Abstract
Whole-genome shuffling (WGS) is a powerful technology of improving the complex traits of many microorganisms. However, the molecular mechanisms underlying the altered phenotypes in isolates were less clarified. Isolates with significantly enhanced stress tolerance and ethanol titer under very-high-gravity conditions were obtained after WGS of the bioethanol Saccharomyces cerevisiae strain ZTW1. Karyotype analysis and RT-qPCR showed that chromosomal rearrangement occurred frequently in genome shuffling. Thus, the phenotypic effects of genomic structural variations were determined in this study. RNA-Seq and physiological analyses revealed the diverse transcription pattern and physiological status of the isolate S3-110 and ZTW1. Our observations suggest that the improved stress tolerance of S3-110 can be largely attributed to the copy number variations in large DNA regions, which would adjust the ploidy of yeast cells and expression levels of certain genes involved in stress response. Overall, this work not only constructed shuffled S. cerevisiae strains that have potential industrial applications but also provided novel insights into the molecular mechanisms of WGS and enhanced our knowledge on this useful breeding strategy.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Mutagenic breeding and protoplast fusion are traditional breeding approaches commonly used to improve the traits of many industrial microorganisms. Whole-genome shuffling (WGS), which combines mutagenesis and protoplast fusion, was first proposed by del Cardayré and colleagues (Patnaik et al. 2002; Zhang et al. 2002). WGS allows the recombination of genotypes related to desired phenotypes in parent strains through recursive protoplast fusion or crossing, and isolates with improved traits are obtained by the repetition of this process (Bajwa et al. 2010; Lv et al. 2013; Otte et al. 2009; Zhang and Geng 2012; Zheng et al. 2012a, b). Previous studies reported that WGS improves stress tolerance and increases fermentative product titer in various microbial strains. However, the detailed molecular mechanisms underlying the improved phenotype of mutants remain unknown. For example, the genomic variations that appear during genome shuffling and the mechanism by which these mutations improve the traits in isolates have yet to be elucidated. A thorough understanding of these issues is necessary for the further development and application of WGS.
Bioethanol has attracted the most attention among potential renewable fuels; it is primarily produced through the fermentation of starch, sugars, and cellulosic feedstock by Saccharomyces cerevisiae. Environmental stress factors greatly inhibit the viability of yeast cells in industrial fermentation (Bajwa et al. 2013; Graves et al. 2006, 2007; Rossouw et al. 2009; Yang et al. 2011). The mechanisms underlying stress tolerance have been characterized by the synergism observed between multiple metabolic pathways involving dozens or even hundreds of genes (Brown et al. 2013; Dinh et al. 2009; Hirasawa et al. 2006; Lewis et al. 2010; Mahmud et al. 2010; Rossouw et al. 2012; Yoshikawa et al. 2009). Thus, global breeding strategies, such as WGS, are considered effective in enhancing the stress tolerance and ethanol fermentation efficiency of yeast (Pinel et al. 2011; Shi et al. 2009; Wang et al. 2012).
In the present study, S. cerevisiae strains with improved ethanol fermentation efficiency under very-high-gravity (VHG) conditions were constructed using WGS, and the mechanisms underlying the phenotypes of the isolates were investigated. The S. cerevisiae strain ZTW1 was subjected to WGS to obtain mutants with high stress tolerance and ethanol concentration. Results showed that large genomic structural variations (GSV) appeared frequently during genome shuffling. Comparative functional genomics of ZTW1 and the isolates was carried out to determine how these types of genomic variations contributed to the altered traits of the mutants.
Materials and methods
Strains and culture conditions
S. cerevisiae ZTW1 (CCTCC M 2013061) is an industrial strain used for bioethanol production by the Henan Tianguan Fuel Ethanol Co. Ltd., Nanyang, Henan, China. ZTW1 is an aneuploid strain (near-triploid). The laboratory S. cerevisiae strain BYZ1 (MATa/MATα his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 lys2Δ0/+ met15Δ0/+ ura3Δ0/ura3Δ0) was generated from a cross between BY4741 and BY4742 (these two S288c-isogenic haploid strains were provided by Oliver Valerius, University of Göttingen, Germany) (Zheng et al. 2012a, b). BYZ1 served as a control in the pulsed field gel electrophoresis (PFGE) and array-comparative genomic hybridization (aCGH) experiments. Yeast cells were grown in YPD medium (10 g/L yeast extracts, 20 g/L peptone, and 20 g/L glucose, pH 5.5) or YNB medium (6.7 g/L yeast nitrogen base, and 20 g/L glucose, pH 5.5).
WGS of S. cerevisiae ZTW1
Genome shuffling was performed by treating the strain with antimitotic drug methyl benzimidazole-2-yl-carbamate (MBC) followed by three rounds of sporulation and hybridization. MBC can cause random chromosomal aberrations, such as chromosomal amplification and loss, in S. cerevisiae cells (Wood 1982) and was thus used to produce mutants with GSV from ZTW1 in the present study. In the first round of genome shuffling, ZTW1 cells were grown in 25 mL YPD medium at 30 °C and 200 rpm with an initial density of 1.5 × 106/mL. MBC was added to ZTW1 culture when the cell density reached 3 × 107/mL. MBC (Adamas-beta, Shanghai, China) was first dissolved in dimethyl sulfoxide (50 mg/mL), and the final treatment concentration in YPD was 50 μg/mL (MBC had a cumulative inhibitory effect on yeast growth with increasing concentration, even if MBC was not completely dissolved in the medium; Fig. S1 in Supplementary Material). The ZTW1 cells were treated with MBC for 12 h and then collected by centrifugation at 5,000 rpm for 5 min at 4 °C. The residual MBC was removed by washing the ZTW1 cells with sterile water. The washed cells (8 × 108) were collected by centrifugation at 5,000 rpm for 5 min at 4 °C and then cultured in 30 mL sporulation medium (1 % NaAc, 0.2 % yeast extract, 0.1 % glucose, and 0.2 % KCl, at pH 6.0) for 4 days for sporulation. Then, 8 × 108 cells from the sporulation medium were collected by centrifugation at 12,000 rpm for 10 min at 4 °C and then maintained at 55 °C for 10 min to kill those cells that did not sporulate. The asci were transferred into a 10 mL medium containing 2 % (m/v) snailase (Sigma, St. Louis, MO, USA) and 0.5 M sorbitol (Sigma, St. Louis, MO, USA) and then cultured at 100 rpm for 6 h at 30 °C to release the spores. Approximately 2 × 108 free spores were collected by centrifugation at 12,000 rpm for 10 min at 4 °C and mated randomly in 25 mL YPD medium for 3 days. The hybrids were diluted to the cell density of 3 × 107/mL, and 100 μL of the cells was spread onto an initial screening plate containing 10 g/L yeast extract, 20 g/L peptone, 200 g/L glucose, and 50 g/L ethanol before incubation at 38 °C. This initial screening condition was demonstrated in our previous study to be effective for selecting mutants with high ethanol production under VHG conditions (Zheng et al. 2011). A total of 116, 127, and 122 isolated colonies that first appeared on the selection plates from the first, second, and third rounds of shuffling, respectively, were picked for fermentation tests. Three isolates with the highest ethanol concentrations under VHG conditions were selected and cultured together in 30 mL sporulation medium for the next round of WGS. Procedures for the two succeeding rounds of sporulation and hybridization were similar to those adopted for the first round.
Ethanol fermentation
An ethanol fermentation medium was prepared using corn flour with a two-enzyme (α-amylase and glucoamylase) method. Commercial α-amylase [Liquozyme® Supra; 90 Kilo Novo alpha-amylase Unit (KNU)/g] and glucoamylase [Suhong GA II; 500 Amyloglucosidase Unit (AGU)/mL] were donated by Novozymes (Beijing, China). One KNU was defined as the amount of enzyme that can break down 5.26 g starch per hour at 37 °C (pH 5.6), and one AGU was defined as the amount of enzyme that can hydrolyze 1 μmol maltotriose per minute at 37 °C (pH 4.3). A certain amount of corn flour and water (ratio, 1:2) was mixed, and α-amylase (32 KNU/g corn) was added to the mixture. The corn mash was kept in an autoclave at 95 °C for 2 h for liquefaction. Glucoamylase (1 AGU/g corn) was used to saccharify the mash at 60 °C for 2 h. Urea (NH2CONH2; Sinopharm, Shanghai, China) of 0.2 % (w/v) concentration was added to the corn mash as a nitrogen source for the yeast. Yeast cells were first grown in 250 mL flasks with 50 mL YPD at 200 rpm for 20 h at 30 °C with the initial cell density of 1.5 × 106/mL (OD600 = 0.05). Then, 8 × 109 yeast cells were collected by centrifugation, suspended in 5 mL fermentation medium, and then transferred into 250 mL fermentation medium in 500-mL conical flasks to start fermentation. The initial glucose concentration of the fermentation medium for all the fermentation tests was 285 g/L (fermentation medium used in this study was from the same batch). Fermentation was performed at 80 rpm for 75 h at 34 °C. The concentrations of ethanol, glucose, glycerol, and acetic acid were measured at the end of fermentation (75 h) as previously described (Zheng et al. 2011).
Stress tolerance test
Yeast cells were pregrown in 100-mL flasks containing 30 mL liquid YNB medium (pH 5.5) at 200 rpm for 24 h at 30 °C. The cells were then collected by centrifugation at 5,000 rpm for 5 min at 4 °C for stress tolerance tests. The cell density was adjusted to 1 × 109 cells/mL with sterile water, and 50 μL cells were transferred to 2 mL sterile water (control) or 2 mL sterile water containing 20 % (v/v) ethanol or 15 mM H2O2 for 2 h of treatment at 30 °C. To test the heat tolerance of the strains, 100 μL of the diluted cells was kept at 30 °C (control) and 55 °C in a thermostatic water bath for 5 min. The treated cells were then spread onto YPD plates at 30 °C for 2 days to calculate the survival rates. The yeast cells were transferred to 25 mL YNB medium (control) or 25 mL YNB with 35 % glucose with the initial cell density of 1.5 × 106/mL to determine the tolerance of these strains to high glucose concentrations. The cells were then cultured at 30 °C and 200 rpm, and the biomass formation of each strain was measured using a UV–VIS spectrophotometer (756PC; Spectrum Instruments Co. Ltd, Shanghai, China) at OD600 at the time point of 15 h. The tolerance of a certain strain was represented as the ratio of the biomass formation in YNB with 35 % glucose and that in YNB.
Reactive oxygen species (ROS) detection
To detect the intracellular ROS, 200 μL of the cells exposed to the stressors ethanol, H2O2, heat, and high glucose (described above) was collected by centrifugation at 5,000 rpm for 5 min at 4 °C and then washed twice with 0.1 M phosphate-buffered saline (PBS; pH 7.4) before staining with dichlorodihydrofluorescein diacetate (DCFH-DA) (10 μg/mL) at 30 °C for 1 h. The stained cells were observed and recorded using a confocal laser scanning microscope (Carl Zeiss, Oberkochen, Germany) as previously described (Zheng et al. 2011).
PFGE and aCGH
PFGE on a Bio-Rad DRIII apparatus was used to determine the karyotypes of isolates as previously described (Argueso et al. 2009). NimbleGen S. cerevisiae 385 k Whole-Genome Tiling (NimbleGen, Madison, WI, USA) was used to perform the aCGH analysis of ZTW1 and S3-110 as previously described (Zheng et al. 2012a, b). The raw data of the aCGH experiment can be downloaded from the NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo; GSE41017).
Serial passage of isolate S3-110
Serial passage was performed to determine the genomic stability of the isolate S3-110 from the third round of WGS. S3-110 was first grown in 25 mL YPD medium at 30 °C and 200 rpm with the initial cell density of 1.5 × 106/mL for 18 h. Then, 3.75 × 107 cells were harvested, transferred to 25 mL fresh YPD medium, and then cultured at 200 rpm for 12 h at 30 °C (this culture was defined as the first generation of S3-110). Similarly, 3.75 × 107 cells from the first generation were transferred to another 25 mL YPD medium (this culture was defined as the second generation) and cultured under the same conditions as that of the first generation. Such passage was repeated 60 times, and the cells from the 20th, 40th, and 60th generations were diluted and spread onto YPD plates. Three colonies grown on the plates were randomly picked for PFGE analysis in comparison with S3-110.
RNA-Seq and physiological factor analysis
Yeast cells were cultured in 25-mL YPD medium at 30 °C for 18 h to early stationary stage with the initial cell density of 1.5 × 106/mL. Total RNA of three independently cultured cells of each strain was extracted using the Fungal RNAout Kit (TIANDZ, Bejing, China). Poly(A) containing mRNA from 10 μg total RNA of each sample was purified using Sera-mag Magnetic Oligo(dT) Beads (Illumina, San Diego, CA, USA). The mRNA was fragmented into small pieces using divalent cations under elevated temperature (75 °C for 5 min). The cleaved mRNA fragments were converted to double-stranded cDNA using SuperScript II (Invitrogen, Carlsbad, CA, USA), RnaseH (Invitrogen, Carlsbad, CA, USA) and DNA Pol I (Invitrogen, Carlsbad, CA, USA), primed by random primers. The cDNA segments were subjected to end-repair and phosphorylation using T4 DNA polymerase (Invitrogen, Carlsbad, CA, USA), Klenow DNA polymerase (Invitrogen, Carlsbad, CA, USA) and T4 Polynucleotide Kinase (Invitrogen, Carlsbad, CA, USA). These repaired cDNA fragments were 3′-adenylated using Klenow Exo- (Illumina, San Diego, CA, USA) and purified using a PCR Purification Kit (Axygen, Union City, CA, USA). Gel electrophoresis was performed to separate library DNA fragments from unligated adapters by selecting cDNA fragments between 200 and 300 bp in size. The ligated cDNA fragments were recovered using a Gel Extraction Kit (Axygen, Union City, CA, USA), and were amplified by 18 cycles of PCR with primers complementary to the ends of the adapters. Finally, the three cDNA libraries of each strain were mixed and sequenced using the Illumina HiSeq™ 2000 (Illumina, San Diego, CA, USA) as previously described (Zheng et al. 2012a, b). The raw RNA-Seq data were deposited in NCBI’s GEO database with the accession number GSE41017. Clear reads from each sample were obtained by removing reads with 50 % or more low-quality bases. SOAPAligner (Li et al. 2009) was then used to align the clear reads to the S. cerevisiae genes. Differentially expressed genes (DEGs) between samples were screened as described in our previous study (Zheng et al. 2013). Functional annotations of the DEGs were performed using the Saccharomyces Genome Database (Dwight et al. 2002).
Yeast cells were cultured under the same condition for RNA-Seq experiment; cells were also collected at the point of 18 h for physiological analysis. The content of trehalose and the activity of catalase were measured as previously described (Zheng et al. 2011). Intracellular ATP and ADP of ∼3 × 109 yeast cells were extracted in 1 mL trichloroacetic acid (0.5 M) at 0 °C for 15 min. An Agilent 1100 LC (Agilent, Palo Alto, CA, USA) equipped with a ZORBAX SB-Aq column (250 × 4.6 mm) (Agilent, Palo Alto, CA, USA) was used to measure the ADP and ATP in the extracted solution. The following conditions were used: column temperature, 35 °C; ultraviolet detection wavelength, 254 nm; mobile phase, 14 % acetonitrile and 86 % phosphate buffer solution (pH was adjusted to 6.5); flow rate, 1 mL/min; and injection volume, 10 μL supernatant (Guan et al. 2013).
RT-qPCR
An RT-qPCR experiment was performed to verify the relative copy numbers of certain chromosomal regions of ZTW1 and isolates. A total of 38 genes (on average, each chromosome has two or three representative genes; these genes are listed in Table S1 in Supplementary Material) were selected to detect the copy number variation (CNV) of the DNA segment on which the genes are located. We measured the relative expression levels of 20 genes (these genes showed differently expression levels between YJS329 and ZTW1 detected by RNA-Seq; Table S2 in Supplementary Material) using RT-qPCR to determine the accuracy of RNA-Seq. Yeast cells were grown in 25 mL YPD medium at 30 °C for 18 h with the initial cell density of 1.5 × 106/mL. The DNA and total RNA from each strain were extracted as previously described (Zheng et al. 2012a, b). RT-qPCR was preformed using an ABI Prism 7500 instrument (Applied Biosystems, Foster City, CA, USA). The primers used for RT-qPCR were designed using Primer Premier 5.0. The sequences of the primers are listed in Tables S1 and S2. The expression activity or the relative copy number of genes was quantified using the comparative 2−ΔΔCT method with ACT1 as the reference gene (Tao et al. 2012).
Genetic manipulations
YFL052W and SKN7 were overexpressed in ZTW1 by cloning their open reading frames and promoter sequences into the CEN/ARS plasmids pSH65 (the cre gene in pSH65 was substituted by YFL052W) and pSK (this plasmid originated from pSH65, but the ble r gene was replaced by the kanMX gene obtained from plasmid pUG6; the cre gene in pSK was substituted by SKN7), respectively, to confirm the phenotypic effects of CNV of certain functional genes on ZTW1. Plasmids pUG6 and pSH65 were constructed in a previous study by Gueldener et al (2002). The plasmids were transformed into ZTW1 by the LiAc/SS carrier DNA/PEG method (Gueldener et al. 2002), and the transformants were selected on the YPD plates (pH was adjusted to 7) containing 60 μg/mL Zeocin (Invitrogen, San Diego, CA, USA) or 300 μg/mL G418 (Gibco, Eggenstein, Germany). The primers used to clone the genes are listed in Table S1 in Supplementary Material.
Results
WGS of S. cerevisiae strain ZTW1 to improve its ethanol fermentation efficiency under VHG conditions
Selection of S. cerevisiae strains for bioethanol production under VHG conditions (sugar concentration is usually above 260 g/L) is of significance because high final ethanol concentration would reduce energy consumption during the distillation process downstream and would decrease process wastewater discharge (Laopaiboon et al. 2009; Thomas and Ingledew 1992; Wang et al. 2012). However, VHG ethanol fermentation would impose more stresses, such as osmotic pressure, nitrogen shortage, and high ethanol concentration, and cause sluggish fermentation (Kim et al. 2013; Palma et al. 2013; Zheng et al. 2011). In the present study, WGS was conducted to improve the stress tolerance and increase the ethanol titer of ZTW1 under VHG conditions. After the first round of WGS, some isolates producing more ethanol at the end of ethanol fermentation (75 h) than ZTW1 were obtained (Fig. 1a). Isolates S1-60 (121.6 g/L), S1-62 (123.7 g/L), and S1-76 (122.9 g/L), which had the highest final ethanol concentrations among the 116 isolates from the first round of WGS, were selected to undergo the second round of WGS. Approximately 39 % (49/127) of the isolates from the second round of WGS surpassed ZTW1 (117.9 g/L) in ethanol concentrations (Fig. 1b). The isolates [S2-38 (125.4 g/L), S2-94 (129.1 g/L), and S2-104 (129.8 g/L)] that produced more ethanol than the other 124 isolates from the second round of WGS (Fig. 1b) were selected to undergo the third round of WGS. A total of 122 isolates were selected from the third round, and 75 % of them produced more ethanol than ZTW1 [the isolates showing top three ethanol concentrations were S3-28 (129.8 g/L), S3-43 (128.2 g/L), and S3-110 (130 g/L); Fig. 1c]. The isolates (S1-60, S1-62, S1-76, S2-38, S2-94, S2-104, S3-28, S3-43, and S3-110) with the top three ethanol concentrations from each round of WGS were selected for triplicate independent fermentation tests. Table 1 shows the concentrations of fermentation products (ethanol, glycerol, and acetic acid) and residual sugars of these isolates under VHG conditions. All nine isolates consumed more sugars and produced 3 to 11 % more ethanol (t test, P < 0.05) and slightly higher glycerol and acetic acid than ZTW1. In addition, significantly higher viabilities were observed for these mutants at the later stages of fermentation (t test, P < 0.01; Table 1), suggesting improved adaptability as a result of WGS.
WGS of ZTW1 to improve its ethanol titer under very-high-gravity (VHG) conditions. Vertical bars represent the final ethanol concentration of ZTW1 (green) and the isolates (gray) from the first (a), second (b), and third (c) rounds of WGS. The ethanol concentration was a mean of two duplicated samples in one experiment. The ethanol concentrations of ZTW1 in a, b, and c were 118.7, 117.9, and 118.2 g/L, respectively. The pink dotted line marks the final ethanol concentration from ZTW1. The box plots indicate the average (short violet lines in the box) and ranges of the final ethanol concentration of all isolates obtained from each round of WGS
Compared with ZTW1, all nine selected mutants showed higher tolerance to individual stress factors after treatments with ethanol, heat, and H2O2; however, the isolates cultured in a high-glucose-concentration medium demonstrated no improvement in biomass formation (Fig. 2a). These results demonstrate that WGS can effectively improve the stress tolerance and ethanol concentration of ZTW1 under certain conditions. One of the common physiological features in cells exposed to various stressors is the accumulation of intracellular ROS (Herrero et al. 2008). DCFH-DA is a nonpolar dye which will be converted into the polar derivative DCFH by cellular esterases after incorporation into cells. DCFH can be switched to highly fluorescent DCF (green fluorescent) when oxidized by intracellular ROS. Thus, the cells accumulating intracellular ROS will be stained with green after the application of DCFH-DA (Wu et al. 2009). Although ROS accumulation was detected in all the tested strains after the application of stresses, the ratios of the cells stained with DCFH-DA of the nine isolates were always lower than that of ZTW1 (Fig. 2b). For example, the ratios of S3-110 were 30.7, 30, 31.7, and 33 % lower than those of ZTW1 after the treatment of heat, ethanol, H2O2, and high glucose concentration, respectively.
Stress tolerance and ROS accumulation of ZTW1 and isolates from three rounds of WGS. a Comparison of stress tolerance between ZTW1 and nine isolates. The tolerance of a certain strain against heat (55 °C for 5 min), 20 % ethanol (v/v), and 15 mM H2O2 is represented as the ratio (mean ± SD) of the survival cells under an indicated stress condition and under the control condition described in “Materials and methods”. The tolerance to high glucose concentration was represented as the ratio of the biomass formation in YNB with 35 % glucose and that in YNB. b ROS detection of ZTW1 and isolates after the application of stressors. The yeast cells were stained with DCFH-DA after the application of stressors [heat (55 °C for 5 min), 20 % ethanol (v/v), 15 mM H2O2, and 35 % glucose] as described in “Materials and methods”. Then, the cells were observed using a confocal laser scanning microscope, and five fields of view (randomly chosen) from each strain were photographed. The ratio of the cells stained with DCFH-DA was calculated to show the degree of ROS accumulation for each strain. The data (ratio stained/total cells in %) represent means ± SD of the five fields of view
Genomic structural analysis of ZTW1 and the isolates
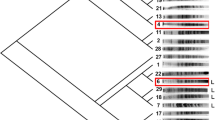
During WGS, MBC treatment and subsequent meiosis are supposed to boost chromosomal aberrations and shuffle the mutated genome of ZTW1 (Wood 1982; Zheng et al. 2011). PFGE revealed that ZTW1 and the nine isolates from the three rounds of WGS (S1-60, S1-62, S1-76, S2-38, S2-94, S2-104, S3-28, S3-43, and S3-110) had all distinctly different chromosomal karyotypes (chromosomal lengths and numbers) (Fig. 3a). In addition, the results of RT-qPCR revealed a CNV in many chromosomal regions in the genomes of ZTW1 and the isolates (Table S3 in Supplementary Material). Some genes in chromosomes 3, 6, 8, 9, 11, and 14 had different copies between ZTW1 and the isolates. For example, ZTW1 had four copies of CLA4 (its location is chromosome 14: 68.4 to 71.5 kb), whereas the nine isolates had three copies of this gene (Table S3 in Supplementary Material). These results indicate that WGS can produce isolates with diverse genomic structures compared with their parent strain. Higher resolution results pertaining to the CNV events in the genomes of ZTW1 and S3-110 were obtained through aCGH analysis (Fig. 3b and c). The copy numbers of some large CNV segments [31 kb to 378 kb on chromosome 11 (four copies), 1 kb to 97 kb on chromosome 14 (four copies), and 727 kb to 777 kb on chromosome 14 (two copies)] in ZTW1 were restored to three in S3-110 (Fig. 3b and c). Moreover, S3-110 had an extra copy of DNA segment (358 kb to 526 kb on chromosome 8; four copies in S3-110) compared with ZTW1. To determine the genomic stability of S3-110, serial passage of this strain was performed (described in “Materials and methods”), and the progenies of S3-110 were randomly selected for PFGE analysis. The serial passage did not cause significant genomic structural variations in the genome of S3-110 (Fig. S2 in Supplementary Material). This result suggests that S3-110 can maintain a stable genome during mitosis.
Genomic structural variation of isolates from three rounds of WGS compared with the parent genome of ZTW1. a Pulse-field gel electrophoresis of the chromosome of ZTW1 and nine isolates from the three rounds of WGS (lanes 1 to 9: the order of the nine isolates is the same as that in Fig. 2). Diploid strain BYZ1 was used as the control indicating the numbers and lengths of chromosomes. The chromosome numbers in this figure refer to those of BYZ1. Comparison of the genomic structures of b ZTW1 and c S3-110 by array-comparative genomic hybridization. Amplified regions and underrepresented regions are shown in red and green, respectively
Comparison of the transcription and physiological metabolism of ZTW1 and S3-110
Comparative studies of transcription between ZTW1 and S3-110 were conducted to determine whether or not certain genomic structural mutations account for the improved stress tolerance of isolates. The total RNA of these two strains was extracted when they were grown to the early stationary phase at which multiple genes involved in stress response are expressed (Gasch et al. 2000; Gasch and Werner-Washburne 2002). Using RNA-Seq, 734 genes were upregulated and 728 genes were expressed at lower levels in S3-110 compared with ZTW1 (P < 0.001; Table S4 in Supplementary Material). A total of 112 genes showed significant changes (change fold > 2 and P < 0.001) in expression between the two strains. Among the DEGs, 20 were randomly chosen to verify the accuracy of RNA-Seq by RT-qPCR. The results of RT-qPCR confirmed that the expression variation ranges of the 20 genes between the two strains had a high consistency in comparison with the data from RNA-Seq experiments (Table S2 in Supplementary Material).
Functional annotations of the DEGs showed that the upregulated genes in S3-110 were involved in the mitotic cell cycle, small molecule metabolism, and stress response (Table S5 in Supplementary Material). These findings showed that the physiological state of S3-110 tended to promote cell reproduction and synthetize antistress factors. S3-110 formed higher cell numbers when entering the stationary phase (Fig. 4a), but their dry weight of biomass was similar (data not shown). In addition, the genes (TPS1, TPS2, TSL1, NTH1, and ATH1) involved in trehalose metabolism and the gene (CTT1) encoding catalase showed higher expression levels in S3-110 than in ZTW1 (Table S4 in Supplementary Material). The trehalose content and catalase activity [two most important physiological factors involved in stress tolerance (Estruch 2000)] of S3-110 were 121 and 56 % higher than those of ZTW1, respectively (t test, P < 0.01; Fig. 4b). Similarly, the other eight isolates also had higher contents of these two antistress factors compared with ZTW1 (Fig. 4b). These results were in agreement with the improved tolerance to multiple stress inducers observed in the selected isolates (Fig. 2a). The downregulated genes were mainly involved in oxidation–reduction, precursor metabolite and energy generation reactions, and transmembrane transport (mainly genes encoding ion and glucose transporters; Table S6 in Supplementary Material). This result suggests higher sugar consumption flux and higher energy production in ZTW1 than in the isolates. In accordance with this speculation, the intracellular ATP/ADP ratio of ZTW1 was higher than those of S3-110 and the other eight isolates (t test, P < 0.05; Fig. 4c). These results suggest that WGS can alter gene expression and physiological metabolism on a global scale, thereby opening the possibility to improve the complex phenotypes of yeast.
Comparison of growth and physiological metabolism of ZTW1 and the nine isolates from three rounds of WGS. a Growth curves of ZTW1 and S3-110. Yeast cells were grown in 25 mL YPD at 30 °C with the initial concentration of 1 × 106 cells/mL. Cells were harvested at the indicated time point and diluted for plating and colony count. b Comparison of trehalose content and catalase activity (CAT) between ZTW1 and isolates. c Intracellular ATP/ADP ratio of ZTW1 and the nine isolates. Lanes 1 to 9 represent the isolates S1-60, S1-62, S1-76, S2-38, S2-94, S2-104, S3-28, S3-43, and S3-110, respectively. Data represent means ± SD of three independent experiments. Letters “a” and “b” indicate the significant difference in CAT activity and trehalose content at P < 0.01, respectively, between ZTW1 and the nine isolates using t test. “c” indicates the significant difference in ATP/ADP ratio at P < 0.05 between ZTW1 and the nine isolates using t test
Effects of CNV on the transcription and phenotypes of the isolates
After mapping the reads from RNA-Seq onto the chromosomes of S. cerevisiae, the transcriptional levels of the DNA segments with CNV were detected. Figure 5a–c shows the relative expression levels (Log2 ratio of S3-110/ZTW1) of genes in chromosomes 8, 11, and 14, respectively. The average expression level of genes on the amplified region (358 to 526 kb on chromosome 8; red dotted line in Fig. 5a) was 39 % higher than that of the normal region (1 to 358 kb on chromosome 8; black dotted line in Fig. 5a), close to the increased DNA dosage in S3-110 (from three copies to four copies). Similarly, the average expression level of genes on the regions with a lower copy (from four copies to three copies), such as 31 to 378 kb on chromosome 11 (red dotted line in Fig. 5b) and 1 to 97 kb on chromosome 14 (red dotted line in Fig. 5c), was decreased by 26.9 and 27.2 %, respectively, compared with that of genes on normal regions on these two chromosomes (black dotted lines in Fig. 5b, c). These results indicate an apparent correlation between the expression levels of genes within regions with CNV and their DNA copy numbers. This correlation suggests that the CNV of certain functional genes in the genomes of ZTW1 and S3-110 directly affects their expression and the phenotypes of strains.
Effects of CNV on transcription and particular phenotypes. Relative expression levels (Log2-ratio of S3-110/ZTW1) of genes on chromosomes a 8, b 11, and c 14. d Changes in the expression levels of SKN7 and YFL052W in the engineered strains compared with ZTW1. e Viability of ZTW1 and engineered strains under ethanol, heat, and H2O2 stresses. Tolerance of engineered strains was significantly higher when the expression levels of SKN7 and YFL052W were upregulated. The data represent means ± SD of three independent experiments. Letters “a” and “b” indicate the significant difference in gene expression at P < 0.01 between ZTW1 and other strains using t test. Letters “c,” “d,” and “e” indicate the significant difference in viability ratio at P < 0.05 after the application of heat, ethanol, and H2O2, respectively, between ZTW1 and other strains using t test. “cc,” “dd,” and “ee” indicate the significant difference in viability ratio at P < 0.01 after the application of heat, ethanol, and H2O2, respectively, between ZTW1 and other strains using t test
YFL052W (a putative transcription factor whose deletion will decrease the tolerance to Calcofluor White, low osmolarity, and heat) (Akache et al. 2001) and SKN7 (a transcription factor required for the optimal induction of heat shock genes in response to oxidative stress that is also involved in osmoregulation) (Raitt et al. 2000) are two genes involved in stress tolerance of yeast. The DNA copy numbers and expression levels of these two genes were lower in ZTW1 than in S3-110 (Table S3 in Supplementary Material and Fig. 5d). YFL052W and SKN7 were overexpressed in ZTW1 by transformation of the low-copy-number plasmids, pSH65 and pSK harboring these genes to assess the phenotypic effects (especially the tolerance to ethanol, heat, and oxidative stresses) of the CNV of these genes. Figure 5d shows that the mRNA expression levels of these two genes in the transformants were 48 and 32 % higher (t test, P < 0.01) than that of ZTW1, respectively. These enhanced levels are close to the levels observed in S3-110. The viability ratio of the engineered strain ZYFL052W was 20, 27, and 55 % higher than that of ZTW1, respectively, after the application of heat, ethanol, and H2O2; the viability ratio of the engineered strain ZSKN7 was 18 and 30 % higher than that of ZTW1, respectively, after the application of heat and H2O2 (t test, P < 0.05; Fig. 5e). However, the tolerance against heat, ethanol, and H2O2 of the two engineered strains (ZYFL052W and ZSKN7) was considerably lower than that of S3-110 (Fig. 5e). The engineered strain ZYS with the overexpression of both YFL052W and SKN7 showed slightly improved tolerance to heat and H2O2 compared with the engineered strain ZYFL052W (Fig. 5e, t test, P < 0.05). This result suggests that the overexpression of these two genes slightly improved the stress tolerance of ZTW1. These results confirmed that the CNV of certain functional genes partly contributed to the improved traits of the isolates.
Discussion
WGS is a popular technology used to improve the desirable performance of microbes, especially the complex traits that are not easily achievable through genetic engineering (Gong et al. 2009). WGS is usually performed in two steps: (1) introducing mutations into the genome of the parent strains by chemical or physical mutagens, followed by (2) several rounds of cell fusion or hybridization to integrate the beneficial mutations from multiple parent strains into the progenies (Bajwa et al. 2010; Lv et al. 2013; Otte et al. 2009; Zhang and Geng 2012; Zheng et al. 2012a, b). In the present study, the bioethanol-producing S. cerevisiae strain ZTW1 was selected as the parent strain to undergo a modified WGS approach to improve its stress tolerance and alcoholic fermentation efficiency under VHG conditions. Boxplot analysis (Fig. 1) of the isolates show the range median, normality, and skew of their ethanol titer distribution. These plots indicate that the average ethanol concentration of the isolates from each round of WGS was gradually enhanced. However, a strain from the fourth round of WGS that produced more ethanol than that of S3-110 was not obtained (data not shown). The nine isolates from the three rounds of WGS were more resistant to stresses from ethanol, high temperature, and H2O2 than ZTW1 but to different degrees (Fig. 2a). One possible physiological mechanism underlying the improved tolerance of the nine isolates is their higher ROS removal capability compared to that of ZTW1 under stressful conditions (Fig. 2b). In summary, these results are consistent with our previous finding that a good correlation exists between complex stress tolerance and ethanol titer under VHG conditions (Zheng et al. 2011).
Disparate from previously reported WGS methods, the present study did not create new gene mutations in the genome of the parent strain through ultraviolet or ethylmethylsulfone mutagenesis. Instead, parent cells were first treated with MBC to cause chromosomal aberrations, and three rounds of sporulation and hybridization were used to shuffle the genomic variations. Thus, the possible theoretical basis underlying the changed traits of isolates obtained in this study mainly includes genomic structural variations, such as insertions, deletions, translocation, and inversions, and DNA sequence variations resulting from homologous recombination during meiosis. The results of PFGE and RT-PCR showed that large chromosomal rearrangements occurred during the WGS of ZTW1 (Fig. 3a and Table S3 in Supplementary Material), which led to the CNV of large DNA regions in the genomes of the isolates. Recent studies have suggested that genomic structural variation is a common phenomenon in yeast populations and might be responsible for their adaptabilities (Borneman et al. 2011; Chang et al. 2013; Lucena et al. 2007). For example, Stambuk et al. (2009) confirmed that the amplification of genes involved in the biosynthesis of vitamins B6 and B1 confers an ability to certain bioethanol-producing yeast strains to grow efficiently in industrial environments. Thus, the genomic structural variations in S3-110 caused by WGS were hypothesized to be an important genetic mechanism responsible for its diverse phenotypes compared with ZTW1.
To confirm this hypothesis, the transcription and physiological factors of ZTW1 and the shuffled strain S3-110 were compared. In line with its higher proliferation capability and improved stress tolerance, the genes involved in the cell cycle and synthesis of some antistress factors were significantly upregulated in the isolates. Meanwhile, ZTW1 showed an increased demand for energy and a weaker capability in cellular proliferation than the isolates. These results suggest that the nine selected isolates shared common physiological and phenotypic traits. For example, all isolates showed improved tolerance to heat, ethanol, and H2O2. Previous studies suggested that compared with euploid cells, aneuploid yeast cells are usually defective in cell cycle progression, produce more energy to maintain a normal physiological state through strengthening the tricarboxylic acid cycle, and are less tolerant to conditions (such as heat) interfering with protein synthesis or folding (Torres et al. 2007, 2010). In the present study, a fewer GSV events in the genome of S3-110 than that of ZTW1 (Fig. 3c) decreased the effects of aneuploidy and increased the proliferation ability and stress tolerance of S3-110. These observations suggested that the different degrees of aneuploidy of ZTW1 and the isolates might be one reason for their different transcription patterns and stress tolerance levels.
Calculation of the mapped reads from RNA-Seq showed that the changed dosage of genes directly affected their transcription activity. Among the DEGs with CNV, YFL052W and SKN7 with increased copy numbers showed enhanced expression level, which increased the stress tolerance of ZTW1 (Fig. 5d,1 e). This finding partly explained the changed tolerance levels of the isolates. These results and similar observations in previous studies (Pavelka et al. 2010; Stambuk et al. 2009) indicated that the CNV of certain functional genes can directly contribute to particular traits of yeast. These analyses were used as bases for the assumption that genomic structural variations cause the CNV of certain large DNA regions in WGS, which would adjust both the ploidy of cells and the expressions levels of certain functional genes. Consequently, the stress tolerance of the isolates might be altered. Overall, this work not only obtained some yeast mutants with industrial application potential but also provided insights into the mechanism by which WGS improves and delivers complex phenotypes of microorganisms.
References
Akache B, Wu K, Turcotte B (2001) Phenotypic analysis of genes encoding yeast zinc cluster proteins. Nucleic Acids Res 29:2181–2190
Argueso JL, Carazzolle MF, Mieczkowski PA, Duarte FM, Netto OV, Missawa SK, Galzerani F, Costa GG, Vidal RO, Noronha MF, Dominska M, Andrietta MG, Andrietta SR, Cunha AF, Gomes LH, Tavares FC, Alcarde AR, Dietrich FS, McCusker JH, Petes TD, Pereira GA (2009) Genome structure of a Saccharomyces cerevisiae strain widely used in bioethanol production. Genome Res 19:2258–2270
Bajwa PK, Pinel D, Martin VJ, Trevors JT, Lee H (2010) Strain improvement of the pentose-fermenting yeast Pichia stipitis by genome shuffling. J Microbiol Methods 81:179–186
Bajwa PK, Ho CY, Chan CK, Martin VJ, Trevors JT, Lee H (2013) Transcriptional profiling of Saccharomyces cerevisiae T2 cells upon exposure to hardwood spent sulphite liquor: comparison to acetic acid, furfural and hydroxymethylfurfural. Antonie Van Leeuwenhoek 103:1–15
Borneman AR, Desany BA, Riches D, Affourtit JP, Forgan AH, Pretorius IS, Egholm M, Chambers PJ (2011) Whole-genome comparison reveals novel genetic elements that characterize the genome of industrial strains of Saccharomyces cerevisiae. PLoS Genet 7:e1001287
Brown NA, de Castro PA, Castro Pimentel Figueiredo B, Savoldi M, Buckeridge MS, Lopes ML, Lima Paullilo SC, Borges EP, Amorim HV, Goldman MH (2013) Transcriptional profiling of Brazilian Saccharomyces cerevisiae strains selected for semi-continuous fermentation of sugarcane must. FEMS Yeast Res 13:277–290
Chang SL, Lai HY, Tung SY, Leu JY (2013) Dynamic large-scale chromosomal rearrangements fuel rapid adaptation in yeast populations. PLoS Genet 9:e1003232
Dinh TN, Nagahisa K, Yoshikawa K, Hirasawa T, Furusawa C, Shimizu H (2009) Analysis of adaptation to high ethanol concentration in Saccharomyces cerevisiae using DNA microarray. Bioprocess Biosyst Eng 32:681–688
Dwight SS, Harris MA, Dolinski K, Ball CA, Binkley G, Christie KR, Fisk DG, Issel-Tarver L, Schroeder M, Sherlock G (2002) Saccharomyces Genome Database (SGD) provides secondary gene annotation using the Gene Ontology (GO). Nucleic Acids Res 30:69–72
Estruch F (2000) Stress-controlled transcription factors, stress-induced genes and stress tolerance in budding yeast. FEMS Microbiol Rev 24:469–486
Gasch AP, Werner-Washburne M (2002) The genomics of yeast responses to environmental stress and starvation. Funct Integr Genomics 2:181–192
Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO (2000) Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell 11:4241
Gong JX, Zheng HJ, Wu ZJ, Chen T, Zhao XM (2009) Genome shuffling: progress and applications for phenotype improvement. Biotechnol Adv 27:996–1005
Graves T, Narendranath NV, Dawson K, Power R (2006) Effect of pH and lactic or acetic acid on ethanol productivity by Saccharomyces cerevisiae in corn mash. J Ind Microbiol Biotechnol 33:469–474
Graves T, Narendranath NV, Dawson K, Power R (2007) Interaction effects of lactic acid and acetic acid at different temperatures on ethanol production by Saccharomyces cerevisiae in corn mash. Appl Microbiol Biotechnol 73:1190–1196
Guan N, Liu L, Shin H-D, Chen RR, Zhang J, Li J, Du G, Shi Z, Chen J (2013) Systems-level understanding how Propionibacterium acidipropionici respond to propionic acid stress at the microenvironment levels: mechanism and application. J Biotechnol 167:56–63
Gueldener U, Heinisch J, Koehler G, Voss D, Hegemann J (2002) A second set of loxP marker cassettes for Cre-mediated multiple gene knockouts in budding yeast. Nucleic Acids Res 30:e23–e23
Herrero E, Ros J, Bellí G, Cabiscol E (2008) Redox control and oxidative stress in yeast cells. Biochim Biophys Acta 1780:1217–1235
Hirasawa T, Nakakura Y, Yoshikawa K, Ashitani K, Nagahisa K, Furusawa C, Katakura Y, Shimizu H, Shioya S (2006) Comparative analysis of transcriptional responses to saline stress in the laboratory and brewing strains of Saccharomyces cerevisiae with DNA microarray. Appl Microbiol Biotechnol 70:346–357
Kim NR, Yang J, Kwon H, An J, Choi W, Kim W (2013) Mutations of the TATA-binding protein confer enhanced tolerance to hyperosmotic stress in Saccharomyces cerevisiae. Appl Microbiol Biotechnol 97:8227–8238
Laopaiboon L, Nuanpeng S, Srinophakun P, Klanrit P, Laopaiboon P (2009) Ethanol production from sweet sorghum juice using very high gravity technology: effects of carbon and nitrogen supplementations. Bioresour Technol 100:4176–4182
Lewis JA, Elkon IM, McGee MA, Higbee AJ, Gasch AP (2010) Exploiting natural variation in Saccharomyces cerevisiae to identify genes for increased ethanol resistance. Genetics 186:1197–1205
Li RQ, Yu C, Li YR, Lam TW, Yiu SM, Kristiansen K, Wang J (2009) SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics 25:1966–1967
Lucena BT, Silva-Filho EA, Coimbra MR, Morais JO, Simões DA, Morais MA Jr (2007) Chromosome instability in industrial strains of Saccharomyces cerevisiae batch cultivated under laboratory conditions. Genet Mol Res 6:1072–1084
Lv XA, Jin YY, Li YD, Zhang H, Liang XL (2013) Genome shuffling of Streptomyces viridochromogenes for improved production of avilamycin. Appl Microbiol Biotechnol 97:641–648
Mahmud SA, Hirasawa T, Furusawa C, Yoshikawa K, Shimizu H (2010) Understanding the mechanism of heat stress tolerance caused by high trehalose accumulation in Saccharomyces cerevisiae using DNA microarray. J Biosci Bioeng 113:526–528
Otte B, Grunwaldt E, Mahmoud O, Jennewein S (2009) Genome shuffling in Clostridium diolis DSM 15410 for improved 1, 3-propanediol production. Appl Environ Microbiol 75:7610–7616
Palma M, Madeira SC, Mendes-Ferreira A, Sá-Correia I (2013) Impact of assimilable nitrogen availability in glucose uptake kinetics in Saccharomyces cerevisiae during alcoholic fermentation. Microb Cell Fact 11:99
Patnaik R, Louie S, Gavrilovic V, Perry K, Stemmer WP, Ryan CM, del Cardayré S (2002) Genome shuffling of Lactobacillus for improved acid tolerance. Nat Biotechnol 20:707–712
Pavelka N, Rancati G, Zhu J, Bradford WD, Saraf A, Florens L, Sanderson BW, Hattem GL, Li R (2010) Aneuploidy confers quantitative proteome changes and phenotypic variation in budding yeast. Nature 468:321–325
Pinel D, D’Aoust F, del Cardayre SB, Bajwa PK, Lee H, Martin VJ (2011) Saccharomyces cerevisiae genome shuffling through recursive population mating leads to improved tolerance to spent sulfite liquor. Appl Environ Microbiol 77:4736–4743
Raitt DC, Johnson AL, Erkine AM, Makino K, Morgan B, Gross DS, Johnston LH (2000) The Skn7 response regulator of Saccharomyces cerevisiae interacts with Hsf1 in vivo and is required for the induction of heat shock genes by oxidative stress. Mol Biol Cell 11:2335–2347
Rossouw D, Olivares-Hernandes R, Nielsen J, Bauer FF (2009) Comparative transcriptomic approach to investigate differences in wine yeast physiology and metabolism during fermentation. Appl Environ Microbiol 75:6600–6612
Rossouw D, Jacobson D, Bauer FF (2012) Transcriptional regulation and the diversification of metabolism in wine yeast strains. Genetics 190:251–261
Shi DJ, Wang CL, Wang KM (2009) Genome shuffling to improve thermotolerance, ethanol tolerance and ethanol productivity of Saccharomyces cerevisiae. J Ind Microbiol Biotechnol 36:139–147
Stambuk BU, Dunn B, Alves SL, Duval EH, Sherlock G (2009) Industrial fuel ethanol yeasts contain adaptive copy number changes in genes involved in vitamin B1 and B6 biosynthesis. Genome Res 19:2271–2278
Tao XL, Zheng DQ, Liu TZ, Wang PM, Zhao WP, Zhu MY, Jiang XH, Zhao YH, Wu XC (2012) A novel strategy to construct yeast Saccharomyces cerevisiae strains for very high gravity fermentation. PLoS One 7:e31235
Thomas K, Ingledew W (1992) Production of 21 % (v/v) ethanol by fermentation of very high gravity (VHG) wheat mashes. J Ind Microbiol 10:61–68
Torres EM, Sokolsky T, Tucker CM, Chan LY, Boselli M, Dunham MJ, Amon A (2007) Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science 317:916–924
Torres EM, Dephoure N, Panneerselvam A, Tucker CM, Whittaker CA, Gygi SP, Dunham MJ, Amon A (2010) Identification of aneuploidy-tolerating mutations. Cell 143:71–83
Wang PM, Zheng DQ, Liu TZ, Tao XL, Feng MG, Min H, Jiang XH, Wu XC (2012) The combination of glycerol metabolic engineering and drug resistance marker-aided genome shuffling to improve very-high-gravity fermentation performances of industrial Saccharomyces cerevisiae. Bioresour Technol 108:203–210
Wood JS (1982) Mitotic chromosome loss induced by methyl benzimidazole-2-yl-carbamate as a rapid mapping method in Saccharomyces cerevisiae. Mol Cell Biol 2:1080–1087
Wu CY, Steffen J, Eide DJ (2009) Cytosolic superoxide dismutase (SOD1) is critical for tolerating the oxidative stress of zinc deficiency in yeast. PLoS One 4:e7061
Yang J, Bae JY, Lee YM, Kwon H, Moon HY, Kang HA, Yee SB, Kim W, Choi W (2011) Construction of Saccharomyces cerevisiae strains with enhanced ethanol tolerance by mutagenesis of the TATA-binding protein gene and identification of novel genes associated with ethanol tolerance. Biotechnol Bioeng 108:1776–1787
Yoshikawa K, Tanaka T, Furusawa C, Nagahisa K, Hirasawa T, Shimizu H (2009) Comprehensive phenotypic analysis for identification of genes affecting growth under ethanol stress in Saccharomyces cerevisiae. FEMS Yeast Res 9:32–44
Zhang W, Geng A (2012) Improved ethanol production by a xylose-fermenting recombinant yeast strain constructed through a modified genome shuffling method. Biotechnol Biofuels 5:1–11
Zhang YX, Perry K, Vinci VA, Powell K, Stemmer WP, del Cardayré SB (2002) Genome shuffling leads to rapid phenotypic improvement in bacteria. Nature 415:644–646
Zheng DQ, Wu XC, Tao XL, Wang PM, Li P, Chi XQ, Li YD, Yan QF, Zhao YH (2011) Screening and construction of Saccharomyces cerevisiae strains with improved multi-tolerance and bioethanol fermentation performance. Bioresour Technol 102:3020–3027
Zheng DQ, Wang PM, Chen J, Zhang K, Liu TZ, Wu XC, Li YD, Zhao YH (2012a) Genome sequencing and genetic breeding of a bioethanol Saccharomyces cerevisiae strain YJS329. BMC Genomics 13:479
Zheng P, Zhang KK, Yan Q, Xu Y, Sun ZH (2012b) Enhanced succinic acid production by Actinobacillus succinogenes after genome shuffling. J Ind Microbiol Biotechnol 40:1–10
Zheng DQ, Liu TZ, Chen J, Zhang K, Li O, Zhu L, Zhao YH, Wu XC, Wang PM (2013) Comparative functional genomics to reveal the molecular basis of phenotypic diversities and guide the genetic breeding of industrial yeast strains. Appl Microbiol Biotechnol 97:2067–2076
Acknowledgments
This work was financially supported by State Key Laboratory of Motor Vehicle Biofuels Technology of China (2013013), and Postdoctoral Science Foundation of China (2013M531450) and Zhejiang Province (BSH1301026).
Author information
Authors and Affiliations
Corresponding author
Additional information
Dao-Qiong Zheng and Jie Chen contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 1803 kb)
Rights and permissions
About this article
Cite this article
Zheng, DQ., Chen, J., Zhang, K. et al. Genomic structural variations contribute to trait improvement during whole-genome shuffling of yeast. Appl Microbiol Biotechnol 98, 3059–3070 (2014). https://doi.org/10.1007/s00253-013-5423-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-013-5423-7