Abstract
The renewed interests in clostridial acetone-butanol-ethanol (ABE) fermentation as a next-generation biofuel source led to significantly intensified research in the past few years. This mini-review focuses on the current status of metabolic engineering techniques available for the model organism of ABE fermentation, Clostridium acetobutylicum. A comprehensive survey of various application examples covers two general issues related to both basic and applied research questions: (i) how to improve biofuel production and (ii) what information can be deduced from respective genotype/phenotype manipulations. Recently developed strategies to engineer C. acetobutylicum are summarized including the current portfolio of altered gene expression methodologies, as well as systematic (rational) and explorative (combinatorial) metabolic engineering approaches.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
General interests in microbial fermentation processes for biofuels as a promising alternative to petrochemical production are steadily increasing, particularly in terms of technology development and economic competitiveness (Chu and Majumdar 2012; Caspeta et al. 2013). One of such advanced biofuels is 1-butanol, a C4 primary alcohol which is also an important commodity chemical used as solvent or precursor for diverse industrial compounds such as butyl acrylate and methacrylate esters (Mascal 2012). The usage of butanol as a fuel or fuel additive has several advantages over ethanol, notably a higher energy density, a lower volatility, and it is less hygroscopic and less corrosive. Therefore, butanol is entirely compatible with the existing infrastructure for liquid fuel transportation and storage, and current gasoline engines do not require any technical modifications (Jin et al. 2011). Besides chemical synthesis, butanol can be obtained biologically, and thus far, microorganisms belonging to the genus Clostridium are the only known organisms capable to naturally synthesize 1-butanol (Papoutsakis 2008; Lee et al. 2008; Jang et al. 2012a).
Already in 1862, this unique fermentative metabolism was described by Louis Pasteur who stated “l'alcool butylique est un produit ordinaire de la fermentation butyrique” (Pasteur 1862). Fifty years later, Chaim Weizmann discovered strain BY, which was later classified as Clostridium acetobutylicum, as a biocatalyst for the conversion of starch into butanol and acetone. The latter solvent was of particular importance, because acetone was needed in vast quantities for the production of cordite during the First World War. Thus, this production process called acetone-butanol-ethanol (ABE) fermentation quickly grew to the commercial scale. After the war, ABE fermentation was a valuable source of butanol to satisfy the increasing demand for this solvent, e.g. in the fast-growing automobile industry in the 1920s. The ammunition-dependent need for acetone re-emerged during the Second World War, and eventually, the ABE fermentation became one of the largest industrial bioprocesses (Jones and Woods 1986). Most of the ABE production plants in Western countries were closed in the 1950s and early 1960s due to cheaper petrochemical production routes of the solvents (Jones and Woods 1986; Kumar and Gayen 2011).
However, ABE fermentation regained much interests in academia and industry in recent years, and several either retrofitted or new industrial plants are currently under construction or even operating in the People's Republic of China (Ni and Sun 2009; Dong et al. 2012a). Fundamental technical improvements in bioprocess optimization such as product recovery and utilization of appropriate feedstocks have been reviewed recently (e.g. Gu et al. 2011; Kumar and Gayen 2011; Jang et al. 2012c; Jurgens et al. 2012; Mariano and Filho 2012; Moholkar et al. 2012; Ranjan and Moholkar 2012; Bankar et al. 2013; Xue et al. 2014).
Metabolic engineering of microorganisms for biofuel production does not only comprise a powerful portfolio for sustainable strain improvement but can also provide beneficial insights into cellular functioning. Basically, two general strategies for biobutanol production can be and have been pursued: (i) implementation of heterologous butanol biosynthesis in a suitable host organism and (ii) enhancement of natural butanol producers (Alper and Stephanopoulos 2009; Gronenberg et al. 2013). The present mini-review will focus exclusively on the latter issue, and recent advances in developing and employing genetic tools to engineer C. acetobutylicum as the model organism of ABE fermentation will be summarized.
Physiology of C. acetobutylicum
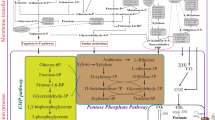
Natural ABE fermentation is exclusively performed by solventogenic clostridia, a group of strictly anaerobic, endospore-forming, Gram-positive bacteria which ubiquitously occur in soil, sludge and wastewater (Berezina et al. 2012). In general, the typical life cycle is tightly associated with two metabolic phases designated as acidogenesis and solventogenesis. During exponential growth, C. acetobutylicum metabolizes carbohydrates to acetic and butyric acids while generating a surplus of adenosine triphosphate (ATP) in addition to the glycolytic ATP in order to produce as much energy as possible under anaerobic conditions (Fig. 1). This extensive glycolytic flux and acid production is accompanied by a severe acidification of both the environment and the cell itself, most likely resulting in a primary stress signal for counteracting the rapid pH decrease. In addition, the non-respiratory metabolism unconditionally requires a balance between formation and utilization of reducing equivalents that necessitates adjustments of the primary metabolism (Jones and Woods 1986; Lütke-Eversloh and Bahl 2011). This is commonly represented by the reduced/oxidized nicotinamide adenine dinucleotide (phosphate) ratio NAD(P)H/NAD(P)+ as a typical cell signal and an important parameter among others to alter the metabolism (Grupe and Gottschalk 1992; Wietzke and Bahl 2012). This metabolic shift occurs during the transition from exponential to stationary growth and initiates solventogenesis. The previously formed acids are re-assimilated, and the solvents acetone, butanol and ethanol are formed, usually at a ratio of 3:6:1 (Fig. 1). The metabolic phases are accompanied by different morphological states of the life cycle. While exponentially growing cells are typically rod-shaped, stationary phase cells become a swollen, cigar-shaped form due to the accumulation of granulose, a starch-like storage polysaccharide which serves as carbon and energy source for the subsequent sporulation process. After maturation, the spores are released from the mother cell and can survive for many years until favourable environmental conditions promote germination (Dürre and Hollergschwandner 2004).
Central metabolism of C. acetobutylicum. Enzymes: Ldh, lactate dehydrogenase; HydA, hydrogenase; Pfor, pyruvate:ferredoxin oxidoreductase; Fnor, ferredoxin:NAD(P) + xidoreductase; Pdc, pyruvate decarboxylase; Pta, phosphotransacetylase; Ack, acetate kinase; AdhE, aldehyde/alcohol dehyrogenase; CtfAB, acetoacetyl-CoA:acyl-CoA transferase; Adc, acetoacetate decarboxylase; Thl, thiolase; Hbd, 3-hydroxybutyryl-CoA dehydrogenase; Crt, crotonase, Bcd, butyryl-CoA dehydrogenase complex; Ptb, phosphotransbutyrylase, Buk, butyrate kinase. Metabolites: Acetyl-P, acetylphosphate; Ac-CoA, acetyl-CoA; AcAld, acetaldehyde; AcAc-CoA, acetoacetyl-CoA; AcAc, acetoacetate; 3HB-CoA, 3-hydroxybutyryl-CoA; Butyryl-P, butyrylphosphate; BuAld, butyraldehyde. The dashed lines indicate solventogenic reactions
Targeted manipulation of gene expression
Plasmids for gene overexpression
The first recombinant strains of C. acetobutylicum for homologous gene overexpression were described in 1992, although construction of suitable cloning vectors and transformation via conjugation or electroporation were reported a few years earlier (Oultram et al. 1988; Truffaut et al. 1989; Young et al. 1989; Willims et al. 1990). Based on a Bacillus subtilis/C. acetobutylicum shuttle vector harbouring the replicon derived from pIM13 of B. subtilis and a macrolide lincosamin resistance gene as selection marker, acetone and butyrate biosynthetic genes were homologously overexpressed (Lee et al. 1992b; Mermelstein et al. 1992, 1993). The main obstacle resulting in low transformation efficiencies and plasmid instability is the native, highly active restriction endonuclease Cac824I of C. acetobutylicum, and multiple recognition sites frequently occur in standard Escherichia coli cloning vectors. Since a methyltransferase for the respective recognition site (5′-GCNGC-3′) was commercially not available, a suitable in vivo methylation protocol was developed which is still commonly used nowadays. Plasmids pAN1 and pAN2, equipped with compatible E. coli replicons and antibiotic resistance genes, express the ϕ3TI methyltransferase gene of the B. subtilis phage ϕ3TI and protects plasmids to be transformed into C. acetobutylicum from hydrolysis (Mermelstein and Papoutsakis 1993; Heap et al. 2007). An alternative strategy to prevent Cac824I restriction of recombinant plasmids is the employment of Cac824I-deficient host strains which allows omission of the additional transformation for in vivo methylation in E. coli and subsequent purification of methylated plasmids prior to electroporation into C. acetobutylicum (Soucaille et al. 2008; Dong et al. 2010).
Due to the limitation of functional antibiotic markers, the number of shuttle vectors established for the use in C. acetobutylicum is quite small, and most overexpression studies were based on derivatives of the pSOS94 and pIMP1 plasmids (Lee et al. 1992a; Mermelstein et al. 1992; Tummala et al. 1999). Hence, N. P. Minton and coworkers developed the pMTL80000 plasmid series, a standardized modular E. coli/Clostridium shuttle vector system which can be adapted as required. It consists of four basic modules comprising different replicons, markers and additional features such as multiple cloning sites or the tra genes for conjugative transfer, which can be assembled in any combination due to the common restriction sites (Heap et al. 2009).
In order to promote plasmid-borne gene expression, native clostridial promoters like the thl, ptb and adc promoters have widely been used, and their expression profiles are well known (Tummala et al. 1999; Feustel et al. 2004). Depending on the type of overexpression, inducible gene expression systems rather than constitutive promoters can be useful is some cases. However, the number of inducible promoter/repressor systems developed for C. acetobutylicum is still limited. Girbal et al. used the xylA promoter and xylR repressor from Staphylococcus xylosus to generate a xylose-inducible expression vector for C. acetobutylicum (Girbal et al. 2003). The renowned lac expression system was fused with the fdx promoter from Clostridium pasteurianum, yielding the artificial lacI-repressed fac promoter, which can be induced in C. acetobutylicum by addition of 1 mM IPTG (Heap et al. 2007). More recently, the portfolio of inducible promoters was complemented by the lactose-inducible bgaL promoter and the anhydrotetracycline-inducible Pcm-2tetO1 promoter, allowing reliable stringency and a broad range of inducibility (Al-Hinai et al. 2012; Dong et al. 2012b).
Chromosomal integration for gene overexpression
With respect to industrial applications of suitable production strains, plasmid-based gene expression is less desirable because of the need for plasmid maintenance involving, for example, addition of expensive antibiotics. Therefore, stable integration of homologous or heterologous genes under the control of a useful promoter is a preferred strategy for the design of potential producers. As described below, chromosomal manipulation of solventogenic clostridia is particularly difficult, and notable breakthroughs in this field were achieved only recently. However, the techniques explained in the last paragraph of this section allow both targeted gene knockout and integration of large DNA fragments of up to 6.5 kbp into the chromosome (Al-Hinai et al. 2012; Heap et al. 2012). Employing this cargo technique, C. acetobutylicum was engineered for isopropanol production by heterologous expression of the secondary alcohol dehydrogenase gene from Clostridium beijerinckii from the chromosome, resulting in the conversion of acetone into isopropanol (Heap et al. 2012). More recently, this method was successfully used to generate a recombinant C. acetobutylicum strain which secreted various synthetic cellulosome components, paving the way for future consolidated bioprocessing strategies (Kovács et al. 2013).
Integrative plasmids for gene inactivation
Already in 1996, the first knockout mutants of C. acetobutylicum with defects in the central metabolic pathways were reported, i.e. mutants of the aldehyde/alcohol dehydrogenase (adhE1), phosphotransacetylase (pta) and butyrate kinase (buk) genes (Green and Bennett 1996; Green et al. 1996). Non-replicative suicide plasmids were used for homologous recombination into the respective genes, but only single-crossover events by a Campbell-like integration of the entire plasmids were obtained. The same strategy was employed to inactivate SolR, a putative regulator of solventogenesis located upstream of the sol locus (Nair et al. 1999). Although the mutant exhibited an improved solventogenic phenotype, the function of SolR as a transcriptional repressor was critically discussed by others (Thormann and Dürre 2001; Thormann et al. 2002). The use of replicating plasmids was also reported for successful integration into a specific gene as demonstrated by inactivation of the spo0A gene encoding the master regulator of sporulation in C. acetobutylicum (Harris et al. 2002). This approach was recently combined with the expression of a recombinant resolvase gene (recU) from B. subtilis in order to delete the sigma factor genes sigE and sigG (Tracy et al. 2011). However, the obtained mutants also comprised only single-crossover events, indicating that heterologous RecU did not promote the Holliday junction resolution during the recombination event.
Antisense RNA for gene downregulation
Gene expression can be non-covalently modulated by in vivo hybridization of complementary messenger RNA (mRNA) transcripts, so-called antisense (as) RNA, which specifically inhibits gene translation (Thomason and Storz 2010). Employing plasmid-encoded asRNA against the butyrate biosynthetic genes ptb and buk, this strategy was shown to successfully reduce respective enzyme activities. However, the butyrate production by the recombinant strains was not reduced (Desai and Papoutsakis 1999). In contrast, targeting the acetone biosynthetic pathway, asRNA constructs against the ctfB gene encoding the acetoacetyl-CoA:acyl-CoA transferase subunit B in fact reduced acetone production in C. acetobutylicum, whereas the asRNA against the acetoacetate decarboxylase gene adc was not effective (Tummala et al. 2003b). Combination of ctfA downregulation and adhE1 overexpression did not only restore butanol formation; the engineered strain exhibited a drastically increased ethanol production (Tummala et al. 2003a; Sillers et al. 2009). Lastly, asRNA was used to investigate the role of the sporulation regulator SpoIIE, and downregulation of spoIIE resulted in prolonged and elevated solventogenesis as well as in a delayed initiation of endospore formation (Scotcher and Bennett 2005).
ClosTron and related techniques for gene inactivation
Since genome manipulations by homologous recombination were particularly difficult in C. acetobutylicum and other clostridia, implementation of mobile group II introns for targeted gene inactivation provided a novel useful tool, probably an overdue breakthrough for clostridial genetics (Kuehne and Minton 2012). Group II introns are catalytically active RNAs comprising a multi-domain intron-encoded protein which mediates sequence recognition and self-splicing. Thus, alteration of only a few bases in the recognition site allows individual re-targeting for virtually any chromosomal gene (Karberg et al. 2001). The TargeTron system, which is commercially available from Sigma-Aldrich, employs this technique for insertional mutagenesis and is based on the L1.LtrB intron from Lactococcus lactis. It has been further engineered by including a retrotransposition-activated marker providing erythromycin resistance after intron insertion, and the functionality has been demonstrated in various clostridial species, including solventogenic strains (Heap et al. 2007; Wang et al. 2013). This so-called ClosTron system was further optimized, most notably by incorporation of flippase recognition target (FRT) sites flanking the marker cassette to allow flippase-mediated excision and thus multiple rounds of intron-mediated mutagenesis (Heap et al. 2010).
Interestingly, the L1.LtrB-intron-based mutagenesis does not necessarily require selective pressure by an antibiotic marker, because intron insertions occur frequently and were shown to be stable and reproducible (Shao et al. 2007; Jiang et al. 2009). Avoiding chromosomal marker excision and plasmid curing also facilitates the procedure of generating multiple-knockout mutants, as demonstrated by Jang et al. (2012b). Alternating use of erythromycin and thiamphenicol resistance genes for the mutagenesis plasmids harbouring the re-targeted group II introns, various gene knockout mutants including three double- and two triple-knockout strains of C. acetobutylicum were generated (Jang et al. 2012b).
Another variant for gene inactivation in C. acetobutylicum which combined both group II intron and homologous recombination techniques was described by Jia et al. (2011). The first step includes the targeted integration of the intron into the respective gene and selection of erythromycin-resistant clones. In the second mutagenesis step, multiple subculturing in the absence of erythromycin is required to identify those mutants in which the intron was deleted by homologous recombination. However, this alternative gene deletion method is very elaborate and time consuming because of successive transfers and large numbers of colonies to be screened; e.g. 648 clones of a chromosomal knockout and 1,998 clones of a pSOL1 knockout mutant had to be analysed to identify positive clones (Jia et al. 2011).
Allelic exchange mutagenesis
Although the ClosTron and related knockout strategies were shown to successfully inactivate a variety of genes in C. acetobutylicum, insertional mutagenesis may also cause polar effects of the integrated group II intron, and intrinsic instability of a ClosTron mutant has also been reported (Steiner et al. 2011). However, double-crossover mutants could be obtained only recently after establishment of a suitable selection system for the second crossover event. Allele-coupled exchange (ACE) was developed to either knockout genes or integrate large DNA fragments into the genome (Heap et al. 2012). Instead of using a plasmid-encoded counterselection marker, the method relies on a selectable phenotype based on the well-known uracil auxotrophy plus antimetabolite toxicity principle: First, uracil-auxotrophic mutants were selected which require uracil supplementation for growth. Second, addition of 5-fluoroorotic acid prevents growth of those mutants with a functional pyrE/pyrF gene, because they are sensitive towards the toxic antimetabolite which results from 5-fluoroorotic acid cleavage, and thus, only double-crossover mutants without integrated plasmid are selected (Heap et al. 2012). An alternative approach to generate stable deletion mutants of C. acetobutylicum was also published recently by E. T. Papoutsakis and coworkers. For the selection of double-crossover mutants, the E. coli mazF toxin gene, coding for an mRNA interferase, was coupled to a lactose-inducible promoter, enabling counterselection in the presence of lactose (Al-Hinai et al. 2012). Furthermore, FRT sites were included adjacent to the thiamphenicol resistance cassette for subsequent excision of the marker and thus allowing ‘clean’ deletions and marker recycling. In order to facilitate plasmid curing, a plasmid carrying an asRNA against the repL replicon fused to the same lactose-inducible promoter was also constructed (Al-Hinai et al. 2012).
Systematic metabolic engineering
Engineering butanol tolerance
Butanol toxicity constitutes an important limiting factor of industrial ABE fermentation, because product tolerance is usually below 20 g/l (Liu and Qureshi 2010). The major stress responses of the cells include changes in the membrane composition to counteract the fluidity increase as well as drastically increased expression of heat shock protein (HSP) encoding genes (Tomas et al. 2004; Ezeji et al. 2010; Nicolaou et al. 2010; Janssen et al. 2012). Therefore, the HSP genes groESL were homologously overexpressed in C. acetobutylicum, leading to significantly improved robustness and solvent titres, and continuative transcriptome analyses indicated that other HSPs might have a similar effect (Tomas et al. 2003, 2004; Alsaker et al. 2010). Overexpression of the HSP genes grpE and htpG did effectively improve butanol tolerance traits, but higher solvent production could not be achieved, indicating that butanol tolerance and production capabilities of C. acetobutylicum are not necessarily linked together (Mann et al. 2012). Similarly, increased expression levels of the cyclopropane fatty acid synthase gene cfa impacted the cell membrane composition and thus exerted a positive effect on acid and solvent tolerance, but the recombinant C. acetobutylicum strain was significantly impaired in solvent production (Zhao et al. 2003).
As a new target for systematic engineering of butanol tolerance, the gshAB genes from E. coli, coding for γ-glutamate-cysteine ligase and glutathione synthetase, were overexpressed in C. acetobutylicum, resulting in a much more robust phenotype in terms of butanol resistance and aerotolerance (Zhu et al. 2011). This strategy was subsequently extended by Hou et al.: while disrupting the adc gene by integration of the gshAB cassette, both glutathione-mediated robustness and reduced acetone formation, an unwanted by-product (see “Engineering reduced by-products” section), were achieved. Further, systematic engineering by overexpression of genes involved in solventogenesis led to high-butanol titres of up to 15 g/l, an almost threefold increase as compared to the wild-type strain C. acetobutylicum ATCC 824 under similar cultivation conditions (Hou et al. 2013).
Interestingly, a negative control factor for butanol resistance was also described: Based on similarities to eukaryotic alcohol-binding regulators, two adjacent genes of unknown functions were identified in a comparative proteomic approach. Inactivation of either or both genes in C. acetobutylicum DSM 1731 clearly lowered the sensitivity towards butanol stress, whereas homologous overexpression of both genes inhibited growth in the presence of 1 vol% butanol (Jia et al. 2012).
Engineering reduced by-products
The fermentative metabolism of C. acetobutylicum and related solventogenic clostridia typically comprises a mixture of end products at varying ratios (Fig. 1). As a classical biotechnological principle of strain optimization, the formation of by-products must be reduced or eliminated to achieve higher yields of the desired product. Therefore, first metabolic engineering attempts implemented this strategy as soon as respective genetic tools were developed. Employing integrative plasmids for gene knockout, key enzymes of the acetate and butyrate biosynthetic pathways were targeted in order to increase the carbon flux towards butanol (Green et al. 1996). A few years later, asRNA constructs were generated to downregulate enzymes of the butyrate and acetone pathways (Desai et al. 1999; Desai and Papoutsakis 1999; Tummala et al. 2003b). However, the vast majority of defined knockout mutants with inactivated enzymes of the central carbon metabolism were generated only recently according to the late development of new gene knockout methodologies. Table 1 shows a survey of all knockout mutants of C. acetobutylicum targeted for altered acid or solvent pathways published thus far, including the major phenotypic characteristics of the mutants.
Interestingly, knockout of either Pta or Ack of the acetate biosynthetic pathway did not result in clear acetate-negative phenotypes, which was similarly true for Buk of the analogous butyrate pathway, suggesting some enzymatic complementation of the corresponding C2- and C4-acidogenic pathways (Table 1 and Fig. 1). With respect to butyrate synthesis, only three studies reported a distinct butyrate-negative phenotype of C. acetobutylicum mutants, comprising an intron-mediated disruption of the hbd or ptb gene, respectively (Lehmann and Lütke-Eversloh 2011; Lehmann et al. 2012b; Cooksley et al. 2012). All single-knockout mutants with disrupted acetone biosynthetic enzymes exhibited reduced or zero, respectively, acetone production with concomitantly elevated acetate titres, indicating a strong relation between acetate re-assimilation and acetone formation (Table 1). Therefore, disruption of Pta was introduced into these acetone-impaired mutants, and the novel double-knockout mutants of C. acetobutylicum exhibited both low-acetate and low-acetone levels. However, instead of an improved butanol production, these mutant strains revealed increased butyrate concentrations, and in some cases significant lactate formation, reflecting the metabolic complexity and general difficulty to manipulate the fermentative pathways by systematic means (Lehmann et al. 2012a; Jang et al. 2012b).
The most notable achievement of the newly developed methods for systematic genetic engineering was probably the comprehensive study by Jang et al., in which multiple knockouts and combinations thereof were constructed and characterized (Jang et al. 2012b). The C. acetobutylicum strains BBEKW and CBEKW represent the first examples of distinct triple-knockout mutants with defects in the acetate, butyrate and acetone formation pathways (Table 1). In another publication, the mutant strain CEKW with inactivated Pta and CtfB was converted to the butyrate producer HCEKW by additional disruption of the adhE1 gene to diminish solvent formation, yielding the third triple-knockout mutant of C. acetobutylicum (Jang et al. 2013c). Very recently, the same authors reported on the generation of novel quadruple and quintuple C. acetobutylicum mutants for improved butyrate production (Jang et al. 2014). It is noteworthy that this work included the first successful knockout of the hydrogenase gene hydA. Inactivation of hydA in the wild-type strain has been attempted by different laboratories (unpublished results, Cooksley et al. 2012), but obviously, this targeted mutation requires a particular genetic background as given by disrupted pta, buk, ctfB and adhE1 genes in the HCBEKW strain (Table 1).
With respect to high-butanol yields, ethanol also constitutes an unwanted by-product, but elimination of ethanol formation by rational means seems to be a currently unsolvable problem, because ethanol and butanol are synthesized by the same enzymes. In order to assess the roles of the different alcohol dehydrogenases encoded in the genome of C. acetobutylicum, Cooksley et al. generated different alcohol dehydrogenase-deficient mutants using the ClosTron mutagenesis system (Cooksley et al. 2012). Among these mutants, only the AdhE1-deficient strain revealed a clear phenotype with drastically reduced solvent production and significant acid accumulation (Table 1). Interestingly, inactivation of the second functionally active aldehyde/alcohol dehydrogenase AdhE2 did not result in any product pattern alterations, although a putative role in redox-dependent solventogenesis was proposed earlier (Fontaine et al. 2002; Hönicke et al. 2012; Wietzke and Bahl 2012).
Understanding the regulatory mechanisms of the solventogenic shift is of great interest, and identification of the molecular triggering machinery still remains an obstacle. The master sporulation factor Spo0A plays a central role in the clostridial life cycle, but the phosphorylation pattern differs from the well-studied Bacillus homolog, and recent work on various sigma factors broadened our knowledge substantially (Paredes et al. 2005; Bi et al. 2011; Jones et al. 2011; Steiner et al. 2011; Tracy et al. 2011; Al-Hinai et al. 2014). With respect to solvent formation, disruption of the putative regulator SolR using the TargeTron technology clearly increased the ABE titre, which was in accordance with the phenotype of the SolR-deficient mutant described earlier (Harris et al. 2001; Shao et al. 2007). More recently, the redox-sensing transcriptional regulator Rex was shown to be de facto involved in the solventogenic shift, and disruption of the rex gene resulted in an enhanced alcohol production accompanied by low-acetone levels (Wietzke and Bahl 2012) (Table 1).
Engineering IBE production
Among the solvents produced by C. acetobutylicum, acetone represents the least attractive by-product, because in contrast to ethanol and butanol, it cannot be used as a biofuel. Therefore, in vivo reduction of acetone to isopropanol converts the native ABE fermentation to isopropanol-butanol-ethanol (IBE) fermentation, an alcohol mixture suitable for direct utilization as fuel blend without previous separation of the solvents. This objective was pursued by four different research groups using different C. acetobutylicum strains or strain backgrounds, respectively. The key metabolic engineering step was the heterologous overexpression of the primary/secondary alcohol dehydrogenase gene sadh from C. beijerinckii. Using the buk-deficient strain PJC4BK as host, expression of sadh and homologous overexpression of adc and ctfAB, encoding the acetone biosynthetic pathway, resulted in 20 g/l IBE in batch fermentations (Lee et al. 2012). Later on, this group employed C. acetobutylicum BKM19, a strain preengineered for ABE production (see “Phenotype screening” section), as host strain and achieved 28 g/l IBE in lab-scale batch fermentations. In addition, the applicability of this recombinant strain for large-scale IBE production was demonstrated by reproducing the lab-scale fermentation performance at a 200-l pilot scale (Jang et al. 2013a).
Dusséaux et al. also chose a host strain with impaired butyrate biosynthesis, C. acetobutylicum 824Δbuk, for overexpression of sadh plus adc and ctfAB and evaluated different expression levels using the native thl and ptb promoters. Application of the ptb promoter, which is highly active during the exponential growth rather than in the stationary phase, resulted in a final biofuel titre of 21 g/l. Moreover, the authors concluded that the elevated intracellular acetate concentrations and the high KM value of the CtfAB enzyme for acetate constitute the major bottlenecks for IBE production in C. acetobutylicum (Dusséaux et al. 2013).
Heterologous overexpression of sadh in the wild-type C. acetobutylicum ATCC 824 yielded 16 g/l IBE, and combination with the native adc and/or ctfAB genes further increased the total alcohol titre with ctfAB overexpression showing the more significant effect. Overexpression of all three acetone biosynthetic genes along with the heterologous sadh gene yielded 24 g/l IBE in addition to small amounts (1.2 g/l) of 2,3-butanediol (Collas et al. 2012). Interestingly, the same IBE titre was reported by Dai et al., who introduced the sadh gene into the preengineered mutant strain C. acetobutylicum Rh8 (see “Random mutagenesis” section) without concomitant adc and ctfAB overexpression (Dai et al. 2012). Chromosomal integration of sadh into the genome of C. acetobutylicum ATCC 824 led to an isopropanol titre of 1.7 g/l, demonstrating the general feasibility of the ACE technique to construct plasmid-independent overexpression strains (Heap et al. 2012).
Engineering substrate utilization
Utilization of cheap non-food feedstocks is the ultimate prerequisite for economically feasible biofuel production (Gu et al. 2011; Tracy et al. 2012; Xue et al. 2013). Since solventogenic clostridia can metabolize a wide variety of hexose and pentose sugars, they are predestined to convert agricultural waste products in the form of pretreated lignocellulosic hydrolysates to bulk chemicals or fuels (Jurgens et al. 2012). In order to promote simultaneous and efficient utilization of these carbohydrate mixtures, metabolic pathways and their regulatory circuits must be known for sustainable metabolic engineering strategies. Components of the phosphoenolpyruvate-dependent phosphotransferase systems (PTS) are suitable targets to enable cofermentation of glucose, xylose and arabinose, the three dominant sugars of lignocellulosic biomass. The glucose PTS of C. acetobutylicum has been identified, and inactivation of the enzyme-II-encoding glcG gene alleviated the glucose-dependent repression of xylose catabolism without affecting glucose utilization (Tangney and Mitchell 2007; Xiao et al. 2011). Additional overexpression of the xylTBA genes, coding for xylose-proton symporter, xylose isomerase and xylulose kinase, in the glcG-negative mutant increased the ABE production by 24 % from a glucose-xylose-arabinose mixture (Xiao et al. 2011). Employing the industrial strain C. acetobutylicum EA 2018 as host for the same genetic modifications, a greater increase of 44 % of the ABE titre was reported (Li et al. 2013). Further, potential pentose phosphate pathway genes were identified in the genome of C. acetobutylicum, and the respective functionalities were experimentally validated. Overexpression of the four genes encoding transketolase, transaldolase, ribose-5-phosphate isomerase and ribulose-5-phosphate epimerase significantly improved xylose utilization, yielding a 42 % higher solvent titre from xylose as compared to the plasmid control strain (Jin et al. 2014). However, xylose and arabinose are not exclusively metabolized via the pentose phosphate pathway; the existence of a functional phosphoketolase pathway in C. acetobutylicum was recently demonstrated by two independent studies (Liu et al. 2012a; Servinsky et al. 2012).
Cofermentation of glucose and xylose was also achieved by disruption of the ccpA gene, encoding the catabolite control protein A, a pleiotropic regulator of carbon catabolite repression. But since CcpA controls a large variety of genes, the CcpA-negative mutant accumulated elevated amounts of acids in pH-uncontrolled cultures, which in turn negatively affected cell growth and solvent formation (Ren et al. 2010). In a follow-up publication, the global role of CcpA was assessed in a detailed transcriptional profiling approach. Interestingly, CcpA does not only control genes involved in sugar uptake and metabolism; solventogenic and sporulation-associated genes are also regulated by CcpA (Ren et al. 2012).
Finally, possible glycolytic limitations in C. acetobutylicum were addressed by homologous overexpression of the pfkA and pykA genes, coding for 6-phosphofructokinase and pyruvate kinase, which increased the final ABE titre by 23 % in batch cultures (Ventura et al. 2013).
Explorative metabolic engineering
Random mutagenesis
In general, the systematic metabolic engineering strategies described above implement the rational design and analysis of particular genetic alterations derived from gene overexpression and/or inactivation. To accomplish this, two indispensable prerequisites must be fulfilled: (i) physiological and genetic knowledge for the ability to define targets and (ii) availability of molecular tools to introduce specific manipulations. In contrast, random mutagenesis can bypass these constraints and broadens the opportunities to obtain useful strains for ABE fermentation independently from sophisticated techniques. Thus, traditional strain breeding was—and still is—quite successful to isolate C. acetobutylicum mutants with enhanced phenotypes. Most commonly, chemical agents such as N-methyl-N'-nitro-N-nitrosoguanidine or ethyl methane sulphonate were used rather than ultraviolet irradiation, which is less effective for clostridia as compared to Gram-negative bacteria, to generate respective mutant populations (Bowring and Morris 1985; Lemmel 1985). Today, less harmful methods are desirable to protect laboratory personnel from exposure to toxic and carcinogen substances and reduce the use of polluting agents. Hence, ionized gas beams for mutagenesis might substitute highly toxic chemicals, and nitrogen ion beam implantation (NIBI) as well as atmospheric and room temperature plasma (ARTP) was shown to generate suitable C. acetobutylicum mutants (Liu et al. 2012b; Li et al. 2014).
Genome shuffling, an evolutionary method based on recursive genomic recombination, allows rapid phenotype improvements. In comparison to conventional strain breeding, genome shuffling requires fewer sequential steps to obtain an optimal phenotype because of minimizing deleterious mutations and maximizing genetic exchange between parental strains (Zhang et al. 2002; Biot-Pelletier and Martin 2014). This two-step procedure of chemical mutagenesis plus selection to generate appropriate parents and subsequent recursive protoplast fusion was shown to generate improved ABE producing mutants of C. acetobutylicum, including strain Rh8 which was subjected to detailed comparative proteomics (Mao et al. 2010, 2011; Gao et al. 2012b).
A more complex synthetic system to generate potential mutant libraries of C. acetobutylicum was developed by Luan et al., and advantageously, the degree of mutagenesis can be adapted easily. For this, the chromosomal mismatch repair operon mutSL was inactivated and complemented by a plasmid harbouring mutSL alleles fused to the anhydrotetracycline-inducible Pcm-2tetO1 promoter as well as two copies of the TetR repressor gene to ensure stringency. Thus, the mutation rates during genome replication can be modulated up to 120-fold as compared to the wild-type by controlling the inducer concentration (Luan et al. 2013).
Phenotype screening
Basically, various options to create mutant libraries of C. acetobutylicum exist, and transfer of novel methods successfully developed for other bacteria such as promoter or transcription factor engineering should be transferable to clostridial species (Kim et al. 2013). Such libraries can be based on single or multiple genomic mutations, mutated global regulators or other specific proteins, in addition to overexpression libraries comprising homologous or heterologous genomic DNA fragments (Borden and Papoutsakis 2007; Borden et al. 2010). However, not the type of library but the availability of new screening methods is currently limiting the application of explorative metabolic engineering strategies. In order to identify desired phenotypes from large populations, the screening method must be conducted in a high-throughput manner, prioritizing technical feasibility, i.e. general handling, expenditure of work, time and costs. Since microbial biofuels are not directly detectable without chromatographic measurements, they must somehow be visualized by colorimetric/fluorescent methods or growth-dependent assays (Dietrich et al. 2010). With the few exceptions listed below, all C. acetobutylicum strains originating from explorative approaches reported so far were isolated due to survival of high butanol concentrations. The phenotype of improved tolerance was generally associated with increased ABE production as compared to the respective parental strains (e.g. Mao et al. 2010; Gao et al. 2012a; Liu et al. 2012b). A possible drawback of randomly mutagenized strains exhibiting improved tolerance and/or solvent production is the unknown genotype. It is self-evident that an ideal butanol-tolerant phenotype is usually determined by multiple genetic alterations, and backtracking these mutations is required to identify novel and interesting target genes for both understanding and engineering cellular robustness towards solvents (Jia et al. 2010; Nicolaou et al. 2010).
Strategies to investigate and modify metabolic pathways during the pregenome era of C. acetobutylicum often included the use of compounds mimicking natural substrates or intermediates. For example, antimetabolites are useful non-metabolizable agents to select deregulated mutants for the overproduction of primary metabolites like amino acids (Lütke-Eversloh and Stephanopoulos 2005; Sanchez and Demain 2008). Suicide substrates represent another group of practical compounds, and since only the cleavage products are toxic, their use allows a positive selection of clones which lost the ability of degradation and thus isolation of mutants with particular enzymatic defects. Employing halogenated carboxylic acid analogues, various mutants of C. acetobutylicum with altered metabolite profiles were isolated (e.g. Junelles et al. 1987; Clark et al. 1989; Medkor et al. 2010). Uptake and conversion of fluoroacetate results in the formation of the highly toxic intermediate fluoroacetyl-CoA and therefore clones able to grow in the presence of fluoroacetate possess mutations in the acetate metabolic pathway (Rothstein 1986). This principle was recently employed by S. Y. Lee and coworkers who generated C. acetobutylicum BKM19. Interestingly, this was the first example of combined systematic and explorative metabolic engineering: The preengineered Buk-negative strain PJC4BK (Table 1) was subjected to chemical mutagenesis, and the resulting library was screened for growth on fluoroacetate-containing agar plates. The selected mutant BKM19 exhibited a significantly improved phenotype; i.e. the ABE titre of batch cultures was increased by 31 and 91 %, as compared to the parental PJC4BK strain and the wild-type, respectively (Jang et al. 2013b).
The peculiarity of the third strategy of phenotype selection is its direct association with alcohol synthesis instead of analysing derived parameters commonly attributed to enhanced butanol formation, for instance, high tolerance, defects in competing pathways or particular cell morphologies. In order to visualize butanol and ethanol in clostridial microtitre cultures, a colorimetric assay using nitroblue tetrazolium derivatization was developed to establish a high-throughput screening system for biofuels. Screening of two chemically mutagenized C. acetobutylicum libraries yielded several mutants with >20 % increased butanol production. On the one hand, this semi-quantitative method was coupled with a preselection step employing butanol tolerance as parameter to reduce the library size for the subsequent microtitre-based screening. On the other hand, a preengineered strain with disrupted adc gene and thus low-acetone production, was used as the parental strain for the library, yet another example of successful implementation of systematic and explorative engineering approaches (Scheel and Lütke-Eversloh 2013).
Finally, another strategy took advantage of the relation between metabolic states and different cell morphologies typically associated with clostridial sporulation. High-throughput flow cytometric analyses comprising different staining methods were used to discriminate and quantify various cell types of C. acetobutylicum (Tracy et al. 2008). Establishment and optimization of single-cell analysing methodologies offer valuable options to investigate basic research objectives such as the correlation of morphological and physiological phenotypes (Tracy et al. 2010). Moreover, flow cytometry was proposed to be a useful tool for process mapping of clostridial ABE fermentation, including differentiation of acidogenic and solventogenic subpopulations and determination of cell viability (Linhová et al. 2012; Patáková et al. 2013).
In summary, four phenotype screening principles are currently available for C. acetobutylicum to select improved biofuel producing mutants: (i) butanol tolerance, (ii) suicide substrates, (iii) colorimetric alcohol detection, and (iv) flow cytometry. Future research will certainly expand this portfolio of high-throughput screening strategies, and researchers can learn from promising methods developed for other microorganisms. For example, a butanol-specific biosensor was recently developed to monitor butanol formation in recombinant E. coli cultures. The transcription-factor-based system was composed of the σ54-dependent activator BmoR and the alcohol-controlled promotor PBMO from the alkane-degrading β-proteobacterium Thauera butanivorans (formerly Pseudomonas butanivorans), fused to the tetracycline resistance gene tetA. This biosensor exhibited a linear correlation of E. coli growth and 1-butanol concentrations in the range of 0.01–40 mM, whereas ethanol was not recognized (Dietrich et al. 2013).
Enzyme engineering
Although solventogenic clostridia represent industrially relevant microbes with unique metabolic capacities, only little work has been conducted on specific enzyme engineering, except for cellulose-degrading proteins (Kellermann and Rentmeister 2014; Thomas et al. 2014; Yang et al. 2014). Most of the central fermentative enzymes have been purified and biochemically characterized (Gheshlaghi et al. 2009), but only two studies on engineering key enzymes of ABE fermentation were published recently. In a rational approach, Jang et al. optimized the AdhE1 activity in the background of strain C. acetobutylicum BEKW (Table 1) for further enhancement of the carbon flux from acetyl-CoA to butanol via butyryl-CoA. According to sequence homologies to the Zymomonas mobilis alcohol dehydrogenase 2, the cofactor specificity of the clostridial AdhE1 was attenuated by a simple D-485-G amino acid substitution, allowing both NADPH and NADH to be used as electron donors (Rellos et al. 1997; Jang et al. 2012b).
Based on the idea of alleviating feedback inhibition of the branchpoint enzyme of the C4 biosynthetic pathway, the thiolase of C. acetobutylicum was specifically engineered to reduce the sensitivity towards its physiological inhibitor free CoA-SH. First, a microtitre high-throughput assay was developed to analyse thiolase activities and CoA-SH inhibition in recombinant E. coli cell extracts. Subsequently, a mutant library of the clostridial thlA gene was generated, whereby only the region of CoA-binding was mutagenized with an average mutation rate of 2.5 amino acid substitutions. Screening of this library resulted in the identification of thiolase derivatives with a more than tenfold decreased inhibition by 50 μM CoA-SH, which is clearly below the physiological concentration (Wiesenborn et al. 1988). Finally, overexpression of the engineered thiolase gene in C. acetobutylicum resulted in an 18 % higher butanol titre and a significant delay in ethanol and acetone production (Mann and Lütke-Eversloh 2013).
The thiolase engineering approach belongs to the explorative category of metabolic engineering; however, recombinant E. coli cells were used to facilitate the screening procedure. With respect to future attempts, genetic engineering might also be performed directly in C. acetobutylicum as the native host. Introduction of the Clostridium perfringens phagenic recT gene was recently shown to mediate recombination of synthetic oligonucleotides, i.e. single-strand DNA, in C. acetobutylicum. Further optimization of this method might enable genomic recombineering in the near future and thus expand the opportunities for single-protein engineering or even whole-cell engineering (Dong et al. 2014).
Concluding remarks
The metabolic engineering toolbox for C. acetobutylicum and related solventogenic clostridia has thrived enormously in the past few years, and application of modern genetic techniques has eventually led to competitive results as compared to classical strain breeding. There are several pros and cons relating to whether butanol production is more attractive in native clostridia or in alternate host organisms (Gronenberg et al. 2013; Branduardi et al. 2014). Since redox-mediated limitations such as the non-functionality of the ferredoxin-dependent butyryl-CoA dehydrogenase complex (Bcd/EtfAB) in E. coli have been resolved, heterologous butanol production in well-accessible host organisms is definitely reasonable, and even in vitro synthesis of butanol from glucose is possible (Li et al. 2008; Bond-Watts et al. 2011; Shen et al. 2011; Lim et al. 2013; Krutsakorn et al. 2013). On the other hand, the recent progress in developing applicable molecular technologies for solventogenic clostridia again compensates for the potential advantages of non-native hosts. Together with the bioinformatic networks providing steadily increasing amounts of ‘omics’ data as well as complex in silico models, the availability of suitable experimental protocols for genetic modifications is pivotal for the application of systems biology to effectively engineer C. acetobutylicum (Jang et al. 2012d; Woolston et al. 2013).
References
Al-Hinai MA, Fast AG, Papoutsakis ET (2012) Novel system for efficient isolation of Clostridium double-crossover allelic exchange mutants enabling markerless chromosomal gene deletions and DNA integration. Appl Environ Microbiol 78(22):8112–8121
Al-Hinai MA, Jones MAA, Papoutsakis ET (2014) σK of Clostridium acetobutylicum is the first known sporulation-specific sigma factor with two developmentally separated roles, one early and one late in sporulation. J Bacteriol 196(2):287–299
Alper H, Stephanopoulos G (2009) Engineering for biofuels: exploiting innate microbial capacity or importing biosynthetic potential? Nat Rev Microbiol 7(10):715–723
Alsaker KV, Paredes C, Papoutsakis ET (2010) Metabolite stress and tolerance in the production of biofuels and chemicals: gene-expression-based systems analysis of butanol, butyrate, and acetate stresses in the anaerobe Clostridium acetobutylicum. Biotechnol Bioeng 105(6):1131–1147
Bankar SB, Survase SA, Ojamo H, Granström T (2013) Biobutanol: the outlook of an academic and industrialist. RSC Adv 3(47):24734–24757
Berezina OV, Zakharova NV, Yarotsky CV, Zverlov VV (2012) Microbial producers of butanol. Appl Biochem Microbiol 48(7):625–638
Bi C, Jones SW, Hess DR, Tracy MBP, Papoutsakis ET (2011) SpoIIE is necessary for asymmetric division, sporulation, and expression of σF, σE, and σG but does not control solvent production in Clostridium acetobutylicum ATCC 824. J Bacteriol 193(19):5130–5137
Biot-Pelletier D, Martin VJ (2014) Evolutionary engineering by genome shuffling. Appl Microbiol Biotechnol. doi:10.1007/s00253-014-5616-8
Bond-Watts BB, Bellerose RJ, Chang MCY (2011) Enzyme mechanism as a kinetic control element for designing synthetic biofuel pathways. Nat Chem Biol 7(4):222–227
Borden JR, Papoutsakis ET (2007) Dynamics of genomic-library enrichment and identification of solvent tolerance genes for Clostridium acetobutylicum. Appl Environ Microbiol 73(9):3061–3068
Borden JR, Jones SW, Indurthi D, Chen Y, Terry Papoutsakis E (2010) A genomic-library based discovery of a novel, possibly synthetic, acid-tolerance mechanism in Clostridium acetobutylicum involving non-coding RNAs and ribosomal RNA processing. Metab Eng 12(3):268–281
Bowring SN, Morris JG (1985) Mutagenesis of Clostridium acetobutylicum. J Appl Microbiol 58(6):577–584
Branduardi P, de Ferra F, Longo V, Porro D (2014) Microbial n-butanol production from Clostridia to non-Clostridial hosts. Eng Life Sci 14(1):16–26
Caspeta L, Buijs NAA, Nielsen J (2013) The role of biofuels in the future energy supply. Energy Environ Sci 6(4):1077–1082
Chu S, Majumdar A (2012) Opportunities and challenges for a sustainable energy future. Nature 488(7411):294–303
Clark SW, Bennett GN, Rudolph FB (1989) Isolation and characterization of mutants of Clostridium acetobutylicum ATCC 824 deficient in acetoacetyl-coenzyme A:acetate/butyrate:coenzyme A-transferase (EC 2.8.3.9) and in other solvent pathway enzymes. Appl Environ Microbiol 55(4):970–976
Collas F, Kuit W, Clement B, Marchal R, Lopez-Contreras AM, Monot F (2012) Simultaneous production of isopropanol, butanol, ethanol and 2,3-butanediol by Clostridium acetobutylicum ATCC 824 engineered strains. AMB Express 2(1):45
Cooksley CM, Zhang Y, Wang H, Redl S, Winzer K, Minton NP (2012) Targeted mutagenesis of the Clostridium acetobutylicum Acetone-Butanol-Ethanol fermentation pathway. Metab Eng 14(6):630–641
Dai Z, Dong H, Zhu Y, Zhang Y, Li Y, Ma Y (2012) Introducing a single secondary alcohol dehydrogenase into butanol-tolerant Clostridium acetobutylicum Rh8 switches ABE fermentation to high level IBE fermentation. Biotechnol Biofuels 5(1):44
Desai RP, Papoutsakis ET (1999) Antisense RNA strategies for metabolic engineering of Clostridium acetobutylicum. Appl Environ Microbiol 65(3):936–945
Desai RP, Harris LM, Welker NE, Papoutsakis ET (1999) Metabolic flux analysis elucidates the importance of the acid-formation pathways in regulating solvent production by Clostridium acetobutylicum. Metab Eng 1(3):206–213
Dietrich JA, McKee AE, Keasling JD (2010) High-throughput metabolic engineering: advances in small-molecule screening and selection. Annu Rev Biochem 79:563–590
Dietrich JA, Shis DL, Alikhani A, Keasling JD (2013) Transcription factor-based screens and synthetic selections for microbial small-molecule biosynthesis. ACS Synth Biol 2(1):47–58
Dong H, Zhang Y, Dai Z, Li Y (2010) Engineering Clostridium strain to accept unmethylated DNA. PLoS One 5(2):e9038
Dong H, Tao W, Dai Z, Yang L, Gong F, Zhang Y, Li Y (2012a) Biobutanol. Adv Biochem Eng Biotechnol 128:85–100
Dong H, Tao W, Zhang Y, Li Y (2012b) Development of an anhydrotetracycline-inducible gene expression system for solvent-producing Clostridium acetobutylicum: a useful tool for strain engineering. Metab Eng 14(1):59–67
Dong H, Tao W, Gong F, Li Y, Zhang Y (2014) A functional recT gene for recombineering of Clostridium. J Biotechnol 173:65–67
Dürre P, Hollergschwandner C (2004) Initiation of endospore formation in Clostridium acetobutylicum. Anaerobe 10(2):69–74
Dusséaux S, Croux C, Soucaille P, Meynial-Salles I (2013) Metabolic engineering of Clostridium acetobutylicum ATCC 824 for the high-yield production of a biofuel composed of an isopropanol/butanol/ethanol mixture. Metab Eng 18:1–8
Ezeji T, Milne C, Price ND, Blaschek HP (2010) Achievements and perspectives to overcome the poor solvent resistance in acetone and butanol-producing microorganisms. Appl Microbiol Biotechnol 85(6):1697–1712
Feustel L, Nakotte S, Dürre P (2004) Characterization and development of two reporter gene systems for Clostridium acetobutylicum. Appl Environ Microbiol 70(2):798–803
Fontaine L, Meynial-Salles I, Girbal L, Yang X, Croux C, Soucaille P (2002) Molecular characterization and transcriptional analysis of adhE2, the gene encoding the NADH-dependent aldehyde/alcohol dehydrogenase responsible for butanol production in alcohologenic cultures of Clostridium acetobutylicum ATCC 824. J Bacteriol 184(3):821–830
Gao K, Li Y, Tian S, Yang X (2012a) Screening and characteristics of a butanol-tolerant strain and butanol production from enzymatic hydrolysate of NaOH-pretreated corn stover. World J Microbiol Biotechnol 28(10):2963–2971
Gao X, Zhao H, Zhang G, He K, Jin Y (2012b) Genome shuffling of Clostridium acetobutylicum CICC 8012 for improved production of acetone-butanol-ethanol (ABE). Curr Microbiol 65(2):128–132
Gheshlaghi R, Scharer JM, Moo-Young M, Chou CP (2009) Metabolic pathways of clostridia for producing butanol. Biotechnol Adv 27(6):764–781
Girbal L, Mortier-Barriere I, Raynaud F, Rouanet C, Croux C, Soucaille P (2003) Development of a sensitive gene expression reporter system and an inducible promoter-repressor system for Clostridium acetobutylicum. Appl Environ Microbiol 69(8):4985–4988
Green EM, Bennett GN (1996) Inactivation of an aldehyde/alcohol dehydrogenase gene from Clostridium acetobutylicum ATCC 824. Appl Biochem Biotechnol 57–58:213–221
Green EM, Boynton ZL, Harris LM, Rudolph FB, Papoutsakis ET, Bennett GN (1996) Genetic manipulation of acid formation pathways by gene inactivation in Clostridium acetobutylicum ATCC 824. Microbiology 142(8):2079–2086
Gronenberg LS, Marcheschi RJ, Liao JC (2013) Next generation biofuel engineering in prokaryotes. Curr Opin Chem Biol 17(3):462–471
Grupe H, Gottschalk G (1992) Physiological events in Clostridium acetobutylicum during the shift from acidogenesis to solventogenesis in continuous culture and presentation of a model for shift induction. Appl Environ Microbiol 58(12):3896–3902
Gu Y, Jiang Y, Wu H, Liu X, Li Z, Li J, Xiao H, Shen Z, Dong H, Yang Y, Li Y, Jiang W, Yang S (2011) Economical challenges to microbial producers of butanol: feedstock, butanol ratio and titer. Biotechnol J 6(11):1348–1357
Harris L, Blank L, Desai RP, Welker NE, Papoutsakis ET (2001) Fermentation characterization and flux analysis of recombinant strains of Clostridium acetobutylicum with an inactivated solR gene. J Ind Microbiol Biotechnol 27(5):322–328
Harris LM, Welker NE, Papoutsakis ET (2002) Northern, morphological, and fermentation analysis of spo0A inactivation and overexpression in Clostridium acetobutylicum ATCC 824. J Bacteriol 184(13):3586–3597
Heap JT, Pennington OJ, Cartman ST, Carter GP, Minton NP (2007) The ClosTron: a universal gene knock-out system for the genus Clostridium. J Microbiol Meth 70(3):452–464
Heap JT, Pennington OJ, Cartman ST, Minton NP (2009) A modular system for Clostridium shuttle plasmids. J Microbiol Meth 78(1):79–85
Heap JT, Kuehne SA, Ehsaan M, Cartman ST, Cooksley CM, Scott JC, Minton NP (2010) The ClosTron: mutagenesis in Clostridium refined and streamlined. J Microbiol Meth 80(1):49–55
Heap JT, Ehsaan M, Cooksley CM, Ng YK, Cartman ST, Winzer K, Minton NP (2012) Integration of DNA into bacterial chromosomes from plasmids without a counter-selection marker. Nucleic Acids Res 40(8):e59
Hönicke D, Janssen H, Grimmler C, Ehrenreich A, Lütke-Eversloh T (2012) Global transcriptional changes of Clostridium acetobutylicum cultures with increased butanol:acetone ratios. N Biotechnol 29(4):485–493
Hou X, Peng W, Xiong L, Huang C, Chen X, Chen X, Zhang W (2013) Engineering Clostridium acetobutylicum for alcohol production. J Biotechnol 166(1–2):25–33
Jang YS, Lee J, Malaviya A, Seung DY, Cho JH, Lee SY (2012a) Butanol production from renewable biomass: rediscovery of metabolic pathways and metabolic engineering. Biotechnol J 7(2):186–198
Jang YS, Lee JY, Lee J, Park JH, Im JA, Eom MH, Lee SH, Song H, Cho JH, Seung do Y, Lee SY (2012b) Enhanced butanol production obtained by reinforcing the direct butanol-forming route in Clostridium acetobutylicum. MBio 3(5):e00314–12
Jang YS, Malaviya A, Cho C, Lee J, Lee SY (2012c) Butanol production from renewable biomass by clostridia. Bioresour Technol 123:653–663
Jang YS, Park JM, Choi S, Choi YJ, Seung DY, Cho JH, Lee SY (2012d) Engineering of microorganisms for the production of biofuels and perspectives based on systems metabolic engineering approaches. Biotechnol Adv 30(5):989–1000
Jang YS, Malaviya A, Lee J, Im JA, Lee SY, Eom MH, Cho JH, Seung DY (2013a) Metabolic engineering of Clostridium acetobutylicum for the enhanced production of isopropanol-butanol-ethanol fuel mixture. Biotechnol Prog 29(4):1083–1088
Jang YS, Malaviya A, Lee SY (2013b) Acetone-butanol-ethanol production with high productivity using Clostridium acetobutylicum BKM19. Biotechnol Bioeng 110(6):1646–1653
Jang YS, Woo HM, Im JA, Kim IH, Lee SY (2013c) Metabolic engineering of Clostridium acetobutylicum for enhanced production of butyric acid. Appl Microbiol Biotechnol 97(21):9355–9363
Jang YS, Im JA, Choi SY, Lee JI, Lee SY (2014) Metabolic engineering of Clostridium acetobutylicum for butyric acid production with high butyric acid selectivity. Metab Eng. doi:10.1016/j.ymben.2014.03.004i
Janssen H, Grimmler C, Ehrenreich A, Bahl H, Fischer RJ (2012) A transcriptional study of acidogenic chemostat cells of Clostridium acetobutylicum—solvent stress caused by a transient n-butanol pulse. J Biotechnol 161(3):354–365
Jia K, Zhang Y, Li Y (2010) Systematic engineering of microorganisms to improve alcohol tolerance. Eng Life Sci 10(5):422–429
Jia K, Zhu Y, Zhang Y, Li Y (2011) Group II intron-anchored gene deletion in Clostridium. PLoS One 6(1):e16693
Jia K, Zhang Y, Li Y (2012) Identification and characterization of two functionally unknown genes involved in butanol tolerance of Clostridium acetobutylicum. PLoS One 7:e38815
Jiang Y, Xu C, Dong F, Yang Y, Jiang W, Yang S (2009) Disruption of the acetoacetate decarboxylase gene in solvent-producing Clostridium acetobutylicum increases the butanol ratio. Metab Eng 11(4–5):284–291
Jin C, Yao M, Liu H, Lee CFF, Ji J (2011) Progress in the production and application of n-butanol as a biofuel. Renew Sust Energ Rev 15(8):4080–4106
Jin L, Zhang H, Chen L, Yang C, Yang S, Jiang W, Gu Y (2014) Combined overexpression of genes involved in pentose phosphate pathway enables enhanced D-xylose utilization by Clostridium acetobutylicum. J Biotechnol 173:7–9
Jones DT, Woods DR (1986) Acetone-butanol fermentation revisited. Microbiol Rev 50(4):484–524
Jones SW, Tracy BP, Gaida SM, Papoutsakis ET (2011) Inactivation of σF in Clostridium acetobutylicum ATCC 824 blocks sporulation prior to asymmetric division and abolishes σE and σG protein expression but does not block solvent formation. J Bacteriol 193(10):2429–2440
Junelles AM, Janati-Idrissi R, Kanouni A, Petitdemange H, Gay R (1987) Acetone-butanol fermentation by mutants selected for resistance to acetate and butyrate halogen analogues. Biotechnol Lett 9(3):175–178
Jurgens G, Survase S, Berezina O, Sklavounos E, Linnekoski J, Kurkijärvi A, Väkevä M, van Heiningen A, Granström T (2012) Butanol production from lignocellulosics. Biotechnol Lett 34(8):1415–1434
Karberg M, Guo H, Zhong J, Coon R, Perutka J, Lambowitz AM (2001) Group II introns as controllable gene targeting vectors for genetic manipulation of bacteria. Nat Biotechnol 19(12):1162–1167
Kellermann SJ, Rentmeister A (2014) Current developments in cellulase engineering. ChemBioEng Rev 1(1):6–13
Kim HJ, Turner TL, Jin Y-S (2013) Combinatorial genetic perturbation to refine metabolic circuits for producing biofuels and biochemicals. Biotechnol Adv 31(6):976–985
Kovács K, Willson BJ, Schwarz K, Heap JT, Jackson A, Bolam DN, Winzer K, Minton NP (2013) Secretion and assembly of functional mini-cellulosomes from synthetic chromosomal operons in Clostridium acetobutylicum ATCC 824. Biotechnol Biofuels 6(1):117
Krutsakorn B, Honda K, Ye X, Imagawa T, Bei X, Okano K, Ohtake H (2013) In vitro production of n-butanol from glucose. Metab Eng 20:84–91
Kuehne SA, Minton NP (2012) ClosTron-mediated engineering of Clostridium. Bioeng Bugs 3(4):247–254
Kuit W, Minton NP, López-Contreras AM, Eggink G (2012) Disruption of the acetate kinase (ack) gene of Clostridium acetobutylicum results in delayed acetate production. Appl Microbiol Biotechnol 94(3):729–741
Kumar M, Gayen K (2011) Developments in biobutanol production: new insights. Appl Energy 88(6):1999–2012
Lee SY, Bennett GN, Papoutsakis ET (1992a) Construction of Escherichia coli-Clostridium acetobutylicum shuttle vectors and transformation of Clostridium acetobutylicum strains. Biotechnol Lett 14(5):427–432
Lee SY, Mermelstein LD, Bennett GN, Papoutsakis ET (1992b) Vector construction, transformation, and gene amplification in Clostridium acetobutylicum ATCC 824. Ann N Y Acad Sci 665:39–51
Lee SY, Park JH, Jang SH, Nielsen LK, Kim J, Jung KS (2008) Fermentative butanol production by clostridia. Biotechnol Bioeng 101(2):209–228
Lee J, Jang YS, Choi SJ, Im JA, Song H, Cho JH, Seung do Y, Papoutsakis ET, Bennett GN, Lee SY (2012) Metabolic engineering of Clostridium acetobutylicum ATCC 824 for isopropanol-butanol-ethanol fermentation. Appl Environ Microbiol 78(5):1416–1423
Lehmann D, Lütke-Eversloh T (2011) Switching Clostridium acetobutylicum to an ethanol producer by disruption of the butyrate/butanol fermentative pathway. Metab Eng 13(5):464–473
Lehmann D, Hönicke D, Ehrenreich A, Schmidt M, Weuster-Botz D, Bahl H, Lütke-Eversloh T (2012a) Modifying the product pattern of Clostridium acetobutylicum: physiological effects of disrupting the acetate and acetone formation pathways. Appl Microbiol Biotechnol 94(3):743–754
Lehmann D, Radomski N, Lütke-Eversloh T (2012b) New insights into the butyric acid metabolism of Clostridium acetobutylicum. Appl Microbiol Biotechnol 96(5):1325–1339
Lemmel SA (1985) Mutagenesis in Clostridium acetobutylicum. Biotechnol Lett 7:711–716
Li F, Hinderberger J, Seedorf H, Zhang J, Buckel W, Thauer RK (2008) Coupled ferredoxin and crotonyl coenzyme A (CoA) reduction with NADH catalyzed by the butyryl-CoA dehydrogenase/Etf complex from Clostridium kluyveri. J Bacteriol 190:843–850
Li Z, Xiao H, Jiang W, Jiang Y, Yang S (2013) Improvement of solvent production from xylose mother liquor by engineering the xylose metabolic pathway in Clostridium acetobutylicum EA 2018. Appl Biochem Biotechnol 171(3):555–568
Li HG, Luo W, Wang Q, Yu XB (2014) Direct fermentation of gelatinized cassava starch to acetone, butanol, and ethanol using Clostridium acetobutylicum mutant obtained by atmospheric and room temperature plasma. Appl Biochem Biotechnol. doi:10.1007/s12010-014-0765-x
Lim JH, Seo SW, Kim SY, Jung GY (2013) Model-driven rebalancing of the intracellular redox state for optimization of a heterologous n-butanol pathway in Escherichia coli. Metab Eng 20:56–62
Linhová M, Branská B, Patáková P, Lipovsky J, Fribert P, Rychtera M, Melzoch K (2012) Rapid flow cytometric method for viability determination of solventogenic clostridia. Folia Microbiol 57(4):307–311
Liu S, Qureshi N (2010) How microbes tolerate ethanol and butanol. N Biotechnol 26(3–4):117–121
Liu L, Zhang L, Tang W, Gu Y, Hua Q, Yang S, Jiang W, Yang C (2012a) Phosphoketolase pathway for xylose catabolism in Clostridium acetobutylicum revealed by 13C metabolic flux analysis. J Bacteriol 194(19):5413–5422
Liu XB, Gu QY, Yu XB, Luo W (2012b) Enhancement of butanol tolerance and butanol yield in Clostridium acetobutylicum mutant NT642 obtained by nitrogen ion beam implantation. J Microbiol 50(6):1024–1028
Luan G, Cai Z, Gong F, Dong H, Lin Z, Zhang Y, Li Y (2013) Developing controllable hypermutable Clostridium cells through manipulating its methyl-directed mismatch repair system. Protein Cell 4(11):854–862
Lütke-Eversloh T, Bahl H (2011) Metabolic engineering of Clostridium acetobutylicum: recent advances to improve butanol production. Curr Opin Biotechnol 22(5):634–647
Lütke-Eversloh T, Stephanopoulos G (2005) Feedback inhibition of chorismate mutase/prephenate dehydrogenase (TyrA) of Escherichia coli: generation and characterization of tyrosine-insensitive mutants. Appl Environ Microbiol 71(11):7224–7228
Mann MS, Lütke-Eversloh T (2013) Thiolase engineering for enhanced butanol production in Clostridium acetobutylicum. Biotechnol Bioeng 110(3):887–897
Mann MS, Dragovic Z, Schirrmacher G, Lütke-Eversloh T (2012) Over-expression of stress protein-encoding genes helps Clostridium acetobutylicum to rapidly adapt to butanol stress. Biotechnol Lett 34(9):1643–1649
Mao S, Luo Y, Zhang T, Li J (2010) Proteome reference map and comparative proteomic analysis between a wild type Clostridium acetobutylicum DSM 1731 and its mutant with enhanced butanol tolerance and butanol yield. J Proteome Res 9(6):3046–3061
Mao S, Luo Y, Bao G, Zhang Y, Li Y, Ma Y (2011) Comparative analysis on the membrane proteome of Clostridium acetobutylicum wild type strain and its butanol-tolerant mutant. Mol BioSyst 7(5):1660–1677
Mariano AP, Filho RM (2012) Improvements in biobutanol fermentation and their impacts on distillation energy consumption and wastewater generation. Bioenergy Res 5(2):504–514
Mascal M (2012) Chemicals from biobutanol: technologies and markets. Biofuels Bioprod Bioref 6(4):483–493
Medkor N, Zerdani I, Sattar S (2010) Isolation of Clostridium acetobutylicum ATCC824 mutants using propionic and isovaleric acid halogen analogues as suicide substrates. Int J Microbiol Res 1(1):22–25
Mermelstein LD, Papoutsakis ET (1993) In vivo methylation in Escherichia coli by the Bacillus subtilis phage ϕ3T I methyltransferase to protect plasmids from restriction upon transformation of Clostridium acetobutylicum ATCC 824. Appl Environ Microbiol 59(4):1077–1081
Mermelstein LD, Welker NE, Bennett GN, Papoutsakis ET (1992) Expression of cloned homologous fermentative genes in Clostridium acetobutylicum ATCC 824. Nat Biotechnol 10(2):190–195
Mermelstein LD, Papoutsakis ET, Petersen DJ, Bennett GN (1993) Metabolic engineering of Clostridium acetobutylicum ATCC 824 for increased solvent production by enhancement of acetone formation enzyme activities using a synthetic acetone operon. Biotechnol Bioeng 42(9):1053–1060
Moholkar VS, Ranjan A, Mayank R (2012) Economics of biobutanol: a review. Res J Pharm Biol Chem Sci 3(4):901–913
Nair RV, Green EM, Watson DE, Bennett GN, Papoutsakis ET (1999) Regulation of the sol locus genes for butanol and acetone formation in Clostridium acetobutylicum ATCC 824 by a putative transcriptional repressor. J Bacteriol 181(1):319–330
Ni Y, Sun Z (2009) Recent progress on industrial fermentative production of acetone-butanol-ethanol by Clostridium acetobutylicum in China. Appl Microbiol Biotechnol 83(3):415–423
Nicolaou SA, Gaida SM, Papoutsakis ET (2010) A comparative view of metabolite and substrate stress and tolerance in microbial bioprocessing: from biofuels and chemicals, to biocatalysis and bioremediation. Metab Eng 12(4):307–331
Oultram JD, Loughlin M, Swinfield TJ, Brehm JK, Thompson DE, Minton NP (1988) Introduction of plasmids into whole cells of Clostridium acetobutylicum by electroporation. FEMS Microbiol Lett 56(1):83–88
Papoutsakis ET (2008) Engineering solventogenic clostridia. Curr Opin Biotechnol 19(5):420–429
Paredes CJ, Alsaker KV, Papoutsakis ET (2005) A comparative genomic view of clostridial sporulation and physiology. Nat Rev Microbiol 3(12):969–978
Pasteur L (1862) Quelques résultats nouveaux relatifs aux fermentations acétique et butyrique. Bulletin de la Société Chimique de Paris:52-53
Patáková P, Linhová M, Rychtera M, Paulova L, Melzoch K (2013) Novel and neglected issues of acetone-butanol-ethanol (ABE) fermentation by clostridia: Clostridium metabolic diversity, tools for process mapping and continuous fermentation systems. Biotechnol Adv 31(1):58–67
Ranjan A, Moholkar VS (2012) Biobutanol: science, engineering, and economics. Int J Energy Res 36(3):277–323
Rellos P, Ma J, Scopes RK (1997) Alteration of substrate specificity of Zymomonas mobilis alcohol dehydrogenase-2 using in vitro random mutagenesis. Protein Expr Purif 9(1):83–90
Ren C, Gu Y, Hu S, Wu Y, Wang P, Yang Y, Yang C, Yang S, Jiang W (2010) Identification and inactivation of pleiotropic regulator CcpA to eliminate glucose repression of xylose utilization in Clostridium acetobutylicum. Metab Eng 12(5):446–454
Ren C, Gu Y, Wu Y, Zhang W, Yang C, Yang S, Jiang W (2012) Pleiotropic functions of catabolite control protein CcpA in butanol-producing Clostridium acetobutylicum. BMC Genomics 13(1):349
Rothstein DM (1986) Clostridium thermosaccharolyticum strain deficient in acetate production. J Bacteriol 165:319–320
Sanchez S, Demain AL (2008) Metabolic regulation and overproduction of primary metabolites. Microb Biotechnol 1(4):283–319
Scheel M, Lütke-Eversloh T (2013) New options to engineer biofuel microbes: development and application of a high-throughput screening system. Metab Eng 17C:51–58
Scotcher MC, Bennett GN (2005) SpoIIE regulates sporulation but does not directly affect solventogenesis in Clostridium acetobutylicum ATCC 824. J Bacteriol 187(6):1930–1936
Servinsky MD, Germane KL, Liu S, Kiel JT, Clark AM, Shankar J, Sund CJ (2012) Arabinose is metabolized via a phosphoketolase pathway in Clostridium acetobutylicum ATCC 824. J Ind Microbiol Biotechnol 39(12):1859–1867
Shao L, Hu S, Yang Y, Gu Y, Chen J, Jiang W, Yang S (2007) Targeted gene disruption by use of a group II intron (Targetron) vector in Clostridium acetobutylicum. Cell Res 17(11):963–965
Shen CR, Lan EI, Dekishima Y, Baez A, Cho KM, Liao JC (2011) Driving forces enable high-titer anaerobic 1-butanol synthesis in Escherichia coli. Appl Environ Microbiol 77(9):2905–2915
Sillers R, Al-Hinai MA, Papoutsakis ET (2009) Aldehyde-alcohol dehydrogenase and/or thiolase overexpression coupled with CoA transferase downregulation lead to higher alcohol titers and selectivity in Clostridium acetobutylicum fermentations. Biotechnol Bioeng 102(1):38–49
Soucaille P, Figge R, Croux C (2008) Process for chromosomal integration and DNA sequence replacement in clostridia. International Patent WO 2008/040387
Steiner E, Dago AE, Young DI, Heap JT, Minton NP, Hoch JA, Young M (2011) Multiple orphan histidine kinases interact directly with Spo0A to control the initiation of endospore formation in Clostridium acetobutylicum. Mol Microbiol 80(3):641–654
Tangney M, Mitchell WJ (2007) Characterisation of a glucose phosphotransferase system in Clostridium acetobutylicum ATCC 824. Appl Microbiol Biotechnol 74(2):398–405
Thomas L, Joseph A, Gottumukkala LD (2014) Xylanase and cellulase systems of Clostridium sp.: an insight on molecular approaches for strain improvement. Bioresour Technol. doi:10.1016/j.biortech.2014.01.140
Thomason MK, Storz G (2010) Bacterial antisense RNAs: how many are there, and what are they doing? Annu Rev Genet 44:167–188
Thormann K, Dürre P (2001) Orf5/SolR: a transcriptional repressor of the sol operon of Clostridium acetobutylicum? J Ind Microbiol Biotechnol 27(5):307–313
Thormann K, Feustel L, Lorenz K, Nakotte S, Dürre P (2002) Control of butanol formation in Clostridium acetobutylicum by transcriptional activation. J Bacteriol 184(7):1966–1973
Tomas CA, Welker NE, Papoutsakis ET (2003) Overexpression of groESL in Clostridium acetobutylicum results in increased solvent production and tolerance, prolonged metabolism, and changes in the cell's transcriptional program. Appl Environ Microbiol 69(8):4951–4965
Tomas CA, Beamish J, Papoutsakis ET (2004) Transcriptional analysis of butanol stress and tolerance in Clostridium acetobutylicum. J Bacteriol 186(7):2006–2018
Tracy BP, Gaida SM, Papoutsakis ET (2008) Development and application of flow-cytometric techniques for analyzing and sorting endospore-forming clostridia. Appl Environ Microbiol 74(24):7497–7506
Tracy BP, Gaida SM, Papoutsakis ET (2010) Flow cytometry for bacteria: enabling metabolic engineering, synthetic biology and the elucidation of complex phenotypes. Curr Opin Biotechnol 21(1):85–99
Tracy BP, Jones SW, Papoutsakis ET (2011) Inactivation of σE and σG in Clostridium acetobutylicum illuminates their roles in clostridial-cell-form biogenesis, granulose synthesis, solventogenesis, and spore morphogenesis. J Bacteriol 193(6):1414–1426
Tracy BP, Jones SW, Fast AG, Indurthi DC, Papoutsakis ET (2012) Clostridia: the importance of their exceptional substrate and metabolite diversity for biofuel and biorefinery applications. Curr Opin Biotechnol 23(3):364–381
Truffaut N, Hubert J, Reysset G (1989) Construction of shuttle vectors useful for transforming Clostridium acetobutylicum. FEMS Microbiol Lett 58(1):15–20
Tummala SB, Welker NE, Papoutsakis ET (1999) Development and characterization of a gene expression reporter system for Clostridium acetobutylicum ATCC 824. Appl Environ Microbiol 65(9):3793–3799
Tummala SB, Junne SG, Papoutsakis ET (2003a) Antisense RNA downregulation of coenzyme A transferase combined with alcohol-aldehyde dehydrogenase overexpression leads to predominantly alcohologenic Clostridium acetobutylicum fermentations. J Bacteriol 185(12):3644–3653
Tummala SB, Welker NE, Papoutsakis ET (2003b) Design of antisense RNA constructs for downregulation of the acetone formation pathway of Clostridium acetobutylicum. J Bacteriol 185(6):1923–1934
Ventura JRS, Hu H, Jahng D (2013) Enhanced butanol production in Clostridium acetobutylicum ATCC 824 by double overexpression of 6-phosphofructokinase and pyruvate kinase genes. Appl Microbiol Biotechnol 97(16):7505–7516
Wang Y, Li X, Milne CB, Janssen H, Lin W, Phan G, Hu H, Jin YS, Price ND, Blaschek HP (2013) Development of a gene knockout system using mobile group II introns (Targetron) and genetic disruption of acid production pathways in Clostridium beijerinckii. Appl Environ Microbiol 79(19):5853–5863
Wiesenborn D, Rudolph F, Papoutsakis E (1988) Thiolase from Clostridium acetobutylicum ATCC 824 and its role in the synthesis of acids and solvents. Appl Environ Microbiol 54(11):2717–2722
Wietzke M, Bahl H (2012) The redox-sensing protein Rex, a transcriptional regulator of solventogenesis in Clostridium acetobutylicum. Appl Microbiol Biotechnol 96(3):749–761
Willims DR, Young DI, Young M (1990) Conjugative plasmid transfer from Escherichia coli to Clostridium acetobutylicum. J Gen Microbiol 136(5):819–826
Woolston BM, Edgar S, Stephanopoulos G (2013) Metabolic engineering: past and future. Annu Rev Chem Biomol Eng 4:259–288
Xiao H, Gu Y, Ning Y, Yang Y, Mitchell WJ, Jiang W, Yang S (2011) Confirmation and elimination of xylose-metabolic bottlenecks in glucose-PTS-deficient Clostridium acetobutylicum to realize simultaneous utilization of glucose, xylose and arabinose. Appl Environ Microbiol 77(22):7886–7895
Xue C, Zhao X-Q, Liu C-G, Chen L-J, Bai F-W (2013) Prospective and development of butanol as an advanced biofuel. Biotechnol Adv 31(8):1575–1584
Xue C, Zhao J-B, Chen L-J, Bai F-W, Yang S-T, Sun J-X (2014) Integrated butanol recovery for an advanced biofuel: current state and prospects. Appl Microbiol Biotechnol. doi:10.1007/s00253-014-5561-6
Yang H, Li J, Shin H-d DG, Liu L, Chen J (2014) Molecular engineering of industrial enzymes: recent advances and future prospects. Appl Microbiol Biotechnol 98(1):23–29
Young M, Minton NP, Staudenbauer WL (1989) Recent advances in the genetics of the clostridia. FEMS Microbiol Rev 63(4):301–325
Zhang YX, Perry K, Vinci VA, Powell K, Stemmer WPC, Del Cardayré SB (2002) Genome shuffling leads to rapid phenotypic improvement in bacteria. Nature 415(6872):644–646
Zhao Y, Hindorff LA, Chuang A, Monroe-Augustus M, Lyristis M, Harrison ML, Rudolph FB, Bennett GN (2003) Expression of a cloned cyclopropane fatty acid synthase gene reduces solvent formation in Clostridium acetobutylicum ATCC 824. Appl Environ Microbiol 69(5):2831–2841
Zhu L, Dong H, Zhang Y, Li Y (2011) Engineering the robustness of Clostridium acetobutylicum by introducing glutathione biosynthetic capability. Metab Eng 13(4):426–434
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Lütke-Eversloh, T. Application of new metabolic engineering tools for Clostridium acetobutylicum . Appl Microbiol Biotechnol 98, 5823–5837 (2014). https://doi.org/10.1007/s00253-014-5785-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-014-5785-5