Abstract
It was previously proposed that regiospecific hydroxylation of daidzein at 3′-position is mediated by cytochrome P450 hydroxylase (CYP105D7) in the presence of putidaredoxin (CamB) and putidaredoxin reductase (CamA) as electron transfer proteins from Pseudomonas putida. The genome sequence of Streptomyces avermitilis MA4680 revealed 33 P450 (CYPs) with 6 ferredoxin reductases (Fprs) and 9 ferredoxins (Fdxs) as their putative electron transfer partner proteins. To identify right endogenous electron transfer proteins for CYP105D7 activity, in vitro reconstitution, gene disruption, and quantitative reverse transcription polymerase chain reaction (qRT-PCR) mRNA expression profile analysis were examined. The most effective electron transfer proteins for CYP105D7 appear to be FdxH (SAV7470), which is located downstream to CYP105D7 as a cluster, and FprD (SAV5675). Throughout our overall analysis, we proposed that the primary electron transfer pathway for CYP105D7 follows as such NAD(P)H→FdxH→FprD→CYP105D7.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The cytochrome P450 superfamily is a large and diverse group of enzymes that catalyze the hydroxylation, epoxidation, dealkylation, or sulfoxidation reaction with a broad range of substrate. (Degtyarenko and Archakov 1993). Most P450s depend on additional electron transfer protein to shuttle electrons from NAD(P)H to their active site. Type I (the mitochondrial/bacterial cytochrome P450 system) is predominant in prokaryotes (Bernhardt 2006). In this system, the electrons required for CYP reaction are delivered from NAD(P)H to CYP via ferredoxin reductase (Fpr) and ferredoxin (Fdx) (Werck and Feyereisen 2000). Although there are various orthologs of ferredoxin and ferredoxin reductase present in other bacterial strains, heterologous reconstruction of the electron transfer partners in other host system is often ineffective (McLean et al. 2005; Hussain and Ward 2003), suggesting that employing the right electron transfer partners is critical to obtain a high CYP activity.
To fulfill the role as redox partners, electron transfer reaction from NAD(P)H to CYP via ferredoxin reductase and/or ferredoxin should be valid and effective. Otherwise, the coupling efficiency of the electron transfer reactions among the three components is too low to observe any active CYP reaction. Usually, the Fdx component of any given CYP system is reduced by Fpr, which in turn takes up electrons from NAD(P)H (Hannemann et al. 2007; Lewis and Hlavica 2000). To achieve the maximum CYP activity, amplification of its functional or natural redox partners should be done, if its genomic sequence information is available. However, identification of correct redox partners is still quite challenging, since the numbers of Fdx and Fpr are not the same and one to one correspondent, and functional redox partner genes are often located at the genomic loci quite distant from the target CYP genes (Hanukoglu 1996). Streptomyces avermitilis genome sequence revealed 33 P450s, six ferredoxin reductases (fprs), and nine ferredoxins (fdxs) (Ikeda et al. 2003). Among them, four ferredoxins are of particular interest in the P450s of S. avermitilis. The four ferredoxins encoded by pteE, fdxA, fdxB, and fdxH are immediately adjacent to corresponding P450s (i.e., pteD (CYP105D6), cyp3 (CYP147B1), cyp7 (CYP105Q1), and cyp28 (CYP105D7), respectively). Until now, all the ferredoxins including FdxH have not been characterized.
Relatively few electron transport pathways to P450s have been characterized in bacteria, particularly in Streptomyces species (Chun et al. 2007; Khatri et al. 2010; Zhu et al. 2011). Most determinations of the catalytic activities of Streptomyces CYPs have come from the studies using the commercially available spinach Fpr and in vitro reconstitution of each redox partners (Sevrioukova and Poulos 2011). CYP105D7 which was previously characterized as daidzein 3′-hydroxylase from S. avermitilis was also reported to be involved in the conversion of 1-deoxypentalenic acid to pentalenic acid (Takamatsu et al. 2011). Pentalenic acid is a shunt metabolite in the biosynthesis of the pentalenolactone family of metabolites in S. avermitilis. Owing to such diverse metabolic functions of CYP105D7 enzyme, identification of its natural electron transfer proteins from host cell is critical to optimize its activity either in S. avermitilis native host or in another heterologous host system.
Here, to find out the potential Fdx and Fpr controlling the CYP105D7 activity, we have presented combined approaches using inactivation of corresponding electron transfer genes, in vitro reconstitution assay, and quantitative reverse transcription polymerase chain reaction (qRT-PCR) mRNA expression profile upon addition of substrate. Our results demonstrated that although several Fpr and Fdx appear to be involved in activation of CYP105D7, FprD (SAV5675) would be the major and potent Fpr for CYP105D7.
Material and methods
Bacterial strains and plasmids
S. avermitilis MA4680 was obtained from the Korea Collection for Type Culture (KCTC, Daejeon, South Korea). The genomic DNA of the strain was prepared by G-spin Genomic DNA Extraction Kit (Intron, Seoul, South Korea) and used as a template for PCR. The cyp105D7, fprD, and fdxH genes were amplified by PCR using a set of specific primers (Supplementary Table S1). The shuttle plasmid for transformation into S. avermitilis was constructed using E. coli JM110 (Novagen, Madison, WI, USA). Disruption of fprs was performed using a plasmid pSuperCos1 to deliver the corresponding apramycin resistance gene cassettes (Supplementary Fig. S1).
Protein expression, purification, and in vitro reconstitution of CYP105D7
S. avermitilis CYP105D7, Fpr, and Fdx proteins were cloned into E. coli expression vector pET28a+ (Novagen, Madison, WI, USA) with N-terminal His-tag. All the strains were cultured in terrific broth media supplemented with 0.2 % bactopeptone and trace element. Induction was carried out when OD600 reaches to 0.8–1.0 with 1 mM isopropyl β-d-thiogalactopyranoside (IPTG). In the case of FprA, FprB, FprC, and FprF, 1 mM IPTG induction at 25 °C for 24 h gives the highest yield of the soluble proteins, whereas in the case of FprD and FprE, the same induction strategy at 30 °C gives the highest protein expression level. Purification of the CYP105D7, Fpr, and Fdx was performed as described previously (Pandey et al. 2010), in this way purified protein have N-terminal His-tag. All the enzyme assays were carried out with the His-tagged purified protein.
In vitro reconstitution of daidzein hydroxylation to 7,3′,4′-trihydroxyisoflavone (3′-ODI) was carried out with autologous electron transfer proteins. For these experiments, the reaction was performed in a final volume of 0.5 ml that contained 1 μM CYP105D7, 10 μM FdxH, each 3 μM Fpr, and 20 μM daidzein in 50 mM Tris buffer at pH 7.5. In addition to this, NAD(P)H regeneration system, consisting of 5 mM of glucose 6-phosphate, 0.5 unit of glucose-6-phosphate dehydrogenase, and 1 mM of magnesium chloride was also applied. After the preincubation of the mixture at 30 °C in the Eppendorf Thermomixer ,the reaction was started by adding NAD(P)H to 500 μM of final concentration, and the mixture was incubated for 1 h. The reaction was stopped by adding an equal amount of ethyl acetate. The product was extracted with an equal amount of ethyl acetate for three times, and the extractant was vacuum dried.
Recombinant S. avermitilis, culture conditions, and reaction product isolation
S. avermitilis MA4680 was cultivated in R2YE medium, and recombinant strains for the expression of each gene into S. avermitilis were constructed through the polyethyleneglycol (PEG)-associated transformation (Stutzman-Engwall et al. 2006). Isolation of reaction metabolites S. avermitilis cells was carried out as described previously (Pandey et al. 2010). The extracted sample was evaporated in a centrifugal vacuum concentrator (Biotron, South Korea). The reactant was analyzed using HPLC and gas chromatography-mass spectrometry (GC-MS) as previously described (Pandey et al. 2011).
Gene expression analysis by RT-PCR
RNA extraction was carried out with the RNeasy Mini Kit (Qiagen, USA), according to the manufacturer’s instructions. Synthesis of cDNA was carried using the SuperScript III First-Strand Synthesis System for reverse transcription polymerase chain reaction RT-PCR (Invitrogen) with 1 μg of total RNA and oligo dT primers (Supplementary Table S2). Gene expression was analyzed using the real-time RT-PCR with SYBR Green Master Mix reagents. The expression level of P450, Fpr, and Fdx was detected. Real-time RT-PCR was performed with a LightCycler 480 Real-Time PCR System (Roche, Indianapolis, IN, USA).
Result
Effect of Fpr deletion on daidzein ortho-hydroxylation
S. avermitilis MA4680 CYPs are of special interest, as the strain exhibits high activity of regiospecific hydroxylation towards (iso)flavonoids such as daidzein, genistein, apigenin, and chrysin as well as the biosynthesis of pentalenolactone (Pandey et al. 2011). The genome analysis provides some interesting insights into the potential redox partners for the S. avermitilis CYPs. Potential flavodoxin partners corresponding to specific CYPs are not obvious in S. avermitilis genome, since only nine fdxs and six fprs are distributed in the genome (Lamb et al. 2003).
A complete picture of the interactions among Fpr, Fdx, and CYP could be deduced by in vitro activity assay and gene expression analysis under specific reaction conditions, which require quite accurate means of identifying optimal partners. According to the complete S. avermitilis genome analysis, fdxH (sav7470) is located downstream next to cyp105D7 (sav7469), so that, it is likely to be the main functional Fdx to transfer electrons from appropriate Fpr. However, it should be further confirmed in detail.
In order to find out the functional Fpr during daidzein biotransformation, all of the six fpr deletion mutants were constructed, and daidzein biotransformation rates by each deletion mutant were measured. Among the six fpr deletion mutants, five showed no noticeable differences in their phenotypes compared to that of the wild type in complex R2YE agar plate; whereas fprE (sav5675) did not produce black-brown pigment. The cell growth of the six fpr deletion mutants was almost the same.
To evaluate the effect of the fpr deletion mutant on the daidzein biotransformation, all the deletion mutants were cultured in complex R2YE media and compared for their ability for daidzein ortho-dihydroxylation (Table 1). The structural and quantification analyses of oxidative metabolites by each fpr deletion mutant were done using GC-MS and HPLC. Among the six fpr deletion mutants, the two fpr mutants (∆fprD and ∆fprE) showed the lowest activity towards the daidzein for 3′-ODI production, suggesting that either FprD or FprE, would be critical to the daidzein biotransformation using the recombinant S. avermitilis-expressing CYP105D7. In order to confirm that FprD and FprE are the major electron-donating proteins to FdxH, further in vitro reconstitution of CYP105D7-FdxH-FprD and CYP105D7-FdxH-FprE were followed by the measurement of their catalytic activities.
qRT-PCR mRNA expression profile by substrate induction
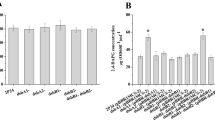
Since daidzein induces the expression of several CYPs, especially sav1611, sav412, and sav7469 which are involved in daidzein hydroxylation reaction (Supplementary Fig. S2), it is possible that different levels of induction profile of CYPs and redox partners would change the contribution of each redox partners in achieving the same final conversion yield of 3′-ODI. To avoid such ambiguities caused by Fpr and Fdx inductions for CYP105D7, mRNA expression levels of Fpr and Fdx genes were compared using qRT-PCR. Cell culture broth was induced with daidzein for a period of time, and total RNA of S. avermitilis cells was prepared from the samples with and without daidzein induction at 0, 6, 12, and 24 h time points. The effects of daidzein induction on the transcription level of fprs and fdxs were quantitatively measured through qRT-PCR analysis. The result showed that the induction of fprs and fdxs were observed after 6 h of the daidzein induction and several fprs and fdxs were induced at the same time. Among the fprs, fprA (sav583), fprD (sav5675), and fprF (sav6956) showed the relatively higher mRNA profile, and fprA showed the highest expressed level, and fprD was also greatly induced under the daidzein addition, whereas fprC (sav1609), fprB (sav1507), and fprE (sav6097) showed almost no changes in their mRNA expression level upon daidzein addition (Fig. 1a). Looking at the time-dependent mRNA profiles, it takes about12 h for the fpr genes to be highly induced, so that 12-h samples showed the highest mRNA profile in all the cases. It is well known that almost one third (11 genes) of all the CYP genes in S. avermitilis are involved in the biosynthesis of secondary metabolites (Martinez et al. 2004). Since Fprs and Fdxs have to support electron transfer systems for the CYPs functions, their expressions are expected to be well coordinated with the expression of CYPs. However, as their numbers are not one to one correspondent, the same families of CYPs might share their redox partners. From that perspective, when cyp105D7 is induced, one of the induced fprs (i.e., fprA, fprD, and fprF) is more likely to become a cognate partner of CYP105D7. However, it was not clear to pinpoint one of them using this result. When we combine this result with the evaluation of the fpr deletion mutants and the reconstitution experiments, it is quite certain that FprD is the most important major electron transfer partner of CYP105D7. In terms of the selection of fdx partner, the genetic locus of fdxH is the most critical clue to be the major partner of CYP105D7 (SAV7469). According to the induction profile, fdxD (sav3129) and fdxH (sav7470) showed the highest mRNA expression profile throughout the induction time points; however, fdxD appeared to be constitutively induced from a 0-h time point both with and without daidzein, whereas fdxH showed a very slight daidzein induction effect on mRNA profile, although the induction level is not that high (Fig. 1b). Summing up, fdxH located just downstream to cyp105D7 appears to be weakly co-induced with CYP105D7.
mRNA expression profile by qRT-PCR analysis of fdx and fpr in S. avermitilis. a Relative mRNA expression profile of ferredoxin reductases (fprs) in S. avermitilis at 0-, 6-, 12-, and 24-h time intervals without and with daidzein induction. b Relative mRNA expression profile of ferredoxins (fdxs) in S. avermitilis at 0-, 6-, 12-, and 24-h time intervals without and with daidzein induction. A, wild-type S. avermitilis MA4680 without daidzein; and B, wild-type S. avermitilis MA4680 with daidzein induction
Expression, purification, and spectral characterization of CYP105D7, Fpr, and Fdx
S. avermitilis CYP105D7, Fpr, and Fdx proteins were expressed in E. coli expression vector pET28a+ (Novagen, Madison, WI, USA) with N-terminal His-tag as mentioned in the “Material and methods.” They were purified for in vitro reconstitution, and the purified proteins were subjected to SDS-PAGE analysis (Supplementary Fig. S3).
The spectral properties of the purified Fpr and CYP105D7 were confirmed through UV-Vis spectra. The spectrum of the oxidized form of CYP105D7 showed a Soret peak at 420 nm, indicating that the heme iron of CYP105D7 is in a low-spin state. The recombinant CYP105D7 showed normal spectral characteristics of a Soret peak at 450 nm after CO gas blowing, indicating the reduced carbon monoxide-bound form. The concentration of the P450s was estimated by CO difference spectra assuming Δ(ε 450–490) = 91 mM−1 cm−1. For the activities of ferredoxin reductases, the purified Fprs were electrophoretically homogeneous and showed the expected flavin spectra (Fig. 2a, b). Spectral properties of ferredoxin reductase showed the local maxima at 369 and 448 nm as well with pronounced shoulder at 473 nm. The concentrations of Fprs were estimated from flavin absorption spectra, using a general ε 450 value of 11.3 mM−1 cm−1.
a Spectral features of CYP105D7. Dotted line, oxidized CYP105D7; long dashed line, reduced form with sodium dithionite; solid line, CO-bound form with dithionite. The insert represents the CO difference spectra. b UV-visible spectra of purified FprD; the spectra were recorded at ambient temperature in 50 mM Tris buffer (pH 7.5)
Dependency of electron donors for Fpr: measurement of electron transfer rate to artificial acceptor
Preference of the Fprs towards NADH or NADPH was determined by measuring the NAD(P)H oxidation rates using nitro blue tetrazolium chloride (NBT) as an artificial electron acceptor and NAD(P)H as an electron donor. For measurement of the rates of NBT reduction, the reactions were done in 50 mM Tris buffer (pH 7.5) containing 100 nM Fpr and 200 μM NBT. Reactions were initiated with the addition of 200 μM NAD(P)H and followed by the increase in absorbance at 535 nm. Concentration changes were calculated using an extinction coefficient of 18.3 mM−1 cm−1 for NBT. The relative efficiencies of the electron transfer rate to artificial electron acceptor NBT are shown in Fig. 3. All the Fprs showed catalytic activities for reducing the artificial electron acceptor NBT and showed a somewhat higher selectivity to NADH over NADPH, except FprD which showed a slightly higher preference towards NADPH. Another interesting thing to remark is that FprE showed much higher selectivity towards NADH, indicating that FprE activity is mainly dependent on the presence of NADH rather than NADPH. However, all the other Fprs appear to accept both NADH and NADPH at similar rates.
In vitro reconstitution of CYP105D7 with S. avermitilis electron transfer proteins
In vitro reconstitution of CYP105D7 and redox proteins for measuring their catalytic activities were examined using the reaction mixture containing CYP105D7 (1 μM), FdxH (10 μM), each Fpr (3 μM), and substrate daidzein (20 μM) in 50 mM Tris buffer having pH 7.5, containing NAD(P)H regeneration system as described in “Material and methods.” The product 3′-ODI was clearly detected by HPLC analysis. However, no product was observed in the absence of NAD(P)H. The daidzein hydroxylation rates by CYP105D7 with its autologous electron transfer counterparts were shown in Fig. 4a. The result showed that the only FprD in combination with FdxH converted daidzein to 3′-ODI in the in vitro reconstitution experiments with CYP105D7, suggesting that FprD is the natural Fpr transferring electrons the most efficiently to CYP105D7.
Reconstitution of CYP105D7, FprD, and FdxH/D for daidzein hydroxylation reactions. a In vitro reconstitution of CYP105D7 for daidzein hydroxylation. Twenty micromolars of daidzein was reacted with the reconstituted enzyme mixtures at a fixed concentration ratio of CYP105D7/Fpr/FdxH, i.e., 1:3:10 (micromolars). b Hydroxylation of daidzein by the in vitro reconstitution of CYP105D7, Fpr, and FdxD at 20 μM of each substrate was reacted with the reconstituted enzyme mixture at an elevated concentration of CYP105D7/Fpr/FdxD, i.e., 1:10:20 (micromolars). As a control, reactions were made without NADPH in the reaction mixture
In order to understand the preference of FprD towards FdxH rather than FdxD for CYP105D7 activity, further reactions were repeated with FdxD, by increasing the concentrations of the corresponding Fpr and FdxD. The reaction carried out enhanced Fpr and FdxD concentration ratio of CYP105D7:Fpr:Fdx (1:10:20 μM). The reason why we only show the case of FdxD is based on the qRT-PCR mRNA expression profile by daidzein induction (shown in the next section), where daidzein hydroxylation activity was observed with FprD and FdxD combination. However, the rate of product formation is still lower than that of FprD and FdxH as shown in Fig. 4b.
This result clearly indicates that FprD interact more efficiently with FdxH than FdxD. One plausible explanation of such difference in the efficiency of electron transfer might come from the difference in their protein structures, such that the FdxH and FdxD are [2Fe–2S] and [4Fe–4S] cluster proteins, respectively. The exact understanding of the mode of interaction of FdxH [2Fe–2S] and FdxD [4Fe–4S] cluster protein with FprD would be possible through the analysis of direct protein–protein interaction study using mass spectrometry or indirect measurement of electron coupling efficiency. However, our reaction study suggests that FprD interaction is more efficient with [2Fe–2S] than [4Fe–4S] cluster protein.
Ortho-hydroxylation of daidzein to 3′-ODI in S. avermitilis
The data of gene inactivation, in vitro reconstitution, and qRT-PCR mRNA profile analysis presented above clearly demonstrated that FprD (SAV5675) is the major reductase to control the CYP105D7 activity in cooperation with FdxH. In result, FprD-FdxH-CYP105D7 was constructed in pIBR25 expression vector under the control of ermE* promoter to be use in S. avermitilis as a host system. Reduced carbon monoxide difference spectra of the soluble fraction of S. avermitilis strain containing each transformant showed a peak absorbance at 450 nm, which was not observed in the extracts of the control culture of wild-type S. avermitilis. This clearly indicated that CYP105D7 was functionally well expressed in S. avermitilis system. Furthermore, the transformation of the construct pIBR25 in wild-type S. avermitilis was confirmed through the real-time PCR analysis of the transformant (Supplementary Fig. S4 (A–B)).
The catalytic activity of CYP105D7 for daidzein in the presence of FprD was investigated in in vivo S. avermitilis cell system. Cells were harvested, and the same amount of the wet weight of the cell was used. Reaction metabolites were quantified using HPLC and GC-MS analysis (Fig. 5). The results showed that the overexpression of CYP105D7-FprD-FdxH together increased the conversion yield of 3′-ODI by 5.5-fold compared to wild-type S. avermitilis (Table 2). In this case, interestingly, substrate and product concentration profiles along the reaction time showed a relatively rapid degradation of daidzein compared to the wild-type strain, and almost 234 μM out of 300 μM of daidzein disappeared in the 24-h reaction time (Supplementary Fig. S5), indicating that either daidzein is degraded through other degradation pathways or 3′-ODI is further metabolized by endogenous enzymes like O-methyltransferase or some other unknown oxidizing enzymes. Further analysis of the reaction metabolites showed that only 4–5 % of final 3′-ODI was detected by the O-methylated from product (data not shown), suggesting that the net 3′-ODI production is higher than the amount of the produced 3′-ODI, but other dominant degrading pathways play a significant role in S. avermitilis cell. Next, we investigated the possible degradation pathways in the S. avermitilis system. It has been well reported that dioxygenase like protocatechuate 3,4-dioxygenase has been involved in the cleavage of the C-ring of the flavonoids through the β-ketoadipate pathway (Arunachalam et al. 2003). The protochatechuate 3,4-dioxygenase gene is encoded by pcaH (sav1703) and pcaG (sav1704) according to S. avermitilis genome data base. The protochatechuate deletion mutant (S. avermitilis MA4680::∆pcaH ∆pcaG) was evaluated towards the same daidzein hydroxylation. The results showed that the β-ketoadipate pathway is not significantly contributing to the degradation pathway of daidzein (data not shown). The inactivation of the corresponding genes cannot completely abolish the degradation of daidzein, and still other pathways for the daidzein degradation are working in the S. avermitilis reaction system.
Representative GC-MS spectra of daidzein metabolites in S. avermitilis. i Gas chromatogram of daidzein reaction metabolites; RT = 20.26 min and RT = 26.12 min correspond to daidzein and 3′-ODI, respectively. ii Electron impact mass spectra of TMS derivatives of daidzein (RT = 20.26 min). iii Electron impact mass spectra of TMS derivatives of 3′-ODI (RT = 26.12 min)
During the whole cell reaction, it was observed that daidzein degradation is relatively high when cell broth tends to become black and brownish in color, i.e., melanin pigment, which is known to be synthesized by tyrosinase. At the same time, there was a report saying that two tyrosinases from Streptomyces coelicolor are active for the degradation of polyphenolic and flavonoid compounds (Yang and Chen 2009). One tyrosinase called, melC2 from S. coelicolor converted polyphenolic compounds to more hydrophobic and membrane permeable quinones. As 3′-ODI has a hydroxyl group in ortho-position of C4, it is very likely to be converted into quinone form by tyrosinase. In addition, it has been reported that isoflavonoids and its hydroxylated metabolites function as suicide substrates for a mushroom tyrosinase function (Chang 2007). In order to find out the effect of tyrosinase on daidzein or 3′-ODI degradation in S. avermitilis, daidzein biotransformation with tyrosinase inhibitors was attempted. In the presence of tyrosinase inhibitors such as catechol, daidzein degradation as well as 3′-ODI degradation was somewhat prevented (Lee et al. 2012). To confirm the effect of tyrosinase on daidzein degradation, melC2 gene from S. avermitilis was deleted. Only the melC2 deletion mutant did not produce melanin pigment, and the daidzein degradation was remarkably reduced. In result, ortho-hydroxylation of daidzein in ΔmelC2 appeared to show intact daidzein hydroxylation activity (Supplementary Fig. S6); however, the low activity is related to the low expression level of cyp105D7, fpr, and fdx (unpublished work). This result clearly demonstrated that the daidzein biotransformation in Streptomyces is mainly carried out by FprD-FdxH-CYP105D7, and tyrosinase is involved in the modification of the 3′-ODI to its quinone form.
Discussion
Identification of right electron transfer system for CYPs in Streptomyces, which contains the large number of CYPs, Fdxs, and Fprs, is a big challenge, and most of these electron transfer protein are distant to each other. Previously, we have reported that CYP105D7 when expressed in S. avermitilis showed the high regiospecific hydroxylation activity towards daidzein to 3′-ODI production, indicating that the CYP105D7 recognize its own endogenous electron transfer protein more effectively than the electron transfer protein CamA and CamB from Pseudomonas putida (Pandey et al. 2010). It is crucial to identify the natural electron transfer partner protein for CYP105D7. S. avermitilis M4680 alone have 33 CYPs, 9 ferredoxins (Fprs), and 6 ferredoxin reductases (Fdrs). The existence of nine Fdx and six Fpr proteins in S. avermitilis generates 54 possible combinations for electron transfer chain to each CYP, which is very hard to figure out by using in vitro interaction. It would be possible, if all the Fprs and Fdxs are in soluble and active form. However, an initial attempt to soluble expression of all the Fdxs in E. coli failed; this might be due to the different codon usage in the mRNA for recombinant gene from that of the host organism E. coli. Even few soluble Fdxs were not stable enough to carry out the in vitro reconstitution. Owing to such limitations, our attempt to in vitro reconstitution was not sufficient enough to pinpoint the target electron transfer protein for CYP105D7. It is necessary to study the effective combination of those Fdx and Fpr from S. avermitilis. The only alternative way to study those effect is using fpr or fdx deletion mutants of S. avermitilis, as this host have already been proven to be efficient for the (iso)flavonoid biotransformation (Pandey et al. 2011). We have examined this possibility using model CYP105D7. The ease of this enzyme is that it has already been proven for daidzein hydroxylase activity, and fdxH is located in the downstream to this cyp105D7 (sav7469), for which we believe to function with CYP105D7, when appropriate Fpr is combined. We applied two strategies, i.e., RT-PCR and fpr deletion from the S. avermitilis chromosome, with a belief that the fpr deletion mutant strain showing the lowest activity towards daidzein hydroxylation would indicate the best combination of functional Fpr with CYP105D7 and FdxH.
Through the evaluation, ferredoxin reductase deletion mutant in S. avermitilis showed FprD and FprE as potent electron transfer proteins. However, FprE deletion completely changed the cell morphology of S. avermitilis without producing black and brownish pigments (i.e., melanin pigment). The low activity of FprE might be associated with the change in the cell morphology. However, the Fpr deletion mutation alone was not clear enough to pinpoint one functional Fpr. In order to further confirm this finding, real-time PCR analysis of all the six fprs and nine fdxs were carried out in the presence of daidzein induction. The results presented here clearly showed significant mRNA expression profile of several fprs and fdxs; however, fprD and fdxH showed the effect of daidzein induction, and remaining several others showed constitutive induction profile. Furthermore, our results were supported by the in vitro reconstitution experiments with Fpr and Fdx for daidzein hydroxylation.
Combined upon substrate addition, we could propose that FprD and FdxH are the major electron transfer partners for CYP105D7, performing hydroxylation reaction of daidzein. Together with this finding, we have also identified that the tyrosinase melC2 (sav1137) from S. avermitilis is deeply involved in the degradation of daidzein as well as the conversion of daidzein into 3′-ODI. In this case, the 3′-ODI product might function as an inhibitor against the tyrosinase, unless it is converted into quinone form. Such hypothesis is currently under investigation in our laboratory. Not only ∆melC2 mutant did not produce any spores, but also, the mutation greatly downregulated the expression levels of cyps, fdxs, and fprs according to RT-PCR data (unpublished work), suggesting that the function of the tyrosinase is somewhat related to do with sporulation and cell stress environment. It would be interesting to understand the possible roles of the tyrosinase in S. avermitilis related to the daidzein metabolism as well as the regulation of cyp expression, which will add up new insights to the biology of tyrosinase and cell metabolism related with of CYPs functions in S. avermitilis.
References
Arunachalam M, Mohan N, Mahadevan A (2003) Cloning of Acinetobacter calcoaceticus chromosomal region involved in catechin degradation. Microbiol Res 158:37–46
Bernhardt R (2006) Cytochromes P450 as versatile biocatalysts. J Biotechnol 124:128–145
Chang TS (2007) Two potent suicide substrates of mushroom tyrosinase: 7,8,4′-trihydroxy isoflavone and 5,7,8,4′-tetrahydroxyisoflavone. J Agric Food Chem 7:55(5)
Chun YJ, Shimada T, Sanchez-Ponce R, Martin MV, Lei L, Zhao B, Kelly SL, Waterman MR, Lamb DC, Guengerich FP (2007) Electron transport pathway for a Streptomyces cytochrome P450: cytochrome P450105D5-catalyzed fatty acid hydroxylation in Streptomyces coelicolor A3 (2). J Biol Chem 282:17486–17500
Degtyarenko KN, Archakov AI (1993) Molecular evolution of P450 super family and P450-containing monooxygenase system. FEBS Lett 332:1–8
Hannemann F, Bichet A, Ewen KM, Bernhardt R (2007) Cytochrome P450 systems—biological variations of electron transport chains. Biochim Biophys Acta 1770:330–344
Hanukoglu I (1996) Electron transfer proteins of cytochrome P450 systems. Adv Mol Cell Biol 14:29–56
Hussain HA, Ward JM (2003) Enhanced heterologous expression of two Streptomyces griseolus cytochrome P450s and Streptomyces coelicolor ferredoxin reductase as potentially efficient hydroxylation catalysts. Appl Environ Microbiol 69:373–382
Ikeda H, Ishikawa J, Hanamoto A, Shinose M, Kikuchi H, Shiba T, Sakaki Y, Hattori M, Omura S (2003) Complete genome sequence and comparative analysis of the industrial microorganism Streptomyces avermitilis. Nat Biotechnol 5:526–531
Khatri Y, Hannemann F, Ewen KM, Pistorius D, Perlova O, Kagawa N, Brachmann AO, Müller R, Bernhardt R (2010) The CYPome of Sorangium cellulosum So ce56 and identification of CYP109D1 as a new fatty acid hydroxylase. Chem Biol 17:1295–1305
Lamb DC, Ikeda H, Nelson DR, Ishikawa J, Skaug T, Jackson C, Omura S, Waterman MR, Kelly SL (2003) Cytochrome complement (CYPome) of the avermectin-producer Streptomyces avermitilis and comparison to that of Streptomyces coelicolor A3(2). Biochem Biophys Res Commun 307:610–619
Lee N, Kim EJ, Kim BG (2012) Regioselective hydroxylation of trans-resveratrol via inhibition of tyrosinase from Streptomyces avermitilis MA4680. ACS Chem Biol 10:1687–1692
Lewis DF, Hlavica P (2000) Interactions between redox partners in various cytochrome P450 systems: functional and structural aspects. Biochim Biophys Acta 1460:353–374
Martinez A, Kolvek SJ, Yip CL, Hopke J, Brown KA, MacNeil IA, Osburne MS (2004) Genetically modified bacterial strains and novel bacterial artificial chromosome shuttle vectors for constructing environmental libraries and detecting heterologous natural products in multiple expression hosts. Appl Environ Microbiol 70:2452–2463
McLean KJ, Sabri M, Marshall KR, Lawson RJ, Lewis DG, Clift D, Balding PR, Dunford AJ, Warman AJ, McVey JP, Quinn AM, Sutcliffe MJ, Scrutton NS, Munro AW (2005) Biodiversity of cytochrome P450 redox systems. Biochem Soc Trans 33:796–801
Pandey BP, Roh C, Choi KY, Lee N, Kim EJ, Ko S, Kim T, Yun H, Kim BG (2010) Regioselective hydroxylation of daidzein using P450 (CYP105D7) from Streptomyces avermitilis MA4680. Biotechnol Bioeng 4:697–704
Pandey BP, Nahum L, Choi KY, Jung E, Jeong D, Kim BG (2011) Screening of bacterial cytochrome P450s responsible for regiospecific hydroxylation of (iso)flavonoids. Enzym Microb Technol 48:386–392
Sevrioukova IF, Poulos TL (2011) Structural biology of redox partner interactions in P450cam monooxygenase: a fresh look at an old system. Arch Biochem Biophys 507:66–74
Stutzman-Engwall K, Krebber A, Gustafsson C, Minshull J, Raillard S, Kim S, Chen Y (2006) Methods related to Streptomyces avermitilis gene directed the ration of B2:B1 avermectin. U.S. patent 7, 029,887 B2
Takamatsu S, Xu LH, Fushinobu S, Shoun H, Komatsu M, Cane DE, Ikeda H (2011) Pentalenic acid is a shunt metabolite in the biosynthesis of the pentalenolactone family of metabolites: hydroxylation of 1-deoxypentalenic acid mediated by CYP105D7 (SAV_7469) of Streptomyces avermitilis. J Antibiot (Tokyo) 64:65–71
Werck RD, Feyereisen R (2000) Cytochrome P450: a success story. Genome Biol 6:3003.1–3003.9
Yang HY, Chen CW (2009) Extracellular and intracellular polyphenol oxidases cause opposite effects on sensitivity of Streptomyces to phenolics: a case of double-edged sword. PLoS One 10:e7462
Zhu D, Seo MJ, Ikeda H, Cane DE (2011) Genome mining in Streptomyces. Discovery of an unprecedented P450-catalyzed oxidative rearrangement that is the final step in the biosynthesis of pentalenolactone. J Am Chem Soc 133:2128–2131
Acknowledgments
This research was supported by WCU (World Class University) program through the National Research Foundation of Korea (NRF) grant funded by the Ministry of Education, Science and Technology (R322012000102130).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 374 kb)
Rights and permissions
About this article
Cite this article
Pandey, B.P., Lee, N., Choi, KY. et al. Identification of the specific electron transfer proteins, ferredoxin, and ferredoxin reductase, for CYP105D7 in Streptomyces avermitilis MA4680. Appl Microbiol Biotechnol 98, 5009–5017 (2014). https://doi.org/10.1007/s00253-014-5525-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-014-5525-x