Abstract
The microbial community compositions of a chemostat enriched in a thermophilic (55 °C) mixed culture fermentation (MCF) for hydrogen production under different operational conditions were revealed in this work by integrating denaturing gradient gel electrophoresis (DGGE), Illumina Miseq high-throughput sequencing, and 16S rRNA clone library sequencing. The results showed that the community structure of the enriched cultures was relatively simple. Clones close to the genera of Thermoanaerobacter and/or Bacillus mainly dominated the bacteria. And homoacetogens and archaea were washed out and not detected even by Illumina Miseq high-throughput sequencing which supported the benefit for hydrogen production. On the other hand, the results revealed that the metabolic shift was clearly associated with the change of dominated bacterial groups. The effects of hydrogen partial pressure (PH2) and pH from 4.0 to 5.5 on the microbial compositions were not notable and Thermoanaerobacter was dominant, thus, the metabolites were also not changed. While Bacillus, Thermoanaerobacter and Propionispora hippei dominated the bacteria communities at neutral pH, or Bacillus and Thermoanaerobacter dominated at high influent glucose concentrations, consequently the main metabolites shifted to acetate, ethanol, propionate, or lactate. Thereby, the effect of microbial composition on the metabolite distribution and shift shall be considered when modeling thermophilic MCF in the future.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
To reduce the dependency on petroleum fuels and lowering the environmental pollution, there is a growing trend in developing clean and biobased fuel processes (Angenent et al. 2004; Kleerebezem and van Loosdrecht 2007). Mixed culture fermentation (MCF), as a worthwhile and well-studied technology, has the merits of having no sterilization requirements, an adaptive capacity to variations in feedstock or conditions, a stable and continuous operation, and so on (Kleerebezem and van Loosdrecht 2007; Liguori et al. 2013; St-Pierre and Wright 2014; Zhang et al. 2014). Consequently, this technology is ubiquitously implemented for converting the degradable organic wastes into bioproducts such as biohydrogen, acetate, ethanol, and even medium chain fatty acids (Kleerebezem and van Loosdrecht 2007; Liguori et al. 2013; Pawar and Niel 2013; Zhang et al. 2014; Zhang et al. 2013a).
Hydrogen production by thermophilic MCF has demonstrated several benefits such as a higher hydrogen yield due to the favorable thermodynamic conditions, high substrate degradation rate and efficient heat utilization for treatment of hot wastewater (Gannoun et al. 2007; Labatut et al. 2014; Pawar and Niel 2013; Zhang et al. 2012a). The metabolites of thermophilic MCF are always a mixture of acetate, butyrate, ethanol, etc., and their distributions are commonly related to the operational conditions such as pH and hydrogen partial pressure (PH2); however, the effect of the microbial community is commonly ignored. For example, Ueno et al. (2006) found a change of butyrate and acetate to acetate and ethanol as pH increased from 5.0 to 8.0. Akutsu et al. (2009) reported that the hydrogen yield increased from 2.3 to 2.8 mol/mol-glucose as the influent concentration of starch increased from 15 to 20 g/L, and then decreased to 2.1 mol/mol-glucose as the starch concentration increased to 70 g/L.
Several recent studies on bacterial communities show that changes in reactor operations are often associated with changes in the composition of bacterial communities (Kröber et al. 2009; O-Thong et al. 2008; Temudo et al. 2008). For example, Temudo et al. (2008) demonstrated the relationship between microbial communities and metabolite distributions in a mesophilic chemostat under different pH and carbon sources by denaturing gradient gel electrophoresis (DGGE) and 16S rRNA clone library. Their results showed that cluster I of the genus Clostridium dominated the microorganism community at acidic pH while at middle pH values, the principal microorganism belonged to the genus Klebsiella. Therefore, the operational variables such as pH have a clear impact on the population established and consequently on the fermentation pattern (Temudo et al. 2008).
Investigating the dominated microbes is necessary to analyze and explain the previously mentioned results and reveal the relationship between the metabolite distribution and microorganism composition in thermophilic MCF. DGGE and 16S rRNA gene clone library sequencing are the typical traditional molecular methods that could identify the major functional microbes (O-Thong et al. 2008; Temudo et al. 2008; Zhang et al. 2012b). For example, Ueno and Ishii (2001) found the hydrogen production yield was around 2.0 mol/mol-hexose in either the thermophilic batch or chemostat cultivation, and most of the isolates belonged to the cluster of the thermophilic Clostridium/Bacillus subphylum of low G + C gram-positive bacteria. O-Thong et al. (2008) recently isolated and identified a thermophilic bacterium, Thermoanaerobacterium thermosaccharolyticum, from a biohydrogen reactor fed with palm oil mill effluent. Except hydrogen producing bacteria in MCF, hydrogenotrophic methanogensis and homoacetogenesis are two main pathways to consume hydrogen, while their percentages are still an open question and needs to be revealed. For example, Siriwongrungson et al. (2007) reported homoacetogenesis in a thermophilic continuously stirred tank reactor (CSTR) with hydraulic retention time (HRT) of 6 days for butyrate degradation, but the functional bacteria were not identified.
Recently, the high-throughput sequencing analysis of bioreactors or environmental samples including 454-pyrosequencing and/or Illumina Miseq high-throughput sequencing can reveal the minority populations that are hard to be explored by traditional molecular methods (Zhang et al. 2012b). But, the 16S rRNA gene length obtained by those high-throughput sequencing technologies were relatively short and around 400 bp; consequently, the phylogenetic characterization could not be assigned to taxonomic affiliations at the species level (Kröber et al. 2009; Zhang et al. 2012b). The 16S rRNA clone library sequencing could offset this drawback. Therefore, an integrated approach that combined high-throughput sequencing reads describing the diversity of the community and clone library sequencing enabling high resolution phylogenetic analysis of abundant taxonomic units shall be an option to reveal the microbial function in thermophilic MCF and may provide deep explanation to the relationship between metabolite shift and microbial composition.
Among the reactor configurations, the chemostat is essential to explain the metabolic distributions and microbial communities (Temudo et al. 2008). However, till now, only few researches have reported the chemostat results in thermophilic MCF. Recently, our group investigated the effect of pH, PH2, and influent glucose concentration on the metabolic distribution in thermophilic chemostat (Zhang et al. 2015). The results showed that the metabolite composition was changed from acetate, butyrate and hydrogen to acetate, ethanol and propionate at neutral pH. Reducing PH2 did not significantly increase the hydrogen yield and consequently other metabolites also did not change much. The maximum hydrogen yield was 2.2 mol/mol-glucose at PH2 0.05 atm. Lactate accumulation was observed only at high influent glucose concentrations. Meanwhile, the results also revealed that the impacts of precedent experimental concentrations on the metabolite distribution were negligible.
Thereby, the aim of the research described in this paper was to investigate the community profiles as a function of pH, glucose concentration and PH2 in a chemostat of thermophilic MCF by integrating DGGE, Illumina Miseq high-throughput sequencing and 16S rRNA clone library sequencing. The outcomes could improve the understanding of the metabolic shift in thermophilic MCF.
Materials and methods
The chemostat of thermophilic MCF for hydrogen production
The detailed information of chemostat was demonstrated in our former work (Zhang et al. 2015), and the operation was briefly described as follows: the anaerobic sludge was collected from a thermophilic (55 °C) CSTR; the volume was 2.0 L, and the working volume was 1.35 L; the feeding glucose concentration was 5 g/L during the enrichment period; the temperature was maintained at 55 ± 0.5 °C with a water bath; the pH was maintained automatically with 2 M NaOH; the stirring velocity was 300 rpm; and the reactor was shaken vigorously every 2 weeks to detach the biofilm on the reactor walls. Through the following experiments, HRT was controlled at 0.7 days. Different pH, PH2, and influent glucose concentrations were conducted and at least five HRTs were operated for each condition.
DNA extraction
The microbial samples were collected from the effluent of chemostat under the conditions of PH2, 0.05 atm; pH of 4.0, 4.5, 5.0, 5.5, 6.0, 6.5, and 7.0; and influent glucose concentration, 50.9, 76.3 and 108.5 mmol/L, and were sequentially named from PH2-L1 to glu-L11, as shown in Table 1. The collected samples were washed with the phosphate-buffered saline solution (PBS solution). The composition of PBS solution (in 1.0 L distilled water) was: NaCl, 8 g; KCl, 0.2 g; Na2HPO4, 1.44 g; and KH2PO4, 0.24 g, the pH was 7.4. Four milliliters of the cultures were collected from the reactor and centrifuged to extract DNA using the PowerSoil DNA isolation kit (MO BIO, USA). The integrality of the DNA was checked with gel electrophoresis in a 1 % agarose gel. The DNA concentration was determined with a spectrophotometer NanoDrop 2000.
Denaturing gradient gel electrophoresis
The amplification of 16S rRNA gene fragments for DGGE was performed by the C1000 thermal Cycler (Bio-Rad, Hercules, USA) using the following program: an initial denaturation at 94 °C for 5 min, followed by 30 cycles of denaturation at 92 °C for 30 s, annealing at 55 °C for 30 s, and extension at 72 °C for 30 s, then the final extension at 72 °C for 15 min. The resulting PCR products were analyzed for their molecular weight and yield on a 1 % agarose gel using ethidium bromide staining.
Denaturing gradient gel electrophoresis (DGGE) was then performed on 8 % polyacrylamide gel using the D-Code system (Bio-Rad Laboratories, Hercules, CA) with a denaturant gradient ranging from 40–70 % (100 % denaturant is defined as 7 M urea and 40 % v/v formamide). Gel was electrophoresed at 60 °C and 75 V for 14 h. Bands were stained for 20 min in 1× TAE containing 1 μg/mL ethidium bromide and photographed using the Gel Doc XR Gel Documentation System (Bio-Rad, Hercules Co., USA).
Illumina Miseq high-throughput sequencing and data analysis
According to the method of Sundberg et al. (2013), DNA for both bacteria and archaea high-throughput sequencing was amplified using the 16S rRNA primers that covers the V3 and V4 regions of the 16S rRNA gene: modified 341 F (CCTAYGGGRBGCASCAG) and modified 806R (GGACTACNNGGGTATCTAAT). The amplification was carried out by ABI GeneAmp® 9700 with the following program: 95 °C for 3 min, then 25 cycles of 95 °C for 30 s, 55 °C for 30 s, and 72 °C for 45 s, followed by a final extension at 72 °C for 10 min (Majorbio Corporation, Shanghai, China).
Sequencing was then carried out on an Illumina Miseq PE 300 high-throughput sequencer according to standard protocols (Majorbio Corporation, Shanghai, China). Paired sequences were joined with software FLASH (Version 1.2.7). The adaptors, barcodes, and primers were trimmed. Sequences either with a low quality or with a length of less than 200 bp were removed. Then, the remaining sequences were denoised and screened for chimera sequences in Mothur program (http://www.mothur.org/wiki/Main_Page). The resulting effective sequences were used for the subsequent information analysis. To analyze bacterial diversity and microbial community structure, the operational taxonomical units (OTUs) were picked at 97 % sequence identity using the Usearch program (Version 7.1, http://derive5.com/uparse/), the representative sequences obtained were phylogenetically allocated down to the domain, phylum, class, family, and genus level. The relative abundance of a given phylogenetic group was calculated by dividing the number of sequences affiliated to that group with the total number of obtained sequences. The OTU-based analyses of the alpha diversity indices, including community richness of Chao1 and abundance-based coverage estimator (ACE), community diversity of Shannon Index and Simpson, and Coverage, were performed in the Mothur program. To define the relative abundance of a given phylogenetic group, the number of sequences affiliated to that group (with 97 % identity) was divided by the total number of obtained sequences. The sequencing data of the five samples (PH2-L1, pH-L2, pH-L5, pH-L8, and glu-L11) were deposited in the NCBI Sequence Read Archive under the accession numbers SRS1071072, SRS1067180, SRS1067301, SRS1071070, and SRS1071071, respectively.
16S rRNA clone library sequencing and phylogenetic analysis
DNA for 16S rRNA clone library sequencing was amplified using the universal bacteria 16S rRNA primers: 27 F (AGAGTTTGATCCTGGCTCAG) and 1492R (GGTTACCTTGTTACGACTT) (Kröber et al. 2009; Zhang et al. 2013a). The Amplification of bacteria 16S rRNA gene was carried out by ABI GeneAmp® 9700 with the following program: 94 °C for 5 min, then 30 cycles of 94 °C for 30 s, 54 °C for 30 s, and 72 °C for 30 s, followed by a final extension at 72 °C for 10 min (Majorbio Corporation, Shanghai, China).
The method of clone library construction and sequencing was according to that of Thomsen et al. (2001), which was briefly described as follows: the amplicons were visualized on 1.5 % agarose gels, the bands were cut out and then recovered directly with the gel DNA recovery kit (Majorbio Corporation, Shanghai, China). The recovered products were ligated into a PMD18-T vector (TAKARA, Dalian, China) and then transformed into Escherichia coli DH5a competent cells according to the manufacturer’s directions. The white clones were obtained by the blue-white screening and were checked by the PCR methods using the M13F (CGCCAGGGTTTTCCCAGTC ACGAC) and M13R (AGCGGATAACAATTTCACACAGGA) primers. The positive clones were selected and sequenced by the ABI PRISMTM 3730XL DNA Analyzer (Majorbio Corporation, Shanghai, China).
The previously mentioned obtained 16S rRNA gene sequences were compared with sequences in the GenBank database using the NCBI Blast search program (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Closest cultured and uncultured relatives were retrieved from the database. A neighbor-joining tree was made based on the 16S rRNA gene sequences determined in this study and related reference sequences. Alignment and phylogenetic analysis were performed with the MEGA 4.1 (Beta) software. Sequences retrieved in this study were accessible under the accession numbers: KT185026-KT185052.
Results
The DGGE gel profiles of microorganisms in a thermophilic chemostat
The effects of PH2, pH, and influent glucose concentration on the enriched microorganisms in thermophilic chemostat can be reflected by their DGGE gel profiles. As shown in Fig. 1, the DGGE gel demonstrated that the bands of 16S rRNA gene fragment were relatively low, and the dominated bands were not changed much, while the minor bands emerged at a neutral pH and disappeared at high influent glucose concentrations. In our former work (Zhang et al. 2015), the metabolic distributions were systemic determined with good COD balances and results reproducibility that the metabolite composition mainly were acetate, butyrate, and hydrogen and were not changed at acidic pH (4.0-6.0, corresponding to DNA samples from pH-L2 to pH-L6), or at constant pH 5.5 when reducing PH2 from 0.62 (pH-L5) to 0.05 atm (PH2-L1) or increasing influent glucose concentration from 27.5 mmol/L (pH-L5) to 76.3 mmol/L (glu-L10). But, those metabolites were changed to acetate, ethanol, propionate and format when pH increased from 6.0 (pH-L6) to 7.0 (pH-L8), and lactate accumulation was observed at a higher influent glucose concentration of 108.5 mmol/L (glu-L11). Therefore, after combing DGGE gel and previous experimental results, the samples of PH2-L1, pH-L2, pH-L5, pH-L8, and glu-L11 were selectively analyzed furthermore by the 16S-rRNA-gene-based Illumina high-throughput sequencing technology.
DGGE analysis of the 16S rRNA gene fragment from microbial communities in a thermophilc chemostat. The sampling conditions in lanes from left to right were PH2 of 0.05 atm (PH2-L1); pH of 4.0 (pH-L2), 4.5 (pH-L3), 5.0 (pH-L4), 5.5 (pH-L5), 6.0 (pH-L6), 6.5 (pH-L7), 7.0 (pH-L8); influent glucose concentration of 50.9 mmol/L (glu-L9), 76.3 mmol/L (glu-L10) and 108.5 (glu-L11) mmol/L, respectively
The Illumina high-throughput sequencing of chemostat microorganisms
To reveal the microbial communities shifting and diversities, the samples, including PH2-L1, pH-L2, pH-L5, pH-L8, and glu-L11, were chosen for the Illumina high-throughput sequencing, their sequencing indices are summarized in Table 2. Over 32,000 effective sequences were obtained for each sample and the maximum value was an even amount of 43,000, and the mean lengths were above 422 bp. Rather high coverages were achieved in this study and the values were all above 0.9999 for each sample. The OTU numbers of each sample were all below 15, which meant that simple microbial communities were enriched in the thermophilic chemostat.
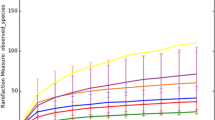
The curves of Shannon index in Fig. 2a showed that when the sequence number of each sample was greater than 2000, Shannon diversity for the isolated microorganism did not increase clearly, indicating that the sequencing depth in this work was sufficient to analyze the archaea and bacteria communities. The bacterial richness levels were determined using Rarefaction analysis in Fig. 2b, which also indicated the highest bacterial richness in reactors. Meanwhile, the microbial diversity indexes of ACE, Chao1, Shannon, and Simpson also indicated that relative simple populations were enriched (Table 2). As shown in Table 2, especially for the sample of PH2-L1, the Shannon index was 0.27 and Simpson index reached to 0.8788.
The microbial community structures by the Illumina high-throughput sequencing at the domain (A), phylum (B), class (C), family (D), and genus (E) levels are summarized in Fig. 3. It was interesting that the percentages of archaea were 0 % at the domain level, which meant that methanogens were completely washed out in the thermophilic chemostat and consequently, this result shall support the benefit of hydrogen production in thermophilic MCF. The Firmicutes dominated the bacteria at the phylum level and the percentages of PH2-L1, pH-L2, pH-L5, and glu-L11 were all above 99.9 %, while the percentage of pH-L8 was 95.0 %, as shown in Fig. 3b. The percentages of another phylum, Proteobacteria, were rather low and even approached 0.03 % in pH-L2.
In Fig. 3c, the sequencing results showed that Clostridia was the main Firmicutes at the class level and the percentages of L1, pH-L2, and pH-L5 were 99.5, 99.9, and 99.8 %, respectively, which was due to its high-resistance nature especially in acidic conditions (Tracy et al. 2012; Zhang et al. 2013b). At a neutral pH (pH-L8) and high glucose concentration (glu-L11), the bacteria diversities also increased as shown in both Figs. 2 and 3c, and Clostridia percentages decreased to 41.5 and 69.4 %, respectively. Bacilli were the second dominated bacteria, and the percentages were 37.5 and 26.9 %, respectively. Other bacteria at the class level including Betaproteobacteria and Negativicutes were also established in sample L8, and the percentage was 16.7 and 16.0 %, respectively.
The results in Fig. 3d and e showed that the dominated bacteria of samples PH2-L1, pH-L2, and pH-L5 were close to the Thermoanaerobacterales Family III at the family level and Thermoanaerobacterium at the genus level, and the percentages were all above 99.5 %. While their percentages of sample pH-L8 and glu-L11 reduced to 35.2 and 69.4 % at the genus levels. Other main bacteria such as Bacillaceae (37.5 and 26.7 %) and Veillonellaceae (16.0 and 3.6 %) were enriched at the family level and Bacillus (37.5 and 26.7 %) and Propionispora (16.0 and 3.6 %) were enriched at the genus level in samples of L8 and L11. The percentages of other minor bacteria such as Leclercia were rather low and below 0.02 %, except for L8 (4.0 %).
16 s rRNA clone library sequencing of chemostat microorganisms
To offset the disadvantage of low sequencing lengths for the Illumina high-throughput sequencing, the typical samples of pH-L2 and pH-L8 were further analyzed in the species level by 16 s rRNA clone library sequencing and the results are shown in Fig. 4, and clone percentages in Tables 3 and 4. Similar to the genus level results (above 99.5 % of Thermoanaerobacterium) of pH-L2 in Fig. 3e, the percentage of clones at the species level related to Thermoanaerobacterium thermosaccharolyticum with similarity of 99 % was up to 100 % and other bacteria were not detected in a total of 30 clones as shown in Table 3.
Bacterial neighbor-joining trees of microorganisms (A, pH-L2 and B, pH-L8) in a thermophilic chemostat for hydrogen production. The scale bars in a and b indicate a 5 % sequence divergence. GenBank accession numbers were given in parentheses. The accession numbers of bacteria (KT185026-KT185052) were the nucleotide sequences in this work
At a neutral pH, microbial communities changed notably and several sub-groups of microorganisms including Bacillus sp., Thermoanaerobacter sp., Propionispora sp., and Leclercia sp. were enriched in pH-L8 as shown in Fig. 4b and Table 4. The percentage of clones related to Bacillus coagulans with similarity above 99 % was 50.0 % in total 30 clones. The second main bacterium was T. thermosaccharolyticum and the percentage was 20.0 %. Other bacteria were Propionispora hippie, Leclercia adecarboxylata, Acinetobacter bereziniae and Clostridium saccharobutylicum, and their percentages were 13.3, 10.0, 3.3 and 3.3 %, respectively.
These results were also consistent with those of the Illumina high-throughput sequencing in Fig. 3. For example, Bacillus coagulans, T. thermosaccharolyticum, P. hippie and C. saccharobutylicum belonged to Firmicutes at the phylum level and the percentage was 86.7 %, while L. adecarboxylata and A. bereziniae belonged to Proteobacteria at the phylum level and the percentage was 13.3 %. At the class level, B. coagulans belonged to Bacilli, T. thermosaccharolyticum, and C. saccharobutylicum both belonged to Clostridia, P. hippie belonged to Negativicutes, L. adecarboxylata and A. bereziniae belonged to Gammaproteobacteria.
Discussion
The relationship between the functional bacteria and metabolites
Specific bacterial composition in MCF should be one of important factors to determine the hydrogen yield and other metabolite distribution. According to the results of Illumina high-throughput sequencing and 16s rRNA clone library sequencing, bacteria close to T. thermosaccharolyticum were the main functional bacteria. T. thermosaccharolyticum is a functional thermophilic (45–70 °C) bacterium, and can produce hydrogen within a wide range of pH (4–8). The main metabolites of T. thermosaccharolyticum were acetate, butyrate, hydrogen, and/or ethanol from organic wastes and its maximum hydrogen yield was 2.53 mol/mol-hexose (O-Thong et al. 2008; Ren et al. 2008). Those results were consistent with our former work that the metabolites of samples pH-L2 and pH-L5 were a mixture of acetate (0.55 and 0.58 mol/mol-glucose), butyrate (0.60 and 0.55 mol/mol-glucose), and hydrogen (1.94 and 1.92 mol/mol-glucose) at a pH of 4.0 and 5.5, respectively (Zhang et al. 2015). And the metabolite yields changed as PH2 reduced from 0.62 (pH-L5) to 0.05 (PH2-L1) atm, were 0.40 mol/mol-glucose of acetate, 0.54 mol/mol-glucose of butyrate and 2.18 mol/mol-glucose of hydrogen at a pH of 5.5 (Zhang et al. 2015).
At a neutral pH, more functional bacteria were enriched. For example, B. coagulans is a functional thermophilic bacterium (50–55 °C) and can ferment various biomass-derived sugars to mainly produce lactate, acetate and ethanol (Patel et al. 2006; Su and Xu 2014). P. hippie is a gram-negative, spore-forming anaerobe and can ferment sugars to propionic and acetic acids (Abou-Zeid et al. 2004). L. adecarboxylata, as a humic pathogen, produces both hydrogen and acids from both sugars and polycyclic aromatic hydrocarbons (Sarma et al. 2004; Tamura et al. 1986). Thereby, it is reasonable that the metabolites of the sample pH-L8 at pH 7.0 is a mixture of acetate (0.94 mol/mol-glucose), propionate (0.26 mol/mol-glucose), and ethanol (0.68 mol/mol-glucose) (Zhang et al. 2015).
Meanwhile, after combining the sequencing results in Figs. 3e and 4, it was revealed that a mixture of Bacillius and Thermoanaerobacter dominated bacteria-glu-L11 at high influent glucose concentration (108.5 mmol-glucose/L), which consequently resulted in a lactate accumulation (0.81 mol/mol-glucose) and the low hydrogen yield (1.25 mol/mol-glucose).
The metabolic pathway in thermophilic bacteria
Normally, Clostridium and Ethanoligenens species, typical bacteria in mesophilic MCF, only have hydrogen yields lower than 2 mol/mol-glucose (Lee et al. 2009). While some thermophilic bacterium Caldicellulosiruptor saccharolyticus and Thermotoga neapolitana produce a higher hydrogen yield of above 3.2 mol/mol-glucose (de Vrije et al. 2007; Nguyen et al. 2010). Karakashev et al. (2009) reported that the genera of Bacillus and Clostridium dominated the thermophilic (70 °C) MCF enriched from household solid wastes and the hydrogen yield was just 1.9 mol/mol-glucose. In the present work, since a mixed culture of Thermoanaerobacter and/or Bacillus was isolated, the maximum hydrogen yield was around 2.0 mol/mol-glucose, which was also similar to former works of Ueno and Ishii (2001).
Likely the yield discrepancy is due to the specific hydrogen production pathways used by different bacteria. NAD+/NADH (E0, −320 mV) and Fdox/Fdred (E0, −420 mV) are two electron carriers to produce hydrogen (E0 of H+/H2, −420 mV) and conserve the intracellular redox balance (Bar-Even et al. 2012; de Kok et al. 2013; Zhang et al. 2013b). For example, under a rather low hydrogen pressure such as 60 Pa in mesophilic conditions, the production of H2 from NADH is thermodynamically favorable (Angenent et al. 2004; Kleerebezem and van Loosdrecht 2007; Zhang et al. 2013b). Consequently, Fdred is considered to be the sole electron donor for hydrogen production at normal hydrogen-producing reactors, and the produced NADH is consumed to produce other metabolites such as lactate, ethanol, propionate, etc. (Angenent et al. 2004; Kleerebezem and van Loosdrecht 2007; Zhang et al. 2015; Zhang et al. 2013b). Recently, Schut and Adams (2009) found a bifurcating [FeFe] hydrogenase in Thermotoga maritima which could synergistically utilize Fdred and NADH in an approximately 1:1 ratio to produce hydrogen, resulting in higher hydrogen yields close to 4 mol/mol-glucose.
On the other hand, hydrogenotrophic methanogensis and homoacetogenesis are the two main pathways to consume hydrogen in MCF. Though using the 16S rRNA based Illumina Miseq high-throughput sequencing technology, in this work, neither archaea (such as Methanothermobacter thermautotrophicus (Martin et al. 2013)) nor homoacetogens (such as Clostridium thermoaceticum (Pierce et al. 2008)) were detected. However, Siriwongrungson et al. (2007) reported homoacetogenesis in a thermophilic CSTR with HRT of 6 days. Therefore, keeping a low HRT shall be a benefit for hydrogen production.
Finally, constructing a mathematic model of MCF is a common but valuable way to simulate and optimize the fermentation systems, in which the microbial communities shift under different operational conditions is generally ignored (Rodriguez et al. 2006; Zhang et al. 2013b). However, as shown in our work, the functional bacteria communities are indeed changed with the operational conditions including pH and substrate concentration, and consequently affect the metabolite distributions. Thereby, an improved model considering both the operational conditions and the microbial communities is crucial for the operation and control of MCF in the future.
In summary, by integrating DGGE, Illumina Miseq high-throughput sequencing, and 16S rRNA clone library sequencing, the microbial compositions of MCF in a thermophilic chemostat were demonstrated in this work. It was found that relatively simple microbial communities were enriched. Clones close to the genera of Thermoanaerobacter and/or Bacillus dominated the bacteria; however, homoacetogens and archaea were washed out and not detected. The results also revealed that the metabolic shift was clearly associated with the change of dominated bacterial groups. The effects of PH2 and pH from 4.0 to 5.5 on the microbial compositions were not notable and Thermoanaerobacter was dominant. While Bacillus, Thermoanaerobacter, and P. hippei dominated the bacteria communities at neutral pH, Bacillus and Thermoanaerobacter dominated at high influent glucose concentrations.
References
Abou-Zeid DM, Biebl H, Spröer C, Müller R-J (2004) Propionispora hippei sp. nov., a novel gram-negative, spore-forming anaerobe that produces propionic acid. Int J Syst Bacteriol Evol Microbiol 54(3):951–954
Akutsu Y, Li Y-Y, Harada H, Yu H-Q (2009) Effects of temperature and substrate concentration on biological hydrogen production from starch. Int J Hydrog Energy 34(6):2558–2566
Angenent LT, Karim K, Al-Dahhan MH, Wrenn BA, Domíguez-Espinosa R (2004) Production of bioenergy and biochemicals from industrial and agricultural wastewater. Trends Biotechnol 22(9):477–485
Bar-Even A, Flamholz A, Noor E, Milo R (2012) Thermodynamic constraints shape the structure of carbon fixation pathways. Biochim Biophys Acta-Bioenergetics 1817(9):1646–1659
de Kok S, Meijer J, van Loosdrecht MCM, Kleerebezem R (2013) Impact of dissolved hydrogen partial pressure on mixed culture fermentations. Appl Microbiol Biotechnol 97(6):2617–2625
de Vrije T, Mars A, Budde M, Lai M, Dijkema C, de Waard P, Claassen P (2007) Glycolytic pathway and hydrogen yield studies of the extreme thermophile Caldicellulosiruptor saccharolyticus. Appl Microbiol Biotechnol 74(6):1358–1367
Gannoun H, Othman NB, Bouallagui H, Moktar H (2007) Mesophilic and thermophilic anaerobic co-digestion of olive mill wastewaters and abattoir wastewaters in an upflow anaerobic filter. Ind Eng Chem Res 46(21):6737–6743
Karakashev D, Kotay SM, Trably E, Angelidaki I (2009) A strict anaerobic extreme thermophilic hydrogen-producing culture enriched from digested household waste. J Appl Microbiol 106(3):1041–1049
Kleerebezem R, van Loosdrecht MCM (2007) Mixed culture biotechnology for bioenergy production. Curr Opin Biotechnol 18(3):207–212
Kröber M, Bekel T, Diaz NN, Goesmann A, Jaenicke S, Krause L, Miller D, Runte KJ, Viehöver P, Pühler A, Schlüter A (2009) Phylogenetic characterization of a biogas plant microbial community integrating clone library 16S-rDNA sequences and metagenome sequence data obtained by 454-pyrosequencing. J Biotechnol 142(1):38–49
Labatut RA, Angenent LT, Scott NR (2014) Conventional mesophilic vs. thermophilic anaerobic digestion. A trade-off between performance and stability? Water Res 53:249–258
Lee HS, Krajmalinik-Brown R, Zhang HS, Rittmann BE (2009) An electron-flow model can predict complex redox reactions in mixed-culture fermentative BioH(2): microbial ecology evidence. Biotechnol Bioeng 104(4):687–697
Liguori R, Amore A, Faraco V (2013) Waste valorization by biotechnological conversion into added value products. Appl Microbiol Biotechnol 97(14):6129–6147
Martin MR, Fornero JJ, Stark R, Mets L, Angenent LT (2013) A single-culture bioprocess of Methanothermobacter thermautotrophicus to upgrade digester biogas by CO2-to-CH4 conversion with H2. Archaea 2013:157529
Nguyen T-AD, Han SJ, Kim JP, Kim MS, Sim SJ (2010) Hydrogen production of the hyperthermophilic eubacterium, Thermotoga neapolitana under N2 sparging condition. Bioresour Technol 101(1 Supplement 1):S38–S41
O-Thong S, Prasertsan P, Karakashev D, Angelidaki I (2008) Thermophilic fermentative hydrogen production by the newly isolated Thermoanaerobacterium thermosaccharolyticum PSU-2. Int J Hydrog Energy 33(4):1204–1214
Patel MA, Ou MS, Harbrucker R, Aldrich HC, Buszko ML, Ingram LO, Shanmugam KT (2006) Isolation and characterization of acid-tolerant, thermophilic bacteria for effective fermentation of biomass-derived sugars to lactic acid. Appl Environ Microbiol 72(5):3228–3235
Pawar S, Niel EJ (2013) Thermophilic biohydrogen production: how far are we? Appl Microbiol Biotechnol 97(18):7999–8009
Pierce E, Xie G, Barabote RD, Saunders E, Han CS, Detter JC, Richardson P, Brettin TS, Das A, Ljungdahl LG, Ragsdale SW (2008) The complete genome sequence of Moorella thermoacetica (f. Clostridium thermoaceticum). Environ Microbiol 10(10):2550–2573
Ren N, Cao G, Wang A, Lee D-J, Guo W, Zhu Y (2008) Dark fermentation of xylose and glucose mix using isolated Thermoanaerobacterium thermosaccharolyticum W16. Int J Hydrog Energy 33(21):6124–6132
Rodriguez J, Kleerebezem R, Lema JM, van Loosdrecht MCM (2006) Modeling product formation in anaerobic mixed culture fermentations. Biotechnol Bioeng 93(3):592–606
Sarma PM, Bhattacharya D, Krishnan S, Lal B (2004) Degradation of polycyclic aromatic hydrocarbons by a newly discovered enteric bacterium, Leclercia adecarboxylata. Appl Environ Microbiol 70(5):3163–6
Schut GJ, Adams MWW (2009) The iron-hydrogenase of Thermotoga maritima utilizes ferredoxin and NADH synergistically: a new perspective on anaerobic hydrogen production. J Bacteriol 191(13):4451–4457
Siriwongrungson V, Zeng RJ, Angelidaki I (2007) Homoacetogenesis as the alternative pathway for H2 sink during thermophilic anaerobic degradation of butyrate under suppressed methanogenesis. Water Res 41(18):4204–4210
St-Pierre B, Wright A-D (2014) Comparative metagenomic analysis of bacterial populations in three full-scale mesophilic anaerobic manure digesters. Appl Microbiol Biotechnol 98(6):2709–2717
Su F, Xu P (2014) Genomic analysis of thermophilic Bacillus coagulans strains: efficient producers for platform bio-chemicals. Sci Rep 4:3926
Sundberg C, Al-Soud WA, Larsson M, Alm E, Yekta SS, Svensson BH, Sørensen SJ, Karlsson A (2013) 454 pyrosequencing analyses of bacterial and archaeal richness in 21 full-scale biogas digesters. FEMS Microbiol Ecol 85(3):612–626
Tamura K, Sakazaki R, Kosako Y, Yoshizaki E (1986) Leclercia adecarboxylata Gen. Nov., Comb. Nov., formerly known as Escherichia adecarboxylata. Curr Microbiol 13(4):179–184
Temudo MF, Muyzer G, Kleerebezem R, van Loosdrecht MCM (2008) Diversity of microbial communities in open mixed culture fermentations: impact of the pH and carbon source. Appl Microbiol Biotechnol 80(6):1121–1130
Thomsen TR, Finster K, Ramsing NB (2001) Biogeochemical and molecular signatures of anaerobic methane oxidation in a marine sediment. Appl Environ Microbiol 67(4):1646–1656
Tracy BP, Jones SW, Fast AG, Indurthi DC, Papoutsakis ET (2012) Clostridia: the importance of their exceptional substrate and metabolite diversity for biofuel and biorefinery applications. Curr Opin Biotechnol 23(3):364–381
Ueno H, Ishii I (2001) Microbial community in anaerobic hydrogen-producing microflora enriched from sludge compost. Appl Microbiol Biotechnol 57(4):555–562
Ueno Y, Sasaki D, Fukui H, Haruta S, Ishii M, Igarashi Y (2006) Changes in bacterial community during fermentative hydrogen and acid production from organic waste by thermophilic anaerobic microflora. J Appl Microbiol 101(2):331–343
Zhang F, Zhang Y, Chen M, Zeng RJ (2012a) Hydrogen supersaturation in thermophilic mixed culture fermentation. Int J Hydrogen Energy 37(23):17809–17816
Zhang T, Shao M-F, Ye L (2012b) 454 Pyrosequencing reveals bacterial diversity of activated sludge from 14 sewage treatment plants. ISME J 6(6):1137–1147
Zhang F, Ding J, Zhang Y, Chen M, Ding Z-W, van Loosdrecht MCM, Zeng RJ (2013a) Fatty acids production from hydrogen and carbon dioxide by mixed culture in the membrane biofilm reactor. Water Res 47(16):6122–6129
Zhang F, Zhang Y, Chen M, van Loosdrecht MCM, Zeng RJ (2013b) A modified metabolic model for mixed culture fermentation with energy conserving electron bifurcation reaction and metabolite transport energy. Biotechnol Bioeng 110(7):1884–1894
Zhang F, Chen Y, Dai K, Zeng R (2014) The chemostat study of metabolic distribution in extreme-thermophilic (70°C) mixed culture fermentation. Appl Microbiol Biotechnol 98(24):10267–10273
Zhang F, Chen Y, Dai K, Shen N, Zeng RJ (2015) The glucose metabolic distribution in thermophilic (55°C) mixed culture fermentation: a chemostat study. Int J Hydrog Energy 40(2):919–926
Acknowledgments
The authors would like to acknowledge the financial support from the National Natural Science Foundation of China (51408530, 50978244), the China Postdoctoral Science Foundation (2015 T80233), the Natural Science Foundation of Hebei Province (E2015203306), the Postdoctoral Science Foundation of Hebei Province (B2014003008), the National Hi-Technology Development 863 Program of China (2011AA060901), the Hundred-Talent Program of Chinese Academy of Sciences, the Program for Changjiang Scholars and Innovative Research Team in University, and the Fundamental Research Funds for the Central Universities (wk2060190040).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no competing of interests.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Rights and permissions
About this article
Cite this article
Zhang, F., Yang, JH., Dai, K. et al. Characterization of microbial compositions in a thermophilic chemostat of mixed culture fermentation. Appl Microbiol Biotechnol 100, 1511–1521 (2016). https://doi.org/10.1007/s00253-015-7130-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-015-7130-z