Abstract
Aspergillus niger has an extraordinary potential to produce organic acids as proven by its application in industrial citric acid production. Previously, it was shown that expression of the cis-aconitate decarboxylase gene (cadA) from Aspergillus terreus converted A. niger into an itaconic acid producer (Li et al., Fungal Genet Bio 48: 602–611, 2011). After some initial steps in production optimization in the previous research (Li et al., BMC biotechnol 12: 57, 2012), this research aims at modifying host strains and fermentation conditions to further improve itaconic acid production. Expression of two previously identified A. terreus genes encoding putative organic acid transporters (mttA, mfsA) increased itaconic acid production in an A. niger cis-aconitate decarboxylase expressing strain. Surprisingly, the production did not increase further when both transporters were expressed together. Meanwhile, oxalic acid was accumulated as a by-product in the culture of mfsA transformants. In order to further increase itaconic acid production and eliminate by-product formation, the non-acidifying strain D15#26 and the oxaloacetate acetylhydrolase (oahA) deletion strain AB 1.13 ∆oahA #76 have been analyzed for itaconic acid production. Whereas cadA expression in AB 1.13 ∆oahA #76 resulted in higher itaconic acid production than strain CAD 10.1, this was not the case in strain D15#26. As expected, oxalic acid production was eliminated in both strains. In a further attempt to increase itaconic acid levels, an improved basal citric acid-producing strain, N201, was used for cadA expression. A selected transformant (N201CAD) produced more itaconic acid than strain CAD 10.1, derived from A. niger strain AB1.13. Subsequently, we have focused on the influence of dissolved oxygen (D.O.) on itaconic acid production. Interestingly, reduced D.O. levels (10–25 %) increased itaconic acid production using strain N201 CAD. Similar results were obtained in strain AB 1.13 CAD + HBD2.5 (HBD 2.5) which overexpressed a fungal hemoglobin domain. Our results showed that overexpression of the hemoglobin domain increased itaconic acid production in A. niger at lower D.O. levels. Evidently, the lower levels of D.O. have a positive influence on itaconic acid production in A. niger strains.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Itaconic acid is a C5-di-carboxylic acid which is produced by Aspergillus terreus (A. terreus) for industrial use (Okabe et al. 2009; Willke and Vorlop 2001). Itaconic acid possesses two carboxyl moieties which give it the ability to act as a co-monomer in the manufacture of polymers (Okabe et al. 2009; Willke and Vorlop 2001). Its application areas are mainly in plastics, paper industry, and adhesives (Okabe et al. 2009; Willke and Vorlop 2001).
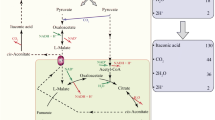
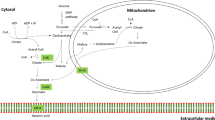
The biological pathway of itaconic acid production in A. terreus occurs in two sub-cellular compartments (Bonnarme et al. 1995; Jaklitsch et al. 1991). In this pathway, pyruvate is converted to acetyl-CoA in the mitochondria and together with oxaloacetate, is converted by citrate synthase to citric acid. Furthermore, in the TCA cycle, citrate is dehydrated to cis-aconitate and then, as cis-aconitate decarboxylase (CAD) is assumed to convert cis-aconitate into itaconate inside the cytosol, mitochondrial transporter is involved in transporting cis-aconitic acid from the mitochondrion to the cytosol. Alternatively, citric acid is first transported to the cytosol by a tri-carboxylic acid transporter and further converted to itaconic acid in two steps. Finally, itaconic acid is exported out of the cell most likely via a di-carboxylic acid transporter. Although the details of tri-/di-carboxylic acid transport in relation to itaconic acid are unknown, the important role of the two transporters in shuttling metabolites required for the itaconic acid metabolic pathway is suggested.
The role of this type of transporters is also suggested from our previous study on the identification of the cis-aconitate decarboxylase; as in this study also, a mitochondrial carrier protein (mttA) and a putative di-carboxylate carrier (mfsA) were found to be expressed in concordance with cadA in a hypothetical itaconic acid gene cluster in A. terreus (Li et al. 2011). These genes lack true orthologous in A. niger, which suggest a unique transport system specific to itaconic acid. Therefore, the expression of these two transporters in A. niger CAD strains seems to be a promising approach to increase the itaconic acid production.
To further increase the production capacity and reduce by-product formation, other parental A. niger strains were evaluated. A. niger was chosen as a production host for itaconic acid due to the high capacity of accumulating citric acid which, as described above, is a precursor of itaconic acid production. We previously showed that one step cadA expression in A. niger converted the parental strain AB 1.13 into an itaconic acid producer (Li et al. 2011). In order to improve the production level of itaconic acid, a combination of genetic modification on the production host and production medium optimization has already been described (Li et al. 2012). Medium optimization led to an improved medium for itaconic acid production in A. niger. However, the batch fermentation of an A. niger transformant showed that oxalic acid as a by-product was still produced in the selected medium condition (Li et al. 2012). Although, also, citric acid was still co-produced with itaconic acid, we have observed that more citric acid production led to higher itaconic acid accumulation (Li et al. 2012). Previous studies in A. terreus showed that reduced dissolved oxygen (D.O.) levels improved itaconic acid production (Li et al. 2011).
From these results, three aspects for further itaconic acid production improvement can be identified, i.e., reduction of oxalic acid by-product formation in the parental strain, use of a host strain with increased citric acid production, and modification of oxygen availability.
Although oxalic acid production can be reduced by using alternative process conditions (Li et al. 2012), here, we designed a molecular/genetic strategy to eliminate oxalic acid. One approach we followed was to delete the gene oahA encoding oxaloacetate hydrolase responsible for cleaving oxaloacetate into oxalate and acetate. An alternative approach to reduce by-product formation was followed by using a previously isolated low-acid-producing mutant from AB 1.13 (Gordon et al. 2000).
Assuming that higher levels of citric acid production in a parental A. niger strain would lead to higher itaconic acid accumulation after introduction of the A. terreus cadA gene, the A. niger strain N201 (ATCC 1015) was selected as parental strain for cadA transformation based on its high citric acid production level. This strain is a fully sequenced wild-type strain used in the first patented citric acid production process (Currie 1917).
As previously shown, itaconic acid production conditions are not identical to citric acid production conditions. In A. terreus, itaconic acid production is influenced by multi-environmental factors such as media components, pH, oxygen availability, temperature, etc. (Gyamerah 1995; Mattey 1992; Milsom and Meers 1985; MuralidharaRao et al. 2007; Okabe et al. 2009; Pfeifer et al. 1952; Riscaldati et al. 2000; Rychtera and Wase 1981; Vassilev et al. 1992; Willke and Vorlop 2001). Here, we considered oxygen availability determined as dissolved oxygen (D.O.) as a potentially important parameter for successful itaconic acid production by A. niger in submerged cultures. Besides altering fermentation conditions, also, a molecular genetic approach was followed to modify oxygen availability. Based on the fact that increased oxygen uptake through overexpression of a bacterial hemoglobin (vgb) was shown to have a strong effect on itaconic acid production in A. terreus (Lin et al. 2004) and overexpression of a fungal hemoglobin domain improved oxygen uptake in Aspergillus oryzae (te Biesebeke et al. 2006), we have overexpressed a fungal hemoglobin domain (hbd1) in A. niger strain previously (Li et al. 2012). Although the resulting transformant (HBD2.5) did not show improved itaconic acid production (Li et al. 2012), we assumed that modification of D.O. levels could have an effect on itaconic acid production in this strain.
Materials and methods
Strain/plasmid/fungal transformation
Aspergillus niger strains AB 1.13 pyrG-, AB 1.13, CAD 10.1(Li et al. 2011), AB 1.13 CAD, HBD 2.5 (Li et al. 2012), D15#26, and N201 (ATCC 1015) (Currie 1917) were used in this study (Table 1). They were all maintained on potato–dextrose agar plates and in 20 % glycerol spore suspensions frozen at −80 °C.
To constitutively express the two transporters mttA (ATEG_09970) and mfsA (ATEG_09972) in strain CAD 10.1, both genes were in vitro synthesized (Geneart) as compatible gene fragments to be cloned into vector pAN52–5doubleNotI using NcoI and BamHI restriction sites. The vector pAN52–5doubleNotI was derived from pAN52–4 (EMBL accession #Z32750) by introducing downstream of the trpC termination an extra NotI site. The plasmid pAB 4–1 (Van Hartingsveldt et al. 1987) containing the pyrG+ gene as a selection marker was used for co-transformation. Then, the mttA (pAN52–5doubleNotI + MTT) and mfsA (pAN52–5doubleNotI + MFS) expression vectors were co-transformed with pAB4–1 into A. niger strain CAD 10.1.All transformants were selected once for their ability to grow on Minimal Medium plates without uridine.
A selected itaconic acid-producing co-transformant carrying mttA (AB 1.13 CAD MTT1.4) was further transformed with mfsA to obtain strain AB 1.13 CAD + MTT + MFS. The Aspergillus vector pAN7–1 containing the Escherichia coli HmB-resistant gene (hph) (Punt and Van Den Hondel 1992) as a selection marker was used to co-transform with mfsA. These transformants were selected on hygromycin containing Minimal Medium agar plates.
To express the cadA gene in strains D15#26, AB 1.13 ∆oahA#76, and N201, the plasmid pAN52-amdS + CAD was used (Li et al. 2011). These transformants were selected on acrylamide containing Minimal Medium agar plates.
Construction of a non-oxalic acid-producing strain
To delete the oxaloacetate hydrolase gene oahA from strain AB 1.13, the deletion plasmid was made as follows: the primers oahA-1535for: 5′-CCATCGGCTTCTCCGTCGG-3′ and oahA + 506rev 5′-GGCAATAAGGTTTTGCTGGGTG-3′ were used for amplifying gene oahA (An10g00820) from chromosomal DNA of A. niger. The resulting 3.2-kb fragment was cloned in pGEM®-T Easy vector using the kit from Promega. From the oahA vector, a 526-bps EheI fragment within oahA was replaced with the pyrG+ selection marker isolated as an EheI fragment. For further fungal transformation, the ΔoahA deletion fragment was cut with NotI from the resulting oahA deletion vector.
To generate a non-oxalic acid-producing strain, the ΔoahA deletion fragment was transformed into A. niger AB 1.13 pyrG-. Ninety six selected transformants were checked via PCR using two oahA-specific primers oahA486for: 5′-CCTGATGGTCGCCCGTTCC-3′ and oahA1099rev: 5′-CACCATTAGCAAACCATCTCC-3′. In total, 33 PCR positive strains were selected for Southern blot analysis as described (Punt et al. 2008). From these, the transformant AB 1.13 ∆oahA#76 was selected for fungal transformation with cadA.
Northern blot analysis
Northern analysis was carried out essentially as described (Sambrook et al. 1989). With the ‘PCR DIG Probe Synthesis Kit’ (Roche, 2006), the DIG (digoxigenin)-labeled fragments derived from cadA and gpdA genes were used as probes for Northern analysis.
PCR was used to amplify cadA probe fragments from plasmid pAN52-4amdS-CAD using primers 5′-GGTCTTAGCCGAGCAAGGC-3′ and 5′-GCGACACTCATCTGCCCTG-3′ and to amplify gpdA probe from plasmid pAB 5–1 (Verdoes et al. 1994) using primers 5′-ATCGAGACCTACGAGGAGGG-3′ and 5′-CCGGGAGTTCCTGCGAAGG-3′ .
Cultivation conditions
For the screening and selection of A. niger CAD or transporter transformants, our previously developed screening assay was used (Li et al. 2012). All plates were placed in a plastic air bag and cultivated in a 33-°C, 850-rpm incubator (Microtron, Infors-ht) for 60 h, except for N201CAD transformants which were incubated for 48 h. In the end of the cultivation, culture medium was harvested and used for HPLC analysis.
For controlled batch fermentations, the production medium (M12) described in our previous study (Li et al. 2012) with the following composition was used (per liter): 100 g glucose, 2.36 g (NH4)2SO4, 0.11 g KH2PO4, 0.5 g MgSO4·7H20, 0.6 mg FeSO4·7H20, 2.5 mg CuSO4·5H20, 0.6 mg ZnSO4·7H20, 0.074 g NaCl, and 0.13 g CaCl2·2H20. This medium was prepared in demineralised water. The production medium M12 + Cu has an extra addition of 2.5 mg CuSO4·5H20 (0.01 mM).
Pre-cultures were prepared by inoculation of 106 spores per milliliter in 2 × 100-mL production medium in two 500-mL baffled Erlenmeyer flasks. After 64 h at 33 °C and shaking at 125 rpm, the pre-cultures were used for inoculation of the fermenters.
Fermentations were performed in 5-L Benchtop Fermentors (BioFlo 3000, New Brunswick Scientific Co., Inc.) at 33 °C. The basic pH regime was initiated at 3.5 and subsequently regulated at 2.3, by addition of 4 M KOH (base). Struktol was applied as antifoam agent (Schill & Seilacher) in all cultures throughout the fermentation.
Air was used for sparging the bioreactor at a constant flow of 0.25 vvm [(vol.liquid)−1 min−1]. The solubility of oxygen in the medium is around 225 μMol at 33 °C. Pure air sparging was calibrated as 100 % D.O., whereas pure nitrogen sparging was calibrated as 0 % D.O. In the basic D.O. regime, D.O. was set at 100 % from the start of the fermentation. As soon as due to mycelial growth D.O. levels dropped below 25 %, stirrer agitation was increased automatically to maintain D.O. at 25 %.
For studying the influence of oxygen availability on itaconic acid production, D.O was fixed throughout the whole fermentation at 10, 15, 20, and 25 % for strain N201 CAD and at 5, 10, and 20 % for strain HBD 2.5. The different percentage of D.O. was obtained by varying the mixture of air/nitrogen in the inlet gas.
Analytical methods
Metabolite analysis
To quantify metabolites, samples collected from fermentation cultures were analyzed by high-performance liquid chromatography (HPLC). Dionex ICS 3000 from Thermo Fisher was used for analyzing organic acids through an organic acids column IonPac® ICE AS6, with 1.6 mM heptafluorobutyric acid as an eluent and a detector of suppressed conductivity CD25. Standard compounds (oxalic acid, citric acid, cis-aconitic acid, and itaconic acid) with concentrations of 100, 200, 500, 750, 1,000, and 2,000 mg/L were used for calibration.
Biomass dry weight
Biomass of the batch cultivation was harvested by filtration of a fixed volume (10 mL) of the culture through the pre-weight dry filters and washed with demineralised water. Subsequently, the harvested biomass with the filter was dried in an oven at 110 °C for 24 h and weighed. The biomass dry weight was established by subtracting the weight of the dry filter.
Results
To further improve the production level of itaconic acid based on our previous research in A. niger, we explored four aspects. The two transporters of the itaconic acid gene cluster from A. terreus (Li et al. 2011) were expressed in A. niger to obtain a better itaconic acid transport system in A. niger. To eliminate accumulation of the by-product oxalic acid in an A. niger itaconic acid-producing host, the low-acid secretion strain D15#26 and the oxaloacetate hydrolase-deficient strain AB 1.13ΔoahA#76 were used. As higher citric acid production was previously shown to lead to higher itaconic acid formation (Li et al. 2012), a basal citric acid-producing strain, A. niger N201, was modified into a new itaconic acid production host by introducing the A. terreus cadA gene as previously described (Li et al. 2011) resulting in strain (N201CAD). Furthermore, oxygen availability was considered as an important parameter in itaconic acid fermentation process. The new host N201CAD together with a hemoglobin domain overexpressing strain (HBD 2.5: Li et al. 2012) was studied in fermentations carried out with various D.O. regimes.
Selection of A. niger itaconic acid transformants
To select the best itaconic acid-producing strains from mttA and mfsA transformants, eight mttA and 24 mfsA transformants taken from Minimal Medium selection plates were cultivated in micro-titer plate. As shown in Fig. 1a, nearly every mfsA transformant produced more itaconic acid than strain AB 1.13 CAD. The best one among them was strain AB 1.13 CAD + MFS (MFS 3.9) which had nearly fivefold higher itaconic acid production (0.292 g/L). Surprisingly, among the mttA transformants, strain AB 1.13 CAD + MTT (MTT 1.4) was the only one producing significantly more itaconic acid (0.492 g/L) than strain AB 1.13 CAD (0.054 g/L) (Fig. 1b), whereas Southern analysis confirmed the presence of the mttA gene in MTT1.4/MTT 1.1 and MTT 1.5. However, as strain MTT 1.4 also produced twofold higher itaconic acid than MFS 3.9, we decided to also introduce mfsA into strain MTT 1.4 to obtain a itaconic acid strain expressing both mttA and mfsA. Unexpectedly, the screening results show no improved itaconic acid production compared to MTT 1.4 (Fig. 2). Among 12 selected transformants, only transformant 3 (CAD + MTT + MFS_3) had a similar production level as the reference; the rest even had somewhat decreased production.
Batch fermentations with transporter strains
To analyze improved itaconic acid levels under more relevant condition, batch fermentation experiments were performed with five A. niger strains (AB 1.13, AB 1.13 CAD, MTT 1.4, MFS 3.9, and CAD + MTT + MFS_3). During fermentation, mycelium was harvested to determine the biomass. Cultivation medium was analyzed for organic acids (Fig. 3).
As shown, the best itaconic acid-producing strains were MTT 1.4 and MFS 3.9, both producing a similar amount of itaconic acid about 1.5 g/L. Both of these two transporter strains produced more itaconic acid then AB 1.13 CAD. The itaconic acid production of the double transporter strain CAD + MTT + MFS3 produced less than MTT 1.4 and MFS 3.9, similar to AB 1.13 CAD (0.9 g/L).
AB 1.13 produced the highest amount of citric acid (22 g/L) and no oxalic acid. Strains MTT 1.4 and AB 1.13 CAD produced less citric acid (16 and 6 g/L) and no oxalic acid. In comparison, strain MFS 3.9 produced far less citric acid (2.2 g/L) but accumulated oxalic acid (1.8 g/L). In case of the double transporter strain CAD + MTT + MFS_3, hardly any citric acid was produced. However, the oxalic acid (4.3 g/L) accumulation in this strain was increased. The mycelium growth of all strains was similar, as presented in Fig. 4, except AB 1.13 CAD having somewhat less biomass than the others.
Comparison of A. niger host strains for itaconic acid production
As mentioned, to improve producing itaconic acid, one aspect was to seek for a better host strain with higher citric acid production, the other aspect was to eliminate producing oxalic acid by-product. Therefore, the basal citric acid-producing strain N201 and the oxaloacetate hydrolase-deficient strain AB 1.13 ∆oahA#76 (∆oahA#76) were selected.
In order to compare citric acid production levels, strains AB 1.13, ∆oahA#76 and N201 were cultivated in controlled batch fermentation with a previously selected itaconic acid production medium (medium 12; Li et al. 2012). As shown in Fig. 5a, in 96 h, strain N201 produced about 0.80 g (cit)/g (biomass) citric acid, which is over three times more than strain AB 1.13 (0.23 g (cit)/g (biomass)). Furthermore, strain N201 did not produce oxalic acid under this condition (Fig. 5b), while glucose consumption was the same and the biomass growth was similar (data not shown). The same production levels of citric acid and oxalic acid were shown with strain ∆oahA#76. Based on this result, we decided to use both of them as production host for itaconic acid.
Similar to the transporter transformants, among 14 cadA transformants from D15#26 and seven from AB 1.13 ∆oahA#76, the best itaconic acid-producing strains (D15#26 CAD7 and AB 1.13 ∆oahA#76 CAD5) were selected using the micro-plate screening assay. All selected itaconic acid strains were compared for itaconic acid production in the micro-titer assay. The results are presented in Table 2. Strain D15#26 CAD7 did not produce higher itaconic acid than CAD 10.1. However, strain ∆oahA#76 CAD5 produced twofold more itaconic acid compared to CAD 10.1, and the production level (0.4 g/L) was similar to MTT 1.4 (0.47 g/L) and MFS 3.9 (0.44 g/L). Moreover, ∆oahA#76 CAD5 also yielded the highest citric acid levels (2.12 g/L). No detectable levels of oxalic acid were produced in the micro-titer cultivations.
Also, strain N201 was transformed with cadA, and the transformants were screened for itaconic acid production. In this case, also cadA expression of several transformants was analyzed by Northern blot analysis (Fig. 6). Strain N201 CAD 02 was selected for its high itaconic acid production and cadA expression. Its itaconic acid production level (135 mg/L) was almost two times higher than strain AB 1.13 CAD (72 mg/L). This strain was used to study the effect of cultivation conditions.
Influence of dissolved oxygen level on organic acids production in A. niger
Based on previous research (Li et al. 2011), it was suggested that D.O. could be an important factor for itaconic acid production. Moreover, as previous results in A. terreus (Lin et al. 2004) have shown that undisturbed aeration is required for optimal itaconic acid production, we have studied the effect of oxygen availability in more detail. Reduced D.O. levels were shown to increase itaconic acid levels in A. terreus batch cultivation (Li et al. 2011). Therefore, we studied the effect of reduced D.O. on itaconic acid production in A. niger. We used strain N201 CAD in controlled batch fermentation with four D.O. levels. The amount of itaconic acid produced by strain N201 CAD is shown in Table 3. The basic cultivation conditions based on a 100–25 % D.O. gradient produced less itaconic acid (0.05 g(ita)/g(biomass)) than the other D.O. cultures (10, 15, 20, and 25 %). The highest production of both itaconic acid (0.20 g(ita)/g(biomass)) and citric acid (0.40 g(cit)/g(biomass)) was observed with 20 % D.O. (Table 3). The production level of strain N201 CAD on oxalic acid was presented in Table 3. The highest oxalic acid was accumulated at 100–25 % D.O. There was no oxalic acid being accumulated in other itaconic acid-producing D.O. conditions. As expected (Table 3), strain ∆oahA#76 CAD5 did not produce any oxalic acid under controlled fermentation conditions. Moreover, this strain produced twofold more itaconic and citric acids in comparison to N201CAD under standard DO conditions (100–25 % D.O.).
Based on these results, we have analyzed the effect of reducing D.O. in an A. niger itaconic acid-producing strain overexpressing a fungal hemoglobin domain in batch fermentations (Table 3). In our previous studies, using A. niger strain HBD 2.5 with overexpressed fungal hemoglobin domain (hbd1) did not improve itaconic acid production in 100–25 % D.O. Here, we analyzed strain HBD 2.5 at reduced D.O. levels. As shown, strain HBD 2.5 produced 0.15 g(ita)/g(biomass) itaconic acid, which is two times higher than the parental strain AB 1.13 CAD at 10 % D.O. (0.07 g(ita)/g(biomass)) (Table 3).
Moreover, in even lower oxygen supply conditions, itaconic acid production level of strain HBD 2.5 was maintained (0.157 g(ita)/g (biomass)). The citric acid production at 5 % D.O. was much lower in comparison to the production of itaconic acid at 5 % D.O. Similar to strain N201CAD, the highest production of both itaconic acid (0.17 g(ita)/g(biomass)) and citric acid (0.68 g(cit)/g(biomass)) was at 20 % D.O. There was no accumulation of oxalic acid under these conditions.
Discussion
Effect of putative transporters from the A. terreus itaconic acid gene cluster on itaconic acid production in batch fermentation experiments with A. niger
In order to increase secreted itaconic acid yields in A. niger production, we expressed two transporters genes from the A. terreus itaconic acid gene cluster gene cluster. The two transporters, mfsA and mttA, lacking orthologs in A. niger were expressed in A. niger strain CAD 10.1. As observed, expression of mfsA increased itaconic acid production secretion in A. niger, indicating their role in the transport of itaconic acid (Fig. 1). The mfsA transporter as a putative di-carboxylate carrier was assumed to facilitate the export of itaconic acid into the medium. Similar as presented in the research of Zelle to improve malic acid production in Saccharomyces cerevisiae (Zelle et al. 2008), we present positive effect on itaconic acid production of the putative pathway-specific di-carboxylate carrier mfsA. A positive effect was observed only in one of the MTT transformants. Moreover, expressing mfsA in this strain did not further improve the production level of itaconic acid (Fig. 3a). Further research is underway to analyze this unexpected phenomenon.
Although metabolite transport across the mitochondrial membrane has been widely investigated, the knowledge about the mechanism of mitochondrion carrier proteins in relation to organic acid transport in fungi is still very limited (Palmieri 2004). From the results we have obtained, it is clear that modifications in the mitochondrial and cytoplasmic membrane transport have profound effects on organic acid transport. As shown, in controlled batch fermentations, the production profiles of citric acid and oxalic acid are not the same as itaconic acid for the different transporter strains. The mttA transformant (MTT 1.4) showed increased accumulation of citric acid, while the mfsA transformant (MFS 3.9) showed decreased accumulation of citric acid (Fig. 3b). Apparently, the mfsA transporter contributes to a better conversion of citric acid toward itaconic acid, possibly by reducing intracellular itaconic acids levels. However, introduction of the mfsA transporter resulted in an accumulation of the by-product oxalic acid, which is not observed with mttA transformant (Fig. 3c). Therefore, both strains MTT 1.4 and MFS 3.9 have advantages and drawbacks in itaconic acid production. The effect of expressing both transporters in one strain (CAD + MTT + MFS_3) seem to be dominated by mfsA as citric acid secretion is completely absent and the accumulation of oxalic acid is highly increased.
As shown in the fermentation with the production strain AB 1.13 CAD, also without any of the two transporters from A. terreus, itaconic acid could also be exported out of the cell. Therefore, we assumed that in A. niger and other transporters, e.g., for citric acid might function as an itaconic acid transporter. As citric acid is secreted in almost every strain, analyzing the citric acid transport system in A. niger is suggested to be very efficient, and several putative homologues for mitochondrial and plasma membrane transport of di- and tri-carboxylic acids can be identified (Li and Punt, submitted for publication). However, until now, detailed knowledge about this transport is lacking, and further research will be required to improve secretion of alternative organic acids in A. niger.
Itaconic acid production test of two non-oxalic acid-producing strains
In our previous studies, we observed that oxalic acid as a by-product was co-produced under preferred itaconic acid production conditions (Li et al. 2011; 2012). To eliminate this by-product, two non-oxalic acid-producing strains from A. niger (D15#26 and AB 1.13 ∆oahA#76) were evaluated in this study. Although D15#26 CAD7 could produce itaconic acid, the levels were clearly lower than in the wild-type strain AB1.13 (Table 2). Strain ∆oahA#76 CAD5 deficient in oxaloacetate hydrolase activity produced even higher itaconic acid especially in fermentation than its reference strain (Table 3). As previous research has shown that the absence of oxaloacetate hydrolase also positively effect citric acid production (Ruijter et al. 1999) and more recently also production of other metabolites was positively influenced by the absence of oxalic acid by-product formation (Pedersen et al. 2000; Gombert et al. 2011), oahA knockout should be considered for further strain improvement in relation to alternative organic acid/building block chemical production..
Influence of D.O. on itaconic acid production under controlled batch fermentation
Another aspect to increase itaconic acid production was to optimize the conditions of A. niger batch fermentations. Here, we focused on the influence of different levels of dissolved oxygen (D.O.), which was based on a previous study that a lower oxygen supply (25 % D.O.) was optimal for itaconic acid production in controlled batch fermentation using A. terreus (Li et al. 2011). Two itaconic acid-producing strains from A. niger were included in this study. One was N201 CAD, expressing cadA in a basal organic acid-producing strain. The other was HBD 2.5 which carried and expressed a fungal hemoglobin domain (hbd1) (Li et al. 2012).
We observed that the highest itaconic acid production was at 20 % D.O. for strain N201 CAD (Table 3). Moreover, lower D.O. conditions (10–25 %) eliminated oxalic acid accumulation in this strain (Table 3). As observed for strain HBD 2.5, overexpressing hbd1 in strain AB 1.13 CAD under low D.O. levels (5–20 %) improved itaconic acid production (Table 3) and also diminished production of oxalic acid. Interestingly, at the lowest D.O. level (5 %), this strain produced considerably reduced levels of citric acid, while itaconic acid levels were retained. Altogether, these results proved our assumption that overexpression of hemoglobin domain could increase itaconic acid production, possibly by improving oxygen sequestration and transport in the cell (te Biesebeke et al. 2006)
In this study, further optimization of A. niger as an itaconic acid production host has been demonstrated. We increased the production level from 0.8 to 2.5 g/L. Expression of the two transporters (mttA or mfsA), overexpression of the hemoglobin domain (hbd1), elimination of oxalic acid by-product (∆oahA), replacement with a basal organic acid production host N201, and lower D.O. (10–20 %) supply in the fermentation culture have all shown to enhance itaconic acid production level in A. niger. To increase itaconic acid production capacity even further, combining these features into one platform will be the target of our ongoing research.
References
Bonnarme P, Gillet B, Sepulchre AM, Role C, Beloeil JC, Ducrocq C (1995) Itaconate biosynthesis in Aspergillus terreus. J Bacteriol 177:3573–3578
Currie JN (1917) The citric acid fermentation of Aspergillus niger. J Biol Chem 31:15–37
Gombert AK, Veiga T, Puig-Martinez M, Lamboo F, Nijland JG, Driessen AJM, Pronk JT, Daran JM (2011) Functional characterization of the oxaloacetase encoding gene and elimination of oxalate formation in the β-lactam producer Penicillium chrysogenum. Fungal Genet Bio 48:831–839
Gordon CL, Khalaj V, Ram AFJ, Archer DB, Brookman JL, Trinci APJ, Jeenes DJ, Doonan JH, Wells B, Punt PJ, Van den Hodel CAMJJ, Robson GD (2000) Glucoamylase: green fluorescent protein fusion to monitor protein secretion in Aspergillus niger. Microbiology 146:415–426
Gyamerah MH (1995) Oxygen requirement and energy relations of itaconic acid fermentation by Aspergillus terreus NRRL 1960. Appl Microbiol Biot 44:20–26
Jaklitsch WM, Kubicek CP, Scrutton MC (1991) The subcellular organization of itaconate biosynthesis in Aspergillus terreus. J Gen Microbiol 137:533–539
Li A, van Luijk N, ter Beek M, Caspers M, Punt P, van der Werf M (2011) A clone-based transcriptomics approach for the identification of genes relevant for itaconic acid production in Aspergillus. Fungal Genet Bio 48:602–611
Li A, Pfelzer N, Zuijderwijk R, Punt P (2012) Enhanced itaconic acid production in Aspergillus niger using genetic modification and medium optimization. BMC biotechnol 12:57
Lin YH, Li YF, Huang MC, Tsai YC (2004) Intracellular expression of Vitreoscilla hemoglobin in Aspergillus terreus to alleviate the effect of a short break in aeration during culture. Biotechnol Lett 26:1067–1072
Mattern IE, Van Noort JM, Van Den Berg P, Archer DB, Roberts IN, Van Den Hondel CAMJ (1992) Isolation and characterization of mutants of Aspergillus niger deficient in extracellular proteases. Mol Gen Genet 234:332–336
Mattey M (1992) The production of organic acids. Crit Rev Biotechnol 12:87–132
Milsom PE, Meers JL (1985) Gluconic and itaconic acids. In: Moo-Young M (ed) Comprehensive biotechnology, vol 3. Pergamon Press, Oxford, pp 681–700
MuralidharaRao D, Jaheer Hussain SMD, Pandu Rangadu V, Subramanyam K, Sivarama Krishna G, Swamy AVN (2007) Fermentatative production of itaconic acid by Aspergillus terreus using Jatropha seed cake. Afr J Biotechnol 6:2140–2142
Okabe M, Lies D, Kanamasa S, Park EY (2009) Biotechnological production of itaconic acid and its biosynthesis in Aspergillus terreus. Appl Microbiol Biot 84:597–606
Palmieri F (2004) The mitochondrial transporter family (SLC25): physiological and pathological implications. Pflügers Arch Eur J Physiol 447:689–709
Pedersen H, Christensen B, Hjort C, Nielsen J (2000) Construction and characterization of an oxalic acid nonproducing strain of Aspergillus niger. Metab Eng 2:34–41
Pfeifer VF, Vojnovich C, Heger EN (1952) Itaconic acid by fermentation with Aspergillus terreus. Ind Eng Chem 44:2975–2980
Punt PJ, Van Den Hondel CAMJ (1992) [39] Transformation of filamentous fungi based on hygromycin B and phleomycin resistance markers. Method Enzymol 216:447–457
Punt PJ, Schuren FHJ, Lehmbeck J, Christensen T, Hjort C, Van Den Hondel CAMJ (2008) Characterization of the Aspergillus niger prtT, a unique regulator of extracellular protease encoding genes. Fungal Genet Bio 45:1591–1599
Riscaldati E, Moresi M, Federici F, Petruccioli M (2000) Effect of pH and stirring rate on itaconate production by Aspergillus terreus. J Biotechnol 83:219–230
Ruijter GJG, Van De Vondervoort PJI, Visser J (1999) Oxalic acid production by Aspergillus niger: an oxalate-non-producing mutant produces citric acid at pH5 and in the presence of manganese. Microbiology 145:2569–2576
Rychtera M, Wase DAJ (1981) Growth of Aspergillus terreus and the production of itaconic acid in batch and continuous cultures. The influence of pH. J Chem Technol Biot 31:509–521
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning. A laboratory manual. Cold Spring Harbor Laboratory Press, Plainview, New York
te Biesebeke R, Boussier A, Van Biezen N, Braaksma M, van den Hondel CA, De Vos WM, Punt PJ (2006) Expression of Aspergillus hemoglobin domain activities in Aspergillus oryzae grown on solid substrates improves growth rate and enzyme production. Biotechnology J 1:822–827
Van Hartingsveldt W, Mattern IE, Van Zeijl CMJ (1987) Development of a homologous transformation system for Aspergillus niger based on the pyrG gene. Mol Gen Genet 206:71–75
Vassilev N, Kautola H, Linko YY (1992) Immobilized Aspergillus terreus in itaconic acid production from glucose. Biotechnol Lett 14:201–206
Verdoes JC, Punt PJ, Van Der Berg P, Debets F, Stouthamer AH, Van Den Hondel CAMJJ (1994) Characterization of an efficient gene cloning strategy for Aspergillus niger based on an autonomously replicating plasmid: cloning of the nicB gene of A. niger. Gene 146:159–165
Willke T, Vorlop KD (2001) Biotechnological production of itaconic acid. Appl Microbiol Biot 56:289–295
Zelle RM, De Hulster E, Van Winden WA, De Waard P, Dijkema C, Winkler AA, Geertman JMA, Van Dijken JP, Pronk JT, Van Maris AJA (2008) Malic acid production by Saccharomyces cerevisiae: engineering of pyruvate carboxylation, oxaloacetate reduction, and malate export. Appl Environ Microbiol 74:2766–2777
Acknowledgments
We thank Dr. Jorg Brunner for his valuable comments to the manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Li, A., Pfelzer, N., Zuijderwijk, R. et al. Reduced by-product formation and modified oxygen availability improve itaconic acid production in Aspergillus niger . Appl Microbiol Biotechnol 97, 3901–3911 (2013). https://doi.org/10.1007/s00253-012-4684-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-012-4684-x