigation of their role in HA production and cell growth. Genome-wide analysis suggests that 11 gene encoding enzymes are expected to be involved in HA production. A semiquantitative reverse transcription polymerase chain reaction (RT-PCR) assay showed that all 11 genes were expressed during an HA fermentation process. All 11 genes were individually deleted, and ΔhasC1ΔhasC2 and Δpgm1Δpgm2 double-mutation strains were constructed. The characterization of these strains indicates that all 11 genes play roles in bacterial growth and/or HA synthesis.
Materials and methods
Bacterial strains and growth conditions
All strains used in this study are listed in Supplementary Material Table S1. S. equi subsp. zooepidemicus ATCC39920 (S. zooepidemicus) wild type (WT) and mutants were grown at 30 or 37 °C in Todd-Hewitt yeast medium (THY) (in g/L: beef extract 10, casein tryptone 20, glucose 2, yeast 2, NaHCO3 2, NaCl 2, Na2HPO4 0.4; pH6.8) as described (Sun et al. 2013). Escherichia coli JM109 and BL21trxB (DE3) were grown at 37 °C in Luria-Bertani (LB) medium supplemented with antibiotics when necessary. The concentrations of antibiotics used in experiments were as follows: for E. coli, ampicillin (100 μg/mL), kanamycin (100 μg/mL), and spectinomycin (50 μg/mL), and for S. zooepidemicus, spectinomycin (100 μg/mL).
Semiquantitative RT-PCR analysis
Total RNA was isolated from S. zooepidemicus ATCC39920 grown in liquid THY medium in late exponential phases (8 h) using the TRIzol method. RNase-free DNase I was added to remove genomic DNA during the isolation process. The quality and the quantity of RNA were examined by 1 % agarose gel electrophoresis and UV spectroscopy at 260 nm. Reverse transcription was performed with 1 μg total RNA using a PrimeScript™ RT reagent kit (TaKaRa, Mountain View, CA, USA) as recommended by the manufacturer. Ten nanograms of cDNA mixture was used for PCR amplification of each gene with specific primer pairs. After 30 cycles, the resultant PCR products were analyzed by 1 % agarose gel electrophoresis. The primers used are listed in Supplementary Material Table S2.
Gene deletion in S. zooepidemicus
Genes were deleted using a markerless gene-deletion system as described previously (Sun et al. 2013). Briefly, using S. zooepidemicus genomic DNA as the template, the upstream and downstream fragments of pgm2 were amplified by PCR with the primer pairs Primer151/Primer152 and Primer153/Primer154, respectively. The two fragments were joined by splicing by overextension (SOE) PCR with the primer pair Primer151/Primer154 to generate a 1730 bp fragment, which was digested and ligated into the EcoRI/PstI sites of the vector pSET4s::sacB to obtain pSET4s::sacB::pgm2LR. S. zooepidemicus containing pSET4s::sacB::pgm2LR was first grown at 30 °C for 12 h and then cultured at 37 °C for another 4 h in THY medium supplemented with 100 μg/mL spectinomycin. The culture was selected on THY medium supplemented with 5 % (w/v) sucrose. The sucrose-resistant and spectinomycin-sensitive clones were isolated, and pgm2 gene-deletion mutants were examined by PCR with the primer pair Primer155/Primer158 and further confirmed by sequencing and RT-PCR. The same strategy as used for pgm2 deletion was followed to construct other single-gene-deficient strains and double mutants. The primers used are listed in Supplementary Material Table S2, and the restriction enzyme sites are underlined.
Expression and purification of 6His-HasC1 and 6His-HasC2 proteins
A standard protocol was followed to express and purify 6His-HasC1 and 6His-HasC2 (Sun et al. 2013). Briefly, pET23b-hasC1 and pET28b-hasC2 vectors were transformed into E. coli BL21trxB (DE3), and the protein expression was induced by 0.1 mM isopropyl-d-thiogalactopyranoside (IPTG) at 30 °C for 12 h. The cultures were chilled on ice, and the cell pellets were harvested by centrifugation at 12,000 rpm at 4 °C for 5 min and then resuspended in a 1× Ni-NTA buffer (50 mM NaH2PO4, 500 mM NaCl, 5 mM imidazole, 10 % glycerin; pH 7.9). The cell suspensions were sonicated (4 s each with 6 s cooling between successive bursts in 20 min). The resulting lysates were centrifuged at 12,000 rpm at 4 °C for 20 min. Fifty milliliters of the supernatant was applied onto a 10 mL column filled with 1.5 mL His-select Ni-chelating affinity gel (Qiagen, Hilden, Germany) preconditioned with 1× Ni-NTA buffer. The elution was performed with 5 mL of 1× binding buffer containing 10, 30, 50, 100, and 250 mM imidazole sequentially. The purity of the recombinant proteins was analyzed by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), and the concentrations were determined by Bradford protein assays kit (Bio-Rad Laboratories, Hercules, CA, USA).
Enzyme assays
UTP-glucose-phosphate uridyltransferase (UGPase) activity was detected as described previously with a slight modification (Daran et al. 1995). Cells were harvested in the midexponential phase and washed twice with 50 mM chilled potassium phosphate buffer (pH 7.4). A cell lysate was prepared using sonication (4 s/6 s, 20 min), and the debris was removed by centrifugation at 4 °C, 15,000 rpm for 15 min. One hundred milligrams of purified protein or 100 μL cell lysate was used in each assay. The standard reaction mixture (1 mL) contained 100 mM tricine buffer (pH 7.9), 2 mM UTP, 5 mM glucose-1-P, 5 mM MgCl2, 2 mM NAD(P)+, and 3.2 units UDPGDH. The reaction ran at 37 °C for 10 min and was terminated by heating to 100 °C for 5 min. The enzymatic activity was measured by coupling enzymatic assays with the reduction of NAD(P)+ to NAD(P)H, which was determined by a UNICO UV-2100 spectrophotometer at a wavelength of 340 nm. One unit of UGPase activity was defined as the amount of the enzyme required to produce 1 μmol UDP-glucose per minute.
PGM activity was determined as described previously (Lu and Kleckner 1994). Briefly, each reaction solution contained final concentrations of 10 mM MgCl2, 0.5 mM NADP+, 2 units/mL glucose 6-phosphate dehydrogenase, 50 μM glucose-1,6-bisphosphate, 5 mM glucose-1-phosphate, and 100 μL cell lysate. The activity of PGM was measured by the conversion of glucose-1-phosphate to glucose-6-phosphate coupled with the reduction of glucose-6-phosphate by glucose-6-phosphate dehydrogenase in 50 mM triethanolamine buffer (pH 7.4), which was determined by spectrophotometer at 340 nm.
Analytic methods
The hyaluronan concentration was determined by the carbazole method as described previously (Sun et al. 2013), where the optical density was measured at 530 nm using d-glucuronic acid as the standard. Cell concentration was determined by measuring the optical density (OD) of the culture at 660 nm using a spectrophotometer (UV-2100 spectrophotometer). The UDP-glucose concentration was determined by HPLC (Agilent, VertexTM C18, 4.6 × 250 mm, 5 μm). Briefly, the cell culture was harvested by centrifugation at 4 °C, 10,000g for 10 min. The pellet was washed with cold distilled water twice and resuspended in 40 mL cold distilled water. The suspension was disrupted by sonication and subsequently centrifuged at 4 °C, 5,000g for 30 min to remove cell debris. Double volumes of anhydrous ethanol were added, and the solution was kept at 4 °C for 1 h and then lyophilized. The dried samples were dissolved in 1 mL distilled water and filtered through a 0.22-μm filter. Quantitative determination of UDP-glucose was performed using an HPLC system with a diode array ultraviolet detector set at 262 nm. The mobile phase, with a flow rate of 1.0 mL/min, contained solution A (mixture of 0.125 M KH2PO4, pH 3.2, and 5 mM tetrabutylammonium hydrogen sulfate) and solution B (chromatographic acetonitrile) in an appropriate ratio. The injection volume of each sample was 10 μL.
Results
Identification of genes involved in HA biosynthetic pathway
Taking advantage of the draft genome sequence of S. zooepidemicus ATCC39920 and the predicted HA biosynthetic pathway (Chong and Nielsen 2003), we identified 11 genes that encode the proteins probably involved in HA production (Fig. 1a). The individual genes and their chromosomal locations are depicted in Fig. 1b. As found for S. zooepidemicus ATCC35246 (Blank et al. 2008), the has operon of S. zooepidemicus ATCC39920 contains a cluster of five genes, hasA, hasB, hasC1, hasD, and hasE in order (Fig. 1b). Single copies each of hasA, hasB, and hasE are identified, and hasE likely encodes a phosphoglucoisomerase in this strain. Genome-wide analysis led to the identification of hasC2 and gcaD, which are homologues of hasC1 and hasD, respectively, and located on distinctive chromosomal loci. Multiple sequence alignment showed that HasC2 and GcaD are 94.6 and 96.5 % identical to HasC1 and HasD, respectively. The genome contains the two genes pgm1 and pgm2, which are predicted to encode phosphoglucomutases, and the polypeptides encoded by these two genes show 30.77 % similarity. We also identified the two genes glmS and glmM, which encode glutamine amidotransferase and phosphoglucosaminemutase, respectively, likely involved in HA biosynthesis. Further analysis revealed no other homologous sequences for either glmS or glmM in the genome. To research the functions of these 11 genes, we first examined their transcription profiles in liquid culture. Semiquantitative RT-PCR assays indicated that all genes were expressed in the exponential growth phase (8-h shake-flask culture) (Fig. 1c). The expression profile of has operon genes is consistent with previous reports (Prasad et al. 2012; Sun et al. 2013). This suggests that all the 11 genes play some roles in cell growth or HA synthesis.
Characterization of deletion mutants
To address the physiological function(s) of these genes in S. zooepidemicus, we deleted each gene individually and constructed the respective single gene-deficient stains (Table S1). The strains were characterized on solid THY plates (Fig. 2a) as well as liquid THY medium (Fig. 2b). As expected, the deletion of hasA and hasB had a limited effect on growth, but greatly affected the mucoid phenotype. The deletion of any of the other genes without a known functional paralogue (ΔhasE, ΔglmM, ΔglmS) caused significant loss of growth and mucoid phenotype. ΔglmS did not grow on the THY medium, but the addition of 0.5 mg/mL glucosamine (GlcN) rescued the ΔglmS growth defect, but not the mucoid colony morphology. The deletion of any one of genes with a functional paralogue had no effect (ΔhasC1, ΔhasC2, Δpgm1, ΔhasD, and ΔgcaD) or only a modest effect (Δpgm2) on growth and the mucoid phenotype. Consistent with the observation of the mucoid phenotype on solid THY plates, the mutants (ΔhasA, ΔhasB, ΔhasC1ΔhasC2, ΔhasE, Δpgm2, Δpgm1Δpgm2, ΔglmM, and ΔglmS) produced less HA than wild type in liquid culture (Fig. 2c), indicating the role of these genes in HA biosynthesis.
Growth profiles of gene-deficient strains. a Wild type (WT) and the indicated mutants were grown on solid Todd-Hewitt yeast (THY) plates with or without glucosamine for 24 h, and the colonies were photographed. b The indicated strains were grown in liquid THY flasks for 24 h, and the cell density was determined by measuring the optical density (OD) of the culture at 660 nm (OD660). c Hyaluronic acid (HA) produced by the indicated strains grown in liquid THY flasks for 24 h was determined by the carbazole method. Data (b, c) represent the mean values from three independent experiments
An attempt was made to produce double mutants for all three paralogue sets (Table S1). However, we failed to obtain the ΔhasDΔgcaD double mutant after screening several hundreds of transformants, even in the presence of GlcN. It appears that the survival of S. zooepidemicus requires the function of either hasD or gcaD, but not both. The ΔhasC1ΔhasC2 double mutant displayed a severe growth defect and the loss of the mucoid phenotype, while the Δpgm1Δpgm2 double mutant displayed a moderate growth defect with a severe colony mucoid morphology (Fig. 2a). We conclude that all 11 genes play role(s) in either the bacterial growth or HA synthesis, although their contributions could be variable.
hasC1 is required for maximum HA biosynthesis
ΔhasC1 and ΔhasC2 displayed no obvious growth or mucoid defect, while the double mutant was severely affected. We further investigated the biochemical mechanism of HasC1 and HasC2 proteins in S. zooepidemicus. HasC1 and HasC2 contain 323 and 322 amino acids, respectively, and are identical except for several amino acids located in the N-terminus and C-terminus (Figure S1). Both HasC1 and HasC2 contain the G-T-R-X-L-P-X-T and V-E-K-P motifs (Figure S1) believed to endow the proteins an UDP-glucose pyrophosphorylase activity to combine UTP and glucose-1-phosphate to form α-type nucleotide sugar UDP-glucose (Daran et al. 1995; Kleczkowski et al. 2004). The in vitro enzyme activity was examined using bacterially expressed 6His-HasC1 and 6His-HasC2 proteins (Fig. 3a), and the activities of the purified 6His-HasC1 and 6His-HasC2 proteins were determined at 1.1 and 0.6 U/mg UGPase activities, respectively (Fig. 3b). To address whether the loss of hasC1 or/and hasC2 gene(s) affects the UGPase activity in S. zooepidemicus, the UGPase activity of the parental strain and the mutants was determined using a cell lysate prepared from the liquid cultures. Compared with the wild type, ΔhasC1 and ΔhasC2 had around 35 and 68 % of UGPase activity, respectively, while ΔhasC1ΔhasC2 showed a negligible UGPase activity (Fig. 3c). This suggests that both hasC1 and hasC2 contribute to cellular UGPase activity in S. zooepidemicus and minimal gene regulatory compensation occurs when one is deleted. Consistent with activity levels, the UDP-glucose level was 40 % lower in ΔhasC1 (169 mg/g cell dry weight [CDW]) compared to that in the wild type (288 mg/g CDW), while the level in ΔhasC2 was barely affected (271 mg/g CDW) (Fig. 3d). The lower level of UDP-glucose translated to lower levels of HA in ΔhasC1 cultures (0.41 g/L) compared to those in the wild type (0.55 g/L), while again the level in ΔhasC2 was barely affected (0.52 g/L) (Fig. 2c). As expected, no UDP-glucose or HA was detected in the ΔhasC1ΔhasC2 sample. Our data suggest that hasC1 is required for maximum UDP-glucose level and HA biosynthesis, while hasC2 activity alone is sufficient for cell growth in S. zooepidemicus.
Biochemical characterization of HasC1 and HasC2. a 6His-tagged HasC1 and HasC2 proteins were expressed in Escherichia coli BL21trxB (DE3) and purified; the protein quality was determined by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). M represents protein marker. b A UDP-glucose pyrophosphorylase activity assay was performed with 6His-HasC1 and 6His-HasC2. The GST protein was used as the control. c A protein lysate was prepared from the indicated strains grown in liquid Todd-Hewitt yeast (THY) medium for 8 h and the UDP-glucose pyrophosphorylase activity of each sample was examined. d The indicated strains were grown in liquid THY medium for 8 h, and the concentration of UDP-glucose in each flask culture was determined using HPLC method. Data (b, d) represent the mean values from three independent experiments. WT wild type
pgm2 is essential for HA biosynthesis
Unlike the phenotype of Δpgm1, Δpgm2 showed defects in both growth and colony mucoid morphology. Moreover, Δpgm1Δpgm2, which was generated by knocking out pgm1 in the Δpgm2 mutant, showed more severe defects than Δpgm2 in terms of growth and colony mucoid morphology. Although Pgm1 and Pgm2 have a low overall sequence similarity (Figure S2), annotation with NCBI Conserved Domain Search Service database indicates that both proteins contain a conserved PGM_PMM domain (Fig. 4a), suggesting that these proteins function as phosphoglucomutases (Lu and Kleckner 1994). In vitro enzyme activity assay with bacterially expressed 6His-Pgm1 and 6His-Pgm2 showed that these two proteins had comparable phosphoglucomutase activity (data not shown). However, analysis of the cell lysate samples showed that the phosphoglucomutase activities of WT, Δpgm1, Δpgm2, and Δpgm1Δpgm2 were 1.53 U/mg, 1.40 U/mg, 0.13 U/mg, and null, respectively (Fig. 4b). The Deletion of pgm2 resulted in the loss of around 90 % of intracellular phosphoglucomutase activity, suggesting that pgm2 plays a more important role than pgm1 in S. zooepidemicus. Consistent with the observations of the colony mucoid phenotype, the level of HA produced by Δpgm1 was comparable to that of the wild type, while HA was hardly detected in the flask cultures of Δpgm2 and Δpgm1Δpgm2 (Fig. 2c). We conclude that pgm2 is essential for optimal growth as well as HA production in S. zooepidemicus, while pgm1 has only a modest function.
pgm2 regulates hyaluronic acid (HA) biosynthesis. a The schematic of domains contained in the polypeptide encoded by pgm1 and pgm2 in Streptococcus zooepidemicus. b A protein lysate was prepared from the indicated strains grown in liquid Todd-Hewitt yeast (THY) medium for 8 h, and the phosphoglucomutase activity of each sample was examined. Data b represent the mean values from three independent experiments. WT wild type
Discussion
Under the standard growth condition, HA constitutes a substantial fraction of S. zooepidemicus biomass and competes with other biomass components for carbon and energy (Chong and Nielsen 2003; Liu et al. 2008). Based on the proposed HA biosynthetic pathway (Fig. 1a) and the genome sequence of S. zooepidemicus ATCC39920 (Fig. 1b), 11 genes were identified and their roles in bacterial growth and HA production were investigated. All 11 genes are expressed in the exponential phase (Fig. 1c).
The UDP-GlcUA branch
Apart from HA, the UDP-GlcUA branch produces precursors for wall polysaccharides and teichoic acids (Fig. 1a). As expected, hasA and hasB are both essential for HA biosynthesis (DeAngelis et al. 1993; Dougherty and van de Rijn 1993), while their deletion has a limited impact on growth (Fig. 2a–c). HA is not an essential biomass component, while UDP-GlcUA is not required in S. zooepidemicus for the synthesis of other components. Thus, the apparent loss in biomass yield is most likely attributable to HA loss.
The genomes of HA-producing bacteria S. pyogenes, S. pneumoniae, and S. zooepidemicus contain two genes; one is located on the has operon and the other at a separate chromosomal locus, predicted to encode uridyltransferase. HasC-encoding uridyltransferase was first identified as an essential gene from the has operon required for HA synthesis in S. pyogenes (Crater et al. 1995). In contrast, the has operon gene cap3C-encoding uridyltransferase in S. pneumoniae was dispensable for HA synthesis, while the deletion of galU, the gene encoding the second uridyltransferase, resulted in the abolishment of HA production (Mollerach et al. 1998). HasC1 and hasC2 of S. zooepidemicus are both expressed during culture, and both proteins has a uridyltransferase activity. The has operon-expressed protein, HasC1, is responsible for 68 % of the activity, while the non-operon HasC2 is responsible for the remaining 35 % (Fig. 3c). HasC1 activity is sufficient for maximum growth and HA production, while HasC2 activity alone causes a 40 % reduction in UDP-Glc and a 20 % decrease in HA production (Fig. 2c) without affecting cell growth (Fig. 2a, b).
Phosphoglucomutase, the enzyme that catalyzes the conversion of glucose-l-phosphate to glucose-6-phosphate, is ubiquitous and plays an important role in carbohydrate metabolism (Brody and Tatum 1967). In S. pneumoniae, cps3M, a PGM homologue present in the type 3 capsule locus, is not essential for capsule production, while the inactivation of pgm, a gene located in a distant locus encoding a second PGM homologue, reduced capsule production to less than 10 % of the parental level (Dillard et al. 1995). The S. zooepidemicus genome encodes two distant related phosphoglucomutase genes, pgm1 and pgm2, neither of which are found in the has operon. The deletion of pgm1 resulted in a marginal reduction in phosphoglucomutase activity, while the knockout of pgm2 led to a 90 % decrease in phosphoglucomutase activity in cell lysate (Fig. 4b). This suggests that pgm2 plays a more important role than pgm1 in S. zooepidemicus, which is supported by the phenotypes of Δpgm1 and Δpgm2 mutants. To further elucidate the difference between pgm1 and pgm2, it will be necessary to investigate the expression profile under various culture conditions, especially on different carbon sources. Moreover, it is also worthwhile to examine whether Pgm1 or Pgm2 preferentially use β-glucose-l-phosphate or another sugar-1-P as a substrate instead of α-glucose-1-P (Mesak and Dahl 2000; Seibold and Eikmanns 2013). The knockout of both hasC or both pgm genes obliterated HA production while significantly inhibiting growth. In other firmicutes, lipoteichoic acids are still formed and anchored to the membranes in the absence of UDP-Glc. Evidently, this process—if it occurs in S. zooepidemicus—is not sufficient to avoid a growth defect.
The UDP-GlcNAc branch
UDP-GlcNAc is essential for the biosynthesis of peptidoglycan, an essential cell wall component (Mengin-Lecreulx and van Heijenoort 1993; Olsen et al. 2007). GlmS catalyzes the conversion of fructose-6-P to glucosamine-6-P, which is subsequently converted to glucosamine-1-P by GlmM (Mengin-Lecreulx and van Heijenoort 1996; Watzele and Tanner 1989). The growth of ΔglmM was severely diminished, while ΔglmS could not grow on THY medium without the addition of GlcN. The dual function of HasD and GcaD is responsible for the final two steps in UDP-NAG biosynthesis, and the failure to obtain a ΔhasDΔgcaD double mutant is most likely due to the essential nature of peptidoglycan. Indeed, it is likely that the ΔglmM is rescued by the nontarget activity of other phosphomutases (EC 5.4.2.–) such as pgm.
HasE catalyzes the reversible isomerization of glucose 6-phosphate to fructose 6-phosphate (Fraenkel and Levisohn 1967). The deletion of hasE caused severe growth defects and the loss of HA production in S. zooepidemicus grown on THY medium, in which glucose is the main source of carbon. The fact that hasE is dispensable for the survival of S. zooepidemicus cultured with glucose as the main carbon source suggests that this bacterium can utilize glucose by the pentose phosphate pathway and/or the Entner-Doudoroff pathway (Fraenkel and Levisohn 1967; Froman et al. 1989). The genome of S. zooepidemicus encodes all the enzymes required for either pathway. The growth defect is not merely due to UDP-GlcNAc deficiency. It was not possible to rescue the growth defect of ΔhasE on solid THY plate supplemented with different concentrations (0.2, 0.5, and 1 mg/mL) of GlcN (see Supplementary Figure S3).
In summary, the analysis of the gene-deficient strains revealed that hasA, hasB, hasE, glmM, glmS, and pgm2 are important for HA biosynthesis, while hasC1, hasC2, pgm1, hasD, and gcaD are not essential for the mucoid phenotype. Moreover, hasE, glmM, glmS, and pgm2 are also important for the optimal growth of this bacterium. This report, together with the previous finding about the effects of overexpressing the five has operon genes on HA biosynthesis and growth of S. zooepidemicus (Chen et al. 2009), may guide the construction of a highly efficient S. zooepidemicus strain for HA production.
References
Beres SB, Sesso R, Pinto S, Hoe NP, Porcella SF, DeLeo FR, Musser JM (2008) Genome sequence of a Lancefield group C Streptococcus zooepidemicus strain causing epidemic nephritis: new information about an old disease. PLoS One 3(8):e3026
Blank LM, Hugenholtz P, Nielsen LK (2008) Evolution of the hyaluronic acid synthesis (has) operon in Streptococcus zooepidemicus and other pathogenic streptococci. J Mol Evol 67(1):13–22
Brody S, Tatum E (1967) Phosphoglucomutase mutants and morphological changes in Neurospora crassa. Proc Natl Acad Sci 58(3):923
Chen WY, Marcellin E, Hung J, Nielsen LK (2009) Hyaluronan molecular weight is controlled by UDP-N-acetylglucosamine concentration in Streptococcus zooepidemicus. J Biol Chem 284(27):18007–18014
Chen WY, Marcellin E, Steen JA, Nielsen LK (2014) The role of hyaluronic acid precursor concentrations in molecular weight control in Streptococcus zooepidemicus. Mol Biotechnol 56(2):147–156
Chong BF, Nielsen LK (2003) Aerobic cultivation of Streptococcus zooepidemicus and the role of NADH oxidase. Biochem Eng J 16(2):153–162
Crater DL, Dougherty BA, van de Rijn I (1995) Molecular characterization of hasC from an operon required for hyaluronic acid synthesis in group A streptococci: demonstration of UDP-glucose pyrophosphorylase activity. J Biol Chem 270(48):28676–28680
Daran JM, Dallies N, Thines‐Sempoux D, Paquet V, François J (1995) Genetic and biochemical characterization of the UGP1 gene encoding the UDP‐glucose pyrophosphorylase from Saccharomyces cerevisiae. Eur J Biochem 233(2):520–530
DeAngelis PL, Papaconstantinou J, Weigel PH (1993) Molecular cloning, identification, and sequence of the hyaluronan synthase gene from group A Streptococcus pyogenes. J Biol Chem 268(26):19181–19184
Dillard JP, Vandersea MW, Yother J (1995) Characterization of the cassette containing genes for type 3 capsular polysaccharide biosynthesis in Streptococcus pneumoniae. J Exp Med 181(3):973–983
Dougherty BA, van de Rijn I (1993) Molecular characterization of hasB from an operon required for hyaluronic acid synthesis in group A streptococci: demonstration of UDP-glucose dehydrogenase activity. J Biol Chem 268(10):7118–7124
Fraenkel D, Levisohn S (1967) Glucose and gluconate metabolism in an Escherichia coli mutant lacking phosphoglucose isomerase. J Bacteriol 93(5):1571–1578
Froman BE, Tait RC, Gottlieb L (1989) Isolation and characterization of the phosphoglucose isomerase gene from Escherichia coli. Mol Gen Genet 217(1):126–131
Izawa N, Serata M, Sone T, Omasa T, Ohtake H (2011) Hyaluronic acid production by recombinant Streptococcus thermophilus. J Biosci Bioeng 111(6):665–670
Kleczkowski LA, Geisler M, Ciereszko I, Johansson H (2004) UDP-glucose pyrophosphorylase: an old protein with new tricks. Plant Physiol 134(3):912–918
Liu L, Wang M, Du G, Chen J (2008) Enhanced hyaluronic acid production of Streptococcus zooepidemicus by an intermittent alkaline-stress strategy. Lett Appl Microbiol 46(3):383–388
Liu L, Liu Y, Li J, Du G, Chen J (2011) Microbial production of hyaluronic acid: current state, challenges, and perspectives. Microb Cell Factories 10:99. doi:10.1186/1475-2859-10-99
Lu M, Kleckner N (1994) Molecular cloning and characterization of the pgm gene encoding phosphoglucomutase of Escherichia coli. J Bacteriol 176(18):5847–5851
Mengin-Lecreulx D, van Heijenoort J (1993) Identification of the glmU gene encoding N-acetylglucosamine-1-phosphate uridyltransferase in Escherichia coli. J Bacteriol 175(19):6150–6157
Mengin-Lecreulx D, van Heijenoort J (1996) Characterization of the essential gene glmM encoding phosphoglucosamine mutase in Escherichia coli. J Biol Chem 271(1):32–39
Mesak LR, Dahl MK (2000) Purification and enzymatic characterization of PgcM: a β-phosphoglucomutase and glucose-1-phosphate phosphodismutase of Bacillus subtilis. Arch Microbiol 174(4):256–264
Mollerach M, López R, García E (1998) Characterization of the galU gene of Streptococcus pneumoniae encoding a uridine diphosphoglucose pyrophosphorylase: a gene essential for capsular polysaccharide biosynthesis. J Exp Med 188(11):2047–2056
Olsen LR, Vetting MW, Roderick SL (2007) Structure of the E. coli bifunctional GlmU acetyltransferase active site with substrates and products. Protein Sci 16(6):1230–1235
Prasad SB, Ramachandran KB, Jayaraman G (2012) Transcription analysis of hyaluronan biosynthesis genes in Streptococcus zooepidemicus and metabolically engineered Lactococcus lactis. Appl Microbiol Biotechnol 94(6):1593–1607
Seibold GM, Eikmanns BJ (2013) Inactivation of the phosphoglucomutase gene pgm in Corynebacterium glutamicum affects cell shape and glycogen metabolism. Biosci Rep 33(4):645–654
Sun X, Yang D, Wang Y, Geng H, He X, Liu H (2013) Development of a markerless gene deletion system for Streptococcus zooepidemicus: functional characterization of hyaluronan synthase gene. Appl Microbiol Biotechnol 97(19):8629–8636
Watzele G, Tanner W (1989) Cloning of the glutamine: fructose-6-phosphate amidotransferase gene from yeast; pheromonal regulation of its transcription. J Biol Chem 264(15):8753–8758
Wessels MR, Moses AE, Goldberg JB, DiCesare TJ (1991) Hyaluronic acid capsule is a virulence factor for mucoid group A streptococci. Proc Natl Acad Sci 88(19):8317–8321
Acknowledgments
This work was supported by the research foundation of Tianjin Science and Technology Commission (13RCGFSY19400) and the Tianjin Municipal High School Science and Technology Development Fund Program (20130602).
Authors’ contributions
L.H. designed the research; L.H., Z.Y., L.K., and Z.Q. performed the research; L.H., Q.Z., and N.L. analyzed the data; and L.H., Q.Z., and N.L. wrote the paper.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interest
The authors declare that they have no competing interests.
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 2037 kb)
Rights and permissions
About this article
Cite this article
Zhang, Y., Luo, K., Zhao, Q. et al. Genetic and biochemical characterization of genes involved in hyaluronic acid synthesis in Streptococcus zooepidemicus . Appl Microbiol Biotechnol 100, 3611–3620 (2016). https://doi.org/10.1007/s00253-016-7286-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-016-7286-1